Abstract
Realizing the therapeutic potential of human induced pluripotent stem (iPS) cells will require robust, precise and safe strategies for genetic modification, as cell therapies that rely on randomly integrated transgenes pose oncogenic risks. Here we describe a strategy to genetically modify human iPS cells at ‘safe harbor’ sites in the genome, which fulfill five criteria based on their position relative to contiguous coding genes, microRNAs and ultraconserved regions. We demonstrate that ~10% of integrations of a lentivirally encoded β-globin transgene in β-thalassemia-patient iPS cell clones meet our safe harbor criteria and permit high-level β-globin expression upon erythroid differentiation without perturbation of neighboring gene expression. This approach, combining bioinformatics and functional analyses, should be broadly applicable to introducing therapeutic or suicide genes into patient-specific iPS cells for use in cell therapy.
The advent of induced pluripotent stem (iPS) cells enables for the first time the derivation of unlimited numbers of patient-specific stem cells1–3 and holds great promise for regenerative medicine4,5. Recent studies have explored the potential of iPS cell generation combined with gene and cell therapy for disease treatment in mice and humans4,5. However, for the promise of iPS cell technology in therapeutic applications to be fully realized, clinically translatable methodologies for the introduction of therapeutic, suicide, drug resistance or reporter genes into human iPS cells will be needed. The foreign genetic material should ideally be delivered into ‘safe harbors’, that is, regions of the genome where the integrated material is adequately expressed without perturbing endogenous gene structure or function, following a process that is amenable to precise mapping and minimizing occult genotoxicity. Retroviruses, such as HIV, efficiently integrate in the human genome with a strong bias toward actively transcribed genes6. This semi-random integration pattern favors expression of retrovirally encoded transgenes but entails a risk of perturbing the expression of neighboring genes, including cancer-related genes7–10. We hypothesized that screening iPS cell clones harboring a single vector copy would enable us to retrieve safe harbor sites that met the following five criteria: (i) distance of at least 50 kb from the 5′ end of any gene, (ii) distance of at least 300 kb from any cancer-related gene, (iii) distance of at least 300 kb from any microRNA (miRNA), (iv) location outside a transcription unit and (v) location outside ultraconserved regions (UCRs) of the human genome11. As the most common insertional oncogenesis event is transactivation of neighboring tumor-promoting genes7,12, the first two criteria exclude the portion of the human genome located near promoters of genes, in particular, cancer-related genes (Supplementary Table 1). The latter were defined as genes functionally implicated in human cancers or the human homologs of genes implicated in cancer in model organisms (available at http://microb230.med.upenn.edu/protocols/cancergenes.html). Proximity to miRNA genes was adopted as an exclusion criterion because miRNAs are implicated in the regulation of many cellular processes, including cell proliferation and differentiation. As vector integration within a transcription unit can disrupt gene function through the loss of function of a tumor suppressor gene or the generation of an aberrantly spliced gene product10, our fourth criterion excludes all sites located inside transcribed genes. Finally, we excluded UCRs—regions that are highly conserved over multiple vertebrates and known to be enriched for enhancers and exons11.
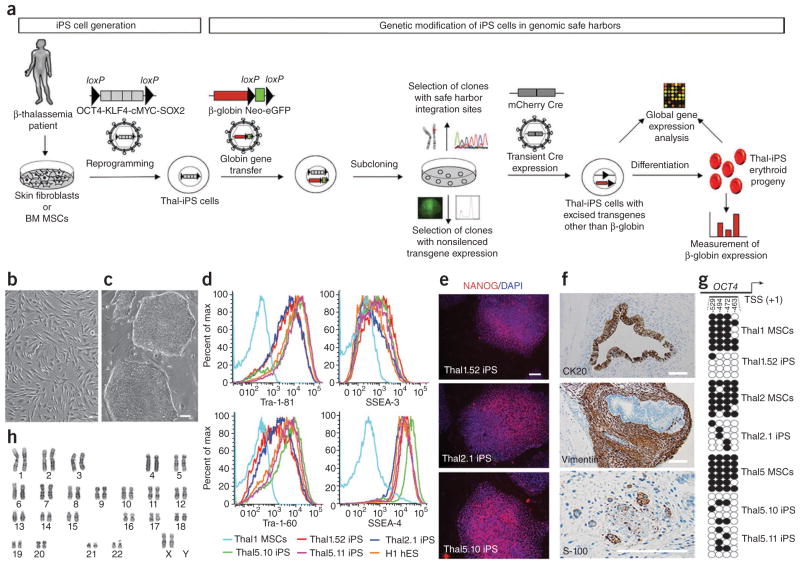
We investigated this approach in an iPS cell model for the genetic correction of β-thalassemia major using a well-characterized globin lentiviral vector13,14 (Fig. 1a). We generated a total of 20 iPS cell lines from skin fibroblasts or bone marrow mesenchymal stem cells (MSCs) (Fig. 1b) from four individuals with β-thalassemia major of various genotypes (Supplementary Table 2). All putative thalassemia iPS cell lines (referred to as thal-iPS) exhibited characteristic human embryonic stem (hES) cell morphology (Fig. 1c and Supplementary Fig. 1). Seven putative thal-iPS cell lines (Supplementary Table 2) were selected for further characterization. They expressed human pluripotent cell markers (Tra-1-81, Tra-1-60, SSEA-3, SSEA-4 and Nanog) and pluripotency-related genes at similar levels to hES cell lines (Fig. 1d–e and Supplementary Figs. 1–3). Their pluripotency was assessed by formation of teratomas comprising tissues derived from all three germ layers after grafting into immunodeficient mice (Fig. 1f and Supplementary Figs. 4 and 5). They could be efficiently differentiated in vitro into mesoderm derivatives, such as beating putative cardiomyocytes (Supplementary Movie 1) and hematopoietic progenitor cells (see below). Genotyping confirmed the β-thalassemia mutations (Supplementary Table 2 and Supplementary Fig. 6). Silencing of all four transgenes was demonstrated by flow cytometry (in thal-iPS cell lines derived using vectors encoding the four reprogramming factors OCT4, SOX2, KLF4 and c-MYC together with distinct fluorescent proteins15, Supplementary Fig. 7), as well as quantitative reverse-transcription (qRT)-PCR (Supplementary Fig. 8). Demethylation of the OCT4 promoter was assessed and confirmed in the thal-iPS cell lines thal1.52, thal2.1, thal5.10 and thal5.11 (Fig. 1g). All seven thal-iPS cell lines tested exhibited normal male or female karyotypes (Fig. 1h and Supplementary Fig. 9). To generate transgene-free thal-iPS cells, we selected two thal-iPS cell lines, thal5.10 and thal5.11, found to contain six copies of the single polycistronic vector flanked by loxP sites (fSV2A) used for reprogramming (Supplementary Fig. 10a), after all six copies of the fSV2A vector they both contained were mapped to the genome (Supplementary Table 3). Several excised thal-iPS cell lines were derived from them after transient Cre expression by an integrase-deficient lentiviral vector (Cre-IDLV). Complete excision of all six copies of the fSV2A vector (Supplementary Fig. 10a–d,f) and absence of integration of the Cre-IDLV vector (Supplementary Fig. 10c,e,f) were thoroughly documented. Altered expression of endogenous genes in the vicinity of the six integrated vectors or of the residual promoterless (U3-deleted) lentiviral long terminal repeats (LTR) before and after vector excision, respectively, was excluded by microarray analysis (Supplementary Fig. 11). Characterization of two vector-excised lines, thal5.10-Cre8 and thal5.11-Cre23 (derived from lines thal5.10 and thal5.11, respectively), confirmed their preserved pluripotency (Supplementary Figs. 1–3, 5). Comparative genomic hybridization (CGH) of the excised line thal5.10-Cre8 and the parental MSCs revealed no genetic abnormalities (Supplementary Fig. 12).
Figure 1.
Safe harbor selection strategy and characterization of thal-iPS cell lines. (a) Following the establishment of patient-specific iPS cell lines, which in this study were generated from skin fibroblasts or bone marrow mesenchymal stem cells (BM MSCs) from β-thalassemia major patients, with an excisable single polycistronic vector co-expressing OCT4, KLF4, cMYC and SOX2 (illustrated in Supplementary Fig. 10a), the genetic rescue strategy is as follows: thal-iPS cells are transduced with a lentiviral vector expressing β-globin and an excisable Neo-eGFP selection cassette and subcloned into single cells, and single-vector-copy integrants are selected according to vector chromosomal position. The levels of β-globin expression afforded by the vector integrated at different genomic positions are analyzed in the erythroid progeny of each selected clone. Microarray analysis is used to examine perturbation of endogenous gene expression by the integrated provirus. The reprogramming vector can be efficiently excised before globin gene transfer or after—together with the Neo-eGFP selection cassette of the globin vector—through transient expression of Cre (Supplementary Fig. 10). (b) MSCs from β-thalassemia major patient 1. (c) Thal1.52 iPS cell line. (d) Expression of pluripotency markers in iPS cell lines thal1.52, thal2.1, thal5.10 and thal5.11. Thal1 MSCs: MSCs from β-thalassemia major patient 1; H1 hES: hES cell line H1. (e) NANOG expression in iPS cell lines. (f) Immunohistochemical analysis of a teratoma derived from line thal1.52. Upper panel: cytokeratin (CK) 20-positive intestine-like epithelium (endoderm); middle panel: vimentin-positive fibroblastic spindle cells (mesoderm); lower panel: S-100–positive peripheral nerve (ectoderm). (g) Bisulfite sequencing analysis of the OCT4 promoter in the indicated thal-iPS cell lines and the MSCs from which they were derived. Each horizontal row of circles represents an individual sequencing reaction with white circles representing unmethylated CpG dinucleotides and black circles representing methylated CpG dinucleotides. The numbers indicate the CpG position relative to the transcriptional start site (TSS). (h) Karyotype analysis of thal2.1 iPS cell line. Scale bars, 50 μm.
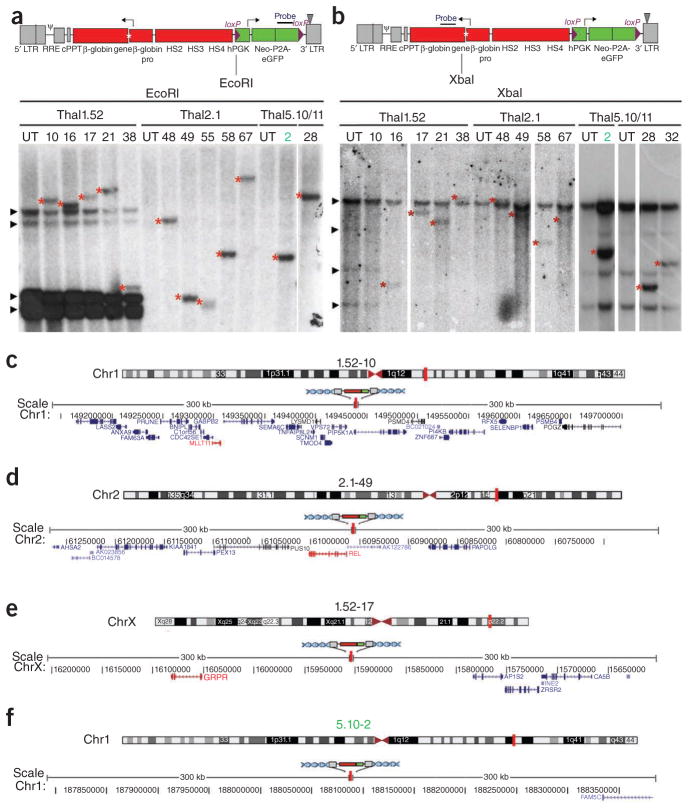
To establish thal-iPS cell clones harboring a therapeutic β-globin gene, we generated a lentiviral vector, TNS9.3/fNG, expressing the human β-globin gene cis-linked to its DNAse I hypersensitive site (HS) 2, HS3 and HS4 locus control region elements, derived from the previously described TNS9 vector13 (Fig. 2a). To determine the probability of retrieving sites that meet the safe harbor criteria, we analyzed 5,840 integration sites of our TNS9.3/fNG vector in the thal5.11-Cre23 iPS cell line. This survey revealed that 17.3% of all integrations met all five criteria (Supplementary Table 4), supporting the feasibility of recovering iPS cell clones harboring vector integrations in safe harbors from a relatively small set of clones. We thus transduced the thal-iPS cell lines thal1.52, thal2.1, thal5.10 and thal5.11 at low multiplicity of infection to isolate thal-iPS cell clones harboring a single TNS9.3/fNG vector copy. Fifteen clones found to harbor a single TNS9.3/fNG copy by quantitative PCR (Supplementary Table 5) were randomly selected. Single-vector integration and clonality could be thoroughly established by Southern blot analysis after digestion using two different restriction enzymes and two different probes (Fig. 2a,b and Supplementary Fig. 13) in 13 of them, and the vector integration sites were mapped to the human genome (Fig. 2c–f and Table 1). One of the 13 clones, clone thal5.10-2, was found to harbor an integration that meets all five safe harbor criteria (Table 1). Two additional safe harbor sites were found among 23 other sites we mapped in multiple-copy thal-iPS cell clones (Supplementary Table 6).
Figure 2.
Single-vector copy, clonality and mapping of the integration site. (a,b) Upper panel: schematic representation of the TNS9.3/fNG lentiviral vector. An asterisk depicts a 4-bp insertion in the 5′ untranslated region (UTR) of the β-globin gene, which allows discrimination of the longer vector-encoded transcript from the endogenous β-globin transcript. TNS9.3/fNG also contains the human phosphoglycerate kinase (hPGK) promoter-driven neomycin phosphoryltransferase (Neo) and enhanced green fluorescent protein (eGFP) genes flanked by loxP sites. LTR: long terminal repeat; RRE: rev-responsive element; cPPT: central polypurine tract; HS: DNAse I hypersensitive site. Lower panels: Southern blot analysis to ascertain single integrations of the TNS9.3/fNG vector and clonality. Genomic DNA was digested with EcoRI (a) or XbaI (b). The probe used in a is eGFP (shown in the upper panel). The probe in b spans exons 1 and 2 of the β-globin gene (shown in the upper panel). The parental thal-iPS cell lines thal1.52, thal2.1, thal5.10 and thal5.11 and the clone number are depicted above the lanes. UT: untransduced. Arrowheads in a indicate bands corresponding to the reprogramming vectors pLM-GO, pLM-YS and pLM-CM (Supplementary Fig. 8d) present in the thal1.52 line and the clones derived from it. Arrowheads in b indicate endogenous bands (corresponding to the endogenous β-globin locus). Asterisks depict unique vector integration bands. (c–f) Examples of chromosome ideograms (upper panels) and graphics (lower panels) depicting 300 kb of human genome on both sides of the globin vector integration site in iPS clones thal1.52-10 (c), thal2.1-49 (d), thal1.52-17 (e) and the safe harbor clone thal5.10-2 (f). A vertical red line depicts the position of the vector insertion. Numbers depict positions in the corresponding human chromosome. All RefSeq genes present in the genomic region spanning 600 kb illustrated in the graphic are shown in blue. Genes implicated in cancer (Supplementary Table 1) are shown in red. Chromosome ideograms and graphics were generated with the UCSC Genome Graphs tool.
Table 1.
Analysis of the globin vector integration site in 13 single-vector-copy thal-iPS cell clones with respect to the five safe harbor criteria
| iPS line | Clone | Chromosome | Position | Orientation | Within 50 kb of 5′ end of any gene | Within 300 kb of cancer gene 5′ or 3′ end | Within 300 kb of miRNA 5′ or 3′ end | Inside a gene transcription unit | Within UCR |
|---|---|---|---|---|---|---|---|---|---|
| Thal1.52 | 1.52-10 | 1 | 149,440,726 | + | + | + | − | + | − |
| Thal1.52 | 1.52-16 | 21 | 43,242,505 | − | + | + | − | − | − |
| Thal1.52 | 1.52-17 | X | 15,909,610 | − | − | + | − | − | − |
| Thal1.52 | 1.52-21 | 11 | 9,936,357 | − | − | + | − | + | − |
| Thal1.52 | 1.52-38 | 2 | 232,144,880 | + | + | + | + | − | − |
| Thal2.1 | 2.1-48 | 15 | 22,575,838 | − | + | + | − | − | − |
| Thal2.1 | 2.1-49 | 2 | 60,964,901 | − | + | + | − | + | − |
| Thal2.1 | 2.1-55 | 19 | 1,183,244 | − | + | + | − | + | − |
| Thal2.1 | 2.1-58 | X | 33,033,291 | − | − | − | − | + | − |
| Thal2.1 | 2.1-67 | 19 | 926,157 | + | + | + | − | − | − |
| Thal5.10 | 5.10-2 | 1 | 188,083,272 | + | − | − | − | − | − |
| Thal5.11 | 5.11-28 | 11 | 45,963,999 | + | − | − | − | + | − |
| Thal5.11 | 5.11-32 | 16 | 1,381,251 | − | + | − | − | − | − |
Clone thal5.10-2 meets all five safe harbor criteria.
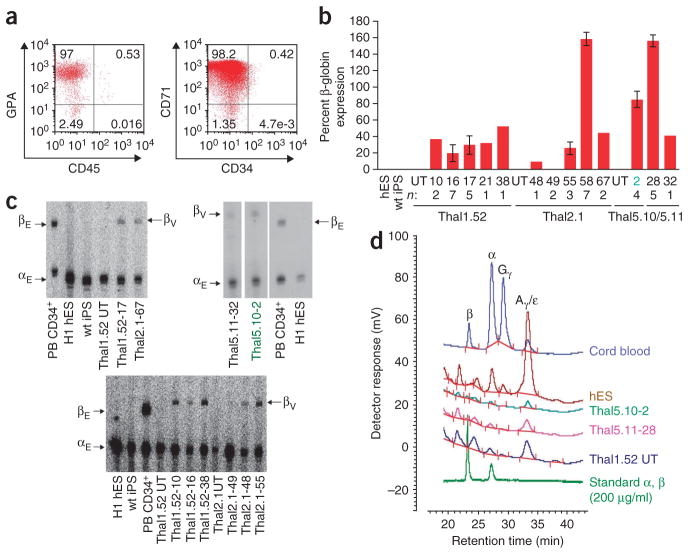
To assess vector-encoded β-globin gene expression, we derived hematopoietic progenitors through embryoid body differentiation of the 13 single-copy thal-iPS cell clones and we further differentiated them along the erythroid lineage (Fig. 3a and Supplementary Fig. 14). By the end of this process, the majority of cells exhibited characteristic hematopoietic cell morphology, expression of the erythroid cell markers glycophorin A and transferrin receptor (CD71) and macroscopic hemoglobinization (Supplementary Figs. 14 and 15). The erythroid nature of these thal-iPS cell derivatives was further corroborated by the marked induction of well-characterized, erythroid-specific genes (Supplementary Fig. 16). Notably, the erythroid progeny of all wild-type and untransduced thal-iPS cell lines expressed α-globin, as well as embryonic and fetal ε- and γ-globins, albeit not the adult β-globin transcript, similarly to the erythroid progeny of the H1 hES cell line (Fig. 3b–d and Supplementary Fig. 17) and in accordance with previous reports16–18. Expression of vector-encoded β-globin was not detected in undifferentiated thal-iPS cell clones, as expected (Supplementary Fig. 17). Upon erythroid differentiation, 12 of the 13 single-copy thal-iPS cell clones expressed detectable vector-encoded β-globin. Expression levels, normalized to endogenous α-globin expression, ranged from 9% to 159% (mean, 53%) of a normal endogenous β-globin allele (Fig. 3b,c), similar to those we and others have obtained by lentiviral-mediated globin gene transfer in murine and human erythroid cells14. β-globin expression was confirmed and quantified at the protein level by high-performance liquid chromatography (HPLC) analysis in four clones (Fig. 3d, Supplementary Table 7 and Supplementary Fig. 18). Notably, clone thal5.10-2, which expressed 85% of the level afforded by a normal endogenous β-globin allele (Fig. 3b), demonstrates that a globin vector, integrated in a site meeting all five of our safe harbor criteria (Table 1) and located >300 kb from the nearest gene 5′ end, is capable of expressing β-globin at a high level.
Figure 3.
β-globin expression in the erythroid progeny of single-vector-copy thal-iPS cell clones. (a) Expression of erythroid cell markers CD71 and glycophorin A (GPA) in the erythroid progeny of thal-iPS cell line 1.52. (b) β-globin expression in the erythroid progeny of 13 single-vector-copy thal-iPS cell clones assessed by qRT-PCR. Expression levels are expressed per gene copy, relative to the average endogenous β-globin expressed in the in vitro differentiated erythroid progeny of peripheral blood CD34+ cells from four healthy individuals and normalized to endogenous α-globin expression. hES: erythroid progeny of hES cell line H1, wt iPS: erythroid progeny of iPS cell line FDCT, derived from fibroblasts of an 11-year-old healthy individual30. Numbers below graphs depict thal-iPS clone numbers derived from lines thal1.52, thal2.1, thal5.10 and thal5.11. n: number of independent differentiations for each clone. UT: untransduced. Error bars denote s.e.m. (c) β-globin expression in the erythroid progeny of a subset of single-vector-copy thal-iPS cell clones and controls assessed by quantitative primer extension. βE: endogenous β-globin (80 bp); αE: endogenous α-globin (60 bp); βV: vector-encoded β-globin (84 bp). PB CD34+: erythroid cell derivatives of in vitro differentiated peripheral blood (PB) CD34+ cells from a normal donor; H1 hES: erythroid cell derivatives of the H1 hES line; wt iPS: erythroid cell derivatives of iPS cell line FDCT, derived from fibroblasts of a healthy individual; thal1.52 UT, thal2.1 UT: erythroid cell derivatives of untransduced lines thal1.52 and thal2.1, respectively; thal1.52-17, thal2.1-67, thal1.52-10, thal1.52-16, thal1.52-38, thal2.1-49, thal2.1-48, thal2.1-55, thal5.11-32, thal5.10-2: erythroid cell derivatives of the respective single-vector-copy thal-iPS clones. (d) Chromatograms of HPLC analysis of α- and β-globin expression in the erythroid progeny of clones thal5.10-2 and thal5.11-28. Cord blood, H1 hES and untransduced (UT) thal1.52 cells were used as controls. For quantification of these data, see Supplementary Table 7.
Expression of genes located within 300 kb of the vector insertion site was assessed in six single-copy thal-iPS cell clones in both the undifferentiated state, as well as in the erythroid progeny by micro-arrays. This analysis revealed that three out of five integrations eliminated by our safe harbor criteria did indeed result in perturbed expression of neighboring genes (Supplementary Figs. 19 and 20). Dysregulated expression was detected in a total of five genes present at a distance ranging from 9 to 275 kb from the vector insertion, whereas we did not detect any genes beyond 300 kb of the insertion to be significantly differentially expressed (P < 0.05). Of note, the safe harbor integration site in clone thal5.10-2 is in a genomic region with no genes within 300 kb on either side. The microarray analysis did not reveal any statistically significant differentially expressed genes elsewhere in the genome in this clone or any other.
Our data demonstrate that the generation and identification of transgene-expressing iPS cell clones, in which transgene expression is obtained at therapeutic levels in iPS cell progeny from selected chromosomal sites, are feasible by screening a limited number of single-copy clones and applying five safe harbor criteria for their selection. Approximately half (47.7%) of the clones we obtained under optimized transduction conditions harbored a single vector copy (Supplementary Table 5), and clonality could be confirmed in 13 out of 15 (86.7%) of them. As the frequency of integrations in sites that meet our five safe harbor criteria is 17.3% (Supplementary Table 4), the overall efficiency of our strategy is 7.1%. Three out of five clones eliminated by our safe harbor criteria showed perturbed expression of neighboring endogenous genes, which was not the case in clone thal5.10-2, demonstrating the usefulness of selecting genetically modified iPS cell clones based on this strategy and these criteria. Notably, applying our criteria to a series of gamma-retroviral and lentiviral integration sites associated with oncogenic events or perturbed endogenous gene expression would effectively eliminate all of these well-characterized deleterious integrations7–10.
This approach has the prospect of broad application in genetic engineering of human iPS cells. Genetic correction through addition of a therapeutic gene into safe harbors in patient-specific iPS cells provides a realistic alternative strategy to targeted gene repair, especially for genetically heterogeneous disorders associated with multiple mutations. In contrast to genome editing strategies, our approach does not require customized targeting vectors with long isogenic ends19 or complex genotoxicity screens that are needed when using endonucleases20,21. In the latter case, the risk of occult genotoxicity mediated by off-target effects of double-stranded DNA cleaving agents needs to be balanced against the long-term experience with risk assessment of retroviral vector integration, which can be thoroughly analyzed, as we demonstrate here. Apart from genetic correction, future clinical applications of iPS cells will likely require addition of drug resistance, reporter or suicide genes to permit in vivo selection, tracking or cell eradication, respectively. To this end, the identification of suitable genomic locations for transgene knock-in is of great importance. Recent studies suggest that genomic sites, such as the adeno-associated virus integration site 1 (AAVS1)20,22 and the human ROSA26 locus23, can support transgene expression, but data on the safety of these sites are lacking. The screening strategy we describe here should prove useful for the de novo discovery and characterization of putative universal genomic safe harbors. The requirements for a safe harbor are (i) avoidance of genotoxicity and (ii) support of the appropriate expression level and regulation of the integrated transgene. Notably, β-globin gene expression in the safe harbor clone thal5.10-2 was in the therapeutic range, which, based on clinical observations in individuals with homozygous β-thalassemia and hereditary persistence of fetal hemoglobin, is on the order of 30% of α-globin expression24.
The potential genotoxicity of the reprogramming process used upstream of our safe harbor strategy also needs to be taken into account. In this study we used an excisable vector system and selected patient-specific iPS cell lines harboring a relatively low number of reprogramming vector copies and determined their position in the genome. Since Cre-mediated excision leaves behind a promoterless, U3-deleted LTR, we propose that lines can be selected—as we demonstrate here—on the basis of (i) exclusion of all integrations within exons, to avoid frame-shift, premature termination of translation or translation of abnormal proteins and (ii) ascertainment of lack of perturbation of gene expression by residual LTR fragments that reside within transcription units. Based on our large integration site data set in human iPS cells, 97% of all lentiviral vector integrations are outside exons. The need to screen thal-iPS cell lines for residual LTR insertions may be eliminated if efficient generation of human iPS cells using nonintegrating systems becomes a realistic option.
Ascertainment of lack of perturbation of gene expression in the host cell in both a local and genome-wide range, as shown here (Supplementary Figs. 19 and 20), provides an important initial safety test. This can be complemented by additional tests for features of neoplastic transformation25 and, eventually, by serial transplantation studies of iPS cell–derived hematopoietic stem cells in immunodeficient mice, currently precluded by the inability to efficiently generate engraftable human hematopoietic stem cells derived from ES and iPS cells5,26. Further evaluation of safe harbors could also include long-term studies in transgenic mice bearing transgenes in syntenic regions, as well as bioinformatics-assisted searches in the cumulated databases of common retroviral integration sites found in patients treated with retroviral vectors and not associated with any side effect27–29, although this information is of limited value in the absence of transgene expression data at these sites.
In conclusion, the present study provides a framework and a strategy combining bioinformatics and functional analyses for identifying safe harbors for transgene integration in the human genome. As our understanding of the function of the human genome and of genome-wide interactions advances, the definition of safe harbors will likely be refined over time, eventually building a registry of dependable genomic locations for the safe and effective genetic engineering of human cells.
ONLINE METHODS
Lentiviral vector construction and production
The four bicistronic vectors pLM-GO, pLM-YS, pLM-RK and pLM-CM, encoding violet excited GFP (vexGFP)-P2A-OCT4, mCitrine-P2A-SOX2, mCherry-P2A-KLF4 and mCerulean-P2A-cMYC, respectively, used to generate iPS cells from subject 1, have been previously described15. The single polycistronic vector pLM-SV2A was constructed as follows: Klf4-P2A-cMYC and cMyc-E2A-SOX2 cassettes were generated by overlapping PCR using Pfu polymerase (Stratagene) with primers introducing the respective intervening 2A peptide preceded by a Gly-Ser-Gly linker and restriction enzyme sites in the ends of each cassette. The Klf4-P2A-cMYC cassette was inserted into NcoI and EcoRI sites of the polylinker of cloning plasmid pSL1180. The cMYC-E2A-SOX2 cassette was digested with ClaI (site within the cMYC cDNA) and EcoRI and ligated downstream of the previous cassette. The OCT4 cDNA and a linker encoding T2A preceded by a Gly-Ser-Gly linker were ligated between AgeI and NcoI sites 3′ to the previously ligated cassettes. The entire OCT4-T2A-KLF4-P2A-cMYC-E2A-SOX2 cassette was transferred as an AgeI/SalI fragment into the pLM lentiviral vector backbone15 downstream of the human phosphoglycerate kinase (hPGK) promoter and upstream of the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) to generate the pLM-SV2A vector. The final vector was sequence-verified by DNA sequencing. Expression and correct processing of all four factors, OCT4, KLF4, SOX2 and c-MYC was confirmed by western blot analysis in transduced human MRC-5 fibroblasts (data not shown). The ‘floxed’ polycistronic vector pLM-fSV2A was derived from SV2A after insertion of annealed oligonucleotides containing a loxP site in a NheI site in the deleted U3 region of the 3′ LTR. The TNS9.3/fNG vector was derived from TNS9 (ref. 13) after insertion of a hPGK-Neo-P2A-eGFP cassette flanked by loxP sites and insertion of a 4-bp sequence in the β-globin 5′ UTR. The bicistronic cassette expressing the neomycin phosphoryltransferase (Neo) gene and eGFP linked by a P2A peptide preceded by a Gly-Ser-Gly linker was generated by overlapping PCR and inserted into AgeI and BglII sites of the pSL1180 cloning vector. The hPGK promoter was inserted between NotI/AgeI sites upstream of the Neo-P2A-eGFP cassette. To introduce loxP sites, we inserted annealed oligonucleotides in a MluI site, upstream of hPGK and in a SalI site downstream of eGFP, respectively. The entire floxed cassette was transferred as a MluI/SalI fragment into TNS9.3. For construction of an integrase-deficient lentiviral vector for Cre-mediated excision, an mCherry-P2A-Cre recombinase cassette was generated by overlapping PCR and inserted into the pLM lentiviral vector backbone under the transcriptional control of the cytomegalovirus immediate early promoter (pCMV). All oligonucleotide sequences are provided in Supplementary Table 8.
Vector production was performed by triple co-transfection of the plasmid DNA encoding the vector, pUCMD.G and pCMVΔR8.91 into 293T cells, as previously described31. For packaging of the integrase-deficient lentiviral vector encoding mCherry and Cre, pCMVΔR8.91 was replaced by pCMVΔR8.91N/N32 (kindly provided by E. Poeschla).
Human iPS cell generation
Skin punch biopsy specimens were obtained after informed consent from patients with β-thalassemia major at the Thalassemia Center at Cornell University. To establish fibroblast cell cultures, we sliced the biopsy into <1 mm fragments, which were transferred into 60 mm plates containing Eagle’s Modified Essential Medium with 10% FBS (FBS) (Hyclone). A cover slip was placed on top of each biopsy fragment and the plates were left undisturbed for 7–10 d to allow migration of cells.
Cryopreserved whole bone marrow specimens obtained from the Bone Marrow Transplantation Center at Memorial Sloan-Kettering Cancer Center (MSKCC) were thawed and, after density gradient separation over Ficoll, mononuclear cells were plated on tissue culture–treated dishes in Complete MesenCult Medium (Stem Cell Technologies). After ~2 weeks, adherent, fibroblast-like cells (Fig. 1b) were harvested by trypsinization and expanded.
iPS cell generation was performed as previously described15. Skin fibroblasts or MSCs at passages 2–7 were plated in gelatin-coated, 6-well plates at a density of 1 × 105 cells per well and transduced 24 h later with lentiviral vectors encoding OCT4, SOX2, KLF4 and c-MYC in the presence of 4 μg/ml polybrene. Media were changed 24 h later and replaced every day thereafter with hES cell media supplemented with 6 ng/ml FGF2 (R&D Systems) and 0.5 mM VPA (Sigma). Fifteen to 25 d after transduction, colonies with hES cell morphology were mechanically dissociated and transferred into plates pre-seeded with mitomycin C–treated mouse embryonic fibroblasts (MEFs) (GlobalStem). Cells were thereafter passaged with dispase and expanded to establish iPS cell lines.
The vector systems used for iPS cell generation were as follows: a combination of four bicistronic lentiviral vectors co-expressing OCT4, KLF4, c-MYC and SOX2 with a distinct fluorescent protein15 (subject 1), a polycistronic vector co-expressing all four factors in a single transcript, SV2A (subject 2) and its derivative fSV2A, containing a loxP site in the 3′ LTR (subjects 4 and 5) (Supplementary Fig. 9d).
iPS cell characterization
Flow cytometry analysis, immunofluorescence, OCT4 promoter methylation analysis, karyotyping and teratoma formation assays were performed as described15,30.
For assessment of expression of pluripotency genes, total RNA from thal-iPS cell lines was isolated with Trizol (Invitrogen). Reverse transcription was performed with Superscript III (Invitrogen) and qPCR was performed with primers shown in Supplementary Table 9 using SYBR Green. Reactions were carried out in duplicate in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Expression was calculated by relative quantification using the ΔΔCt method with actin as endogenous control.
For teratoma formation assays, undifferentiated iPS cells were suspended in hES medium containing 10 μM of the Rho-associated kinase (Rock) inhibitor Y-27632 (Tocris)33. Approximately 2 × 106 cells were injected intramuscularly into NOD-SCID IL2Rg-null mice (Jackson Laboratory). Five to six weeks later, the tumors were surgically dissected and fixed in 4% formaldehyde. Cryosectioned samples were stained with hematoxylin and eosin for histological analysis and with antibodies against cytokeratin (CK) 20, vimentin and S-100 for immunohistochemical analysis. All animal experiments were conducted in accordance with protocols approved by MSKCC Institutional Animal Care and Use Committee (IACUC) and following National Institutes of Health guidelines for animal welfare.
Assessment of reprogramming vector silencing
qRT-PCR was performed with the primers and probes shown in Supplementary Table 9. Reactions were carried out in duplicate in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Expression was calculated by relative quantification using the ΔΔCt method with GAPDH as endogenous control.
Karyotyping
Standard G-banding analysis was performed at the MSKCC molecular cytogenetics core laboratory. Chromosome analysis was performed on a minimum of 10 4,6-diamidino-2-phenylindole (DAPI)-banded meta-phases. All metaphases were fully karyotyped.
β-thalassemia genotyping
Genomic DNA was extracted from thal-iPS cell lines, dermal fibroblasts and MSCs using the DNeasy kit (Qiagen). 200 ng of DNA was used as template in a PCR reaction using the primer pair β-thal-1 (Supplementary Table 9). The 714-bp PCR product was gel-purified and sequenced.
Cre-mediated vector excision
iPS cell lines thal5.10 and thal5.11 were dissociated into single cells with accutase for 30 min at 37 °C. After incubation for 1 h on gelatin-coated plates to allow adherence and subsequent removal of MEFs, cells were plated at 1 × 105 cells per well of a 6-well plate with Matrigel (BD Biosciences) in MEF-conditioned media supplemented with 6 ng/ml FGF2 and 10 μM of Y-27632. The next day the cells were transduced with vector supernatants in the presence of 4 μg/ml polybrene for 16 h. The transduced cells were dissociated with accutase 48 h later, vigorously triturated into single cells and replated at titrated densities (from 100 to 500 cells per cm2) on a layer of mitomycin C–treated MEFs with 10 μM of Y-27632. An aliquot of the cells was used for flow cytometry analysis of mCherry-Cre expression. After 10–15 d, single cell colonies were mechanically dissociated and replated into 6-well dishes on mitomycin C–treated MEFs. One week later, ~100–200 cells from each clone were manually picked under a stereoscope into 0.2 ml tubes and lysed in 25 μl lysis buffer with 100 μg/ml proteinase K, as previously described34. We used 3 μl of cell lysate to screen for excision of the fSV2A vector by multiplex PCR with three primer pairs: fSV2A- 1, fSV2A-2 and LTR (Supplementary Fig. 10b). The PCR products were analyzed by agarose gel electrophoresis. Six out of 47 screened clones from line thal5.10 and 7 out of 42 clones from line thal5.11 were found to no longer contain the integrated provirus. To assess complete vector excision and lack of integration of the Cre-expressing vector, we performed qPCR with primers and probes specific for the gag region of the lentiviral vectors and the human albumin gene (Supplementary Table 9). For Southern blot analysis 5 μg of genomic DNA was digested with XmaI or BglI and probed with a radiolabeled SalI-KpnI fragment spanning the WPRE.
Globin gene transfer and selection of single vector copy thal-iPS cell clones
iPS cell lines thal1.52 and thal2.1 were prepared and transduced with varying MOI of the TNS9.3/fNG vector, as described above. The transduced cells were dissociated 48 h later with accutase and vigorously triturated into single cells. An aliquot of the cells was used for flow cytometry analysis of eGFP expression. Transduced cells with gene transfer <30% (as estimated by the percentage of eGFP+ cells) were replated at a density of 1,500 cells/cm2 on a layer of Neo-resistant mitomycin C–treated MEFs (GlobalStem). G418 (Invitrogen) was added at a concentration of 12.5 μg/ml between days 5 and 9 after transduction. Approximately 20 d post-transduction, Neo-resistant colonies were manually picked and replated into 6-well dishes on mitomycin C-treated MEFs. One week later, ~100–200 cells from each clone were manually picked and lysed as described above. We used 5 μl of cell lysate for measurement of TNS9.3/fNG vector copy number (VCN) with multiplex quantitative PCR (qPCR) using sets of primers and probes specific for the globin vector (GV1) and for the human albumin gene (Supplementary Table 9). To determine absolute VCN, we generated a standard curve using serial dilutions of a plasmid containing both vector and albumin gene amplicons. Reactions were carried out in triplicate in an ABI 7500 detection system (Applied Biosystems). We digested 5–10 μg of genomic DNA extracted from single vector copy iPS clones with NcoI, XbaI or EcoRI and analyzed it by Southern blot analysis, as described13,34, using a radiolabeled NcoI-BamHI fragment spanning exons 1 and 2 of the human β-globin gene or the eGFP cDNA as probe.
Integration site analysis
fSV2A vector integrations were mapped by linear amplification mediated (LAM)-PCR, using digestion with Tsp509I, as described35. PCR products were TOPO cloned and sequenced.
For analysis of TNS9.3/fNG vector integration sites in thal-iPS cells with relation to the 5 safe harbor criteria, line thal5.10-Cre8 was transduced with the vector at high MOI (~100) as described above and genomic DNA was extracted from the polyclonal population 5 d after transduction. Integration sites were isolated by ligation mediated (LM)-PCR, sequenced by 454/Roche pyrosequencing, processed and analyzed, as previously described36.
TNS9.3/fNG vector integration sites in single-copy clones were mapped by inverse PCR (iPCR). We digested 1 μg of genomic DNA with HinP1I or HpyCH4IV, and diluted and incubated it with T4 DNA ligase. After phenol/chloroform extraction and ethanol precipitation, DNA was digested with XbaI or SalI and used as template in a PCR reaction with primers iPCR F and iPCR R (Supplementary Table 9). The PCR product was analyzed on a 3% agarose gel and all bands visualized with ethidium bromide were excised, purified and sequenced.
Integration sites were judged to be authentic if the sequences were adjacent to vector LTR ends and had a unique hit when aligned to the draft human genome (University of California Santa Cruz, UCSC hg18) using BLAT (http://genome.ucsc.edu/)37. Genomic annotations were also obtained from UCSC hg18 Genome Browser and mapped against the integration sites.
Integration sites were confirmed by PCR with LTR universal forward primer for fSV2A vector integrations or GV forward primer for TNS9.3/fNG integrations and reverse primers specific for the genomic sequence adjacent to the integration. All primer sequences are shown in Supplementary Table 9.
For computing the frequencies of integration sites with relation to our safe harbor criteria, gene data were obtained from UCSC RefSeq Gene and wgRna (miRNA) track version 1/31/10. A few of the gene symbols in RefSeq gene track did not match up to the RefSeq gene database from NCBI. Therefore, the mismatched gene symbols from UCSC database were converted to the proper gene symbols as found on NCBI. A few sources used a gene alias instead of gene symbol in which case the corresponding gene symbol was found using NCBI’s gene info table: ftp://ftp.ncbi.nih.gov/gene/DATA/gene_info.gz. UCRs in the human genome were obtained from reference 11 and the data were downloaded from http://users.soe.ucsc.edu/~jill/ultra.html. As the genomic coordinates used in the publication were from an older assembly, we converted the coordinates to the hg18 freeze using UCSC lift genome annotations tool.
Erythroid differentiation
For hematopoietic differentiation of human iPS cells, embryoid bodies were generated and cultured, as previously described30. Briefly, intact human iPS cell colonies were collected with dispase and plated in low-attachment dishes (Corning) in DMEM with 20% FBS, 1% nonessential amino acids (NEAA), 1 mM 3-glutamine and 0.1 mM β-mercaptoethanol (MTG), supplemented with 40 ng/ml bone morphogenetic protein 4 (BMP4) and 40 ng/ml vascular endothelial growth factor (VEGF). Two days later, the medium was switched to X-VIVO 15 (Lonza) with 1% NEAA, 1 mM 3-glutamine and 0.1 mM MTG supplemented with 40 ng/ml BMP4, 40 ng/ml VEGF, 20 ng/ml FGF2, 40 ng/ml stem cell factor (SCF), 40 ng/ml Flt3 ligand (Flt3L) and 40 ng/ml thrombopoietin (TPO) and replaced every 3 d. At day 8 of embryoid body culture cells were dissociated with accutase and passed through a 22G needle 3–4 times. For further erythroid differentiation the cells were plated in low-attachment dishes in X-VIVO 15 supplemented with 20% BIT (Stem Cell Technologies), 1% NEAA, 1 mM 3-glutamine, 0.1 mM MTG, 100 ng/ml SCF, 6 U/ml erythropoietin and 10−6 M dexamethasone. Media were replenished every 3 d for 15 d. Benzidine staining was performed as described38. Erythroid differentiation of CD34+ cells from mobilized peripheral blood of two healthy individuals was performed as previously described38.
Flow cytometry
Undifferentiated iPS cells were dissociated with accutase, stained with Alexa Fluor 647-conjugated anti-Tra-1–81 or anti-Tra-1-60 or anti-SSEA3 or anti-SSEA4 and PE-Cy5-conjugated anti-HLA-ABC antibodies (BD Biosciences). The erythroid progeny of iPS cells were incubated with allophycocyanin (APC)-conjugated anti-CD34 or APC-conjugated anti-glycophorin A (GPA), PerCP-conjugated anti-CD45 and PE-conjugated anti-CD71 (BD Biosciences). Data were acquired in a LSRII cytometer (BD Biosciences) and analyzed with the FlowJo software (version 8.8.4; Tree Star).
Analysis of β-globin expression
Total RNA was isolated with Trizol (Invitrogen). For quantitative RT-PCR, reverse transcription was performed with Superscript III (Invitrogen) and qPCR was performed with primers and probes specific for the human α- and β-globin transcripts (Supplementary Table 9). RNA from human CD34+ cells isolated from mobilized peripheral blood from four healthy individuals and differentiated in vitro along the erythroid lineage, as described above, was used as reference. Reactions were carried out in triplicate in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Vector-encoded β-globin expression per gene copy was calculated by relative quantification using the ΔΔCt method with α-globin as endogenous control, relative to the average expression in four reference samples (accounting for two endogenous β-globin alleles).
The results of quantitative RT-PCR were corroborated in selected samples by quantitative primer extension assay with [32P]dATP end-labeled primers PE-alpha and PE-beta (Supplementary Table 9) specific for the human α- and β-globin transcripts, respectively, as previously described13. Briefly, the radiolabeled primers were annealed to 0.25–1 μg of RNA and reactions were performed using the Primer Extension System-AMV Reverse Transcriptase kit (Promega). The predicted product length is 60 bp for the α-globin transcript, 80 bp for the endogenous β-globin transcript and 84 bp for the vector-encoded β-globin transcript. Radioactive bands were quantified by phosphorimager analysis (BioRad).
Tissue specificity of vector-encoded β-globin expression was assessed by qRT-PCR in undifferentiated thal-iPS cell clones transduced with the TNS9.3/fNG vector and their erythroid progeny using primers and probes specific for β-globin and GAPDH (Applied Biosystems).
HPLC analysis was performed at the MSKCC analytical pharmacology core laboratory. Frozen cord blood and cell pellets were thawed at 24 °C. The blood samples were proportionally diluted volumetrically to be in the calibration standard curve range (10–400 μg/ml). Cell pellets were incubated with 0.1% of sodium lauryl sulfate solution in an ice-water batch for 15 min. All samples were then centrifuged at 14,000g for 5 min and the supernatants were filtered with 0.45 μm polyethersulfone syringe filters before the assay. A gradient elution with a VYDAC Protein C4 column of 250-mm length (inner diameter, 4.6 mm; particle size, 5 μm) and a mobile phase containing acetonitrile and 0.1% trifluoroacetic acid (mobile phase A: 4/1, vol/vol and mobile phase B: 2/3, vol/vol) were used and the mobile phase composition was changed from 10% B to 46% B over 40 min. The separation of the sub-chains of hemoglobin from any potential interference was monitored at 220 nm and the flow rate was set at 1.0 ml/min. Calibration curves were determined for the α- and β-globin chains to permit conversion of peak areas to individual sub-chain amounts against the external reference standards.
Expression microarray analysis
Whole genome gene expression analysis was performed on Illumina BeadArrays at the MSKCC genomics core laboratory. The summarized data from the chips were normalized by variance stabilization normalization (vsn) using the vsn package in Bioconductor39. We used the vsn method to correct for possible batch effects in the data. The differential gene expression analysis was performed using limma Bioconductor package40. The limma package uses linear models to assess differential expression and uses empirical Bayesian methods to provide stable results even when the number of arrays is small. Multiple hypothesis correction was performed using the Benjamini-Hochberg method. Expression of each gene within 300 kb on either side of the globin vector integration site was compared to expression in all other clones with different vector insertions, as well as in untransduced lines.
CGH array analysis
Comparative genomic hybridization analysis of iPS line thal5.10-Cre8 (vector-excised) and the thal5 MSCs this line was derived from was performed on the Agilent 1M CGH platform at the MSKCC genomics core laboratory. The data were normalized using GC-RMA normalization. Circular Binary Segmentation41 from the DNAcopy package of Bioconductor was used to determine any significant copy number alterations. A segment mean of < −0.3 or > +0.3 is generally considered to be an aberration and we did not find any aberrations in the analyzed sample using this threshold.
Supplementary Material
Acknowledgments
We thank X. Wang and N. Wu for assistance with HPLC analysis; L. Ferro, E. Reed, J. Miller, M. Leversha and M. Tomishima for technical assistance; F. Boulad, Memorial Sloan-Kettering Cancer Center New York for bone marrow specimens; and A. Athanassiadou for advice on β-thalassemia genotyping. pCMVΔR8.91N/N was kindly provided by E. Poeschla, Mayo Clinic, Rochester, Minnesota. This work was supported by the Starr Foundation (Tri-Institutional Stem Cell Initiative, Tri-SCI-018), the New York State Stem Cell Science, NYSTEM (N08T-060) and National Heart, Blood, and Lung Institute grant HL053750 (M.S.). F.D.B., S.L.R. and N.M. were supported by National Institutes of Health grants AI052845 and AI082020 (F.D.B.). G.L. was supported by a New York Stem Cell Foundation Druckenmiller fellowship.
Footnotes
Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
E.P.P. conceived and designed the study, designed and performed experiments, analyzed data and wrote the manuscript; G.L. performed iPS cell differentiation experiments; N.M. performed bioinformatics analyses; M.S. and C.L. analyzed microarray data; L.M.S.T. provided technical assistance; K.K. performed histological analyses of teratomas; S.L.R. generated and analyzed integration site data; P.G. provided skin biopsy samples from β-thalassemia patients; A.V. generated microarray data; I.R., F.D.B. and L.S. analyzed data; M.S. conceived and designed the study, analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 5.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 8.Ott MG, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1–EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 9.Howe SJ, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavazzana-Calvo M, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 12.Kustikova O, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 13.May C, et al. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 14.Sadelain M, Boulad F, Lisowki L, Moi P, Riviere I. Stem cell engineering for the treatment of severe hemoglobinopathies. Curr Mol Med. 2008;8:690–697. doi: 10.2174/156652408786241357. [DOI] [PubMed] [Google Scholar]
- 15.Papapetrou EP, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang KH, et al. Definitive-like erythroid cells derived from human embryonic stem cells coexpress high levels of embryonic and fetal globins with little or no adult globin. Blood. 2006;108:1515–1523. doi: 10.1182/blood-2005-11-011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu C, Olivier EN, Velho M, Bouhassira EE. Globin switches in yolk sac-like primitive and fetal-like definitive red blood cells produced from human embryonic stem cells. Blood. 2008;111:2400–2408. doi: 10.1182/blood-2007-07-102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang KH, et al. Globin phenotype of erythroid cells derived from human induced pluripotent stem cells. Blood. 2010;115:2553–2554. doi: 10.1182/blood-2009-11-252650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou J, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JR, et al. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 23.Irion S, et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 24.Safaya S, Rieder RF, Dowling CE, Kazazian HH, Jr, Adams JG., 3rd Homozygous beta-thalassemia without anemia. Blood. 1989;73:324–328. [PubMed] [Google Scholar]
- 25.Werbowetski-Ogilvie TE, et al. Characterization of human embryonic stem cells with features of neoplastic progression. Nat Biotechnol. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
- 26.Ji J, et al. OP9 stroma augments survival of hematopoietic precursors and progenitors during hematopoietic differentiation from human embryonic stem cells. Stem Cells. 2008;26:2485–2495. doi: 10.1634/stemcells.2008-0642. [DOI] [PubMed] [Google Scholar]
- 27.Deichmann A, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aiuti A, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarzwaelder K, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papapetrou EP, Kovalovsky D, Beloeil L, Sant’angelo D, Sadelain M. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J Clin Invest. 2009;119:157–168. doi: 10.1172/JCI37216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saenz DT, et al. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J Virol. 2004;78:2906–2920. doi: 10.1128/JVI.78.6.2906-2920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 34.Papapetrou EP, Ziros PG, Micheva ID, Zoumbos NC, Athanassiadou A. Gene transfer into human hematopoietic progenitor cells with an episomal vector carrying an S/MAR element. Gene Ther. 2006;13:40–51. doi: 10.1038/sj.gt.3302593. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 36.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2009;28:287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
- 39.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarray data. Stat Appl Genet Mol Biol. 2003;2:Article 3. doi: 10.2202/1544-6115.1008. [DOI] [PubMed] [Google Scholar]
- 40.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 41.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.