Abstract
Objectives
To evaluate the association of genetic variation with late-onset Alzheimer disease (AD) in African Americans, including genes implicated in recent genome-wide association studies of whites.
Design
We analyzed a genome-wide set of 2.5 million imputed markers to evaluate the genetic basis of AD in an African American population.
Subjects
Five hundred thirteen well-characterized African American AD cases and 496 cognitively normal African American control subjects.
Setting
Data were collected from multiple sites as part of the Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study and the Henry Ford Health System as part of the Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans (GenerAAtions) Study.
Results
Several significant single-nucleotide polymorphisms (SNPs) were observed in the region of the apolipoprotein E gene (APOE). After adjusting for the confounding effects of APOE genotype, one of these SNPs, rs6859 in PVRL2, remained significantly associated with AD (P=.0087). Association was also observed with SNPs in CLU, PICALM, BIN1, EPHA1, MS4A, ABCA7, and CD33, although the effect direction for some SNPs and the most significant SNPs differed from findings in data sets consisting of whites. Finally, using the African American genome-wide association study data set as a discovery sample, we obtained suggestive evidence of association with SNPs for several novel candidate genes.
Conclusions
Some genes contribute to AD pathogenesis in both white and African American cohorts, although it is unclear whether the causal variants are the same. A larger African American sample will be needed to confirm novel gene associations, which may be population specific.
Alzheimer disease (AD) IS the most common form of dementia. Environmental and host risk factors for common late-onset AD (LOAD) include low educational level, diabetes mellitus, hypertension, and head trauma. Genetic factors also influence LOAD risk, evidenced by heritability estimates as high as 75%1 and analyses showing transmission of a major gene for the disease in families.2 Until recently, the apolipoprotein E gene (APOE [OMIM +107741]) was the only one generally recognized to influence LOAD risk.3 In whites, homozygosity for the ε4 variant is associated with an increased risk by as much as 15 times that of the most common APOE genotype (ε3/ε3).4
Genome-wide association studies (GWASs) have reported genome-wide significant single-nucleotide polymorphisms (SNPs) across a 70-kilobase (kb) region that includes APOE and several neighboring genes,5 namely, poliovirus receptor–related 2 (PVRL2 [OMIM *600798]), translocase of outer mitochondrial membrane 40 yeast homologue (TOMM40 [OMIM *608061]), and apolipoprotein C-I (APOC1 [OMIM *107710]). Both the TOMM40 and APOC1 genes have been considered possible risk factors for AD independent of APOE. Several lines of inquiry have implicated TOMM40 as having an effect on AD risk, including evidence of a role of mitochondria in AD pathogeneisis,6 association of an intronic TOMM40 repeat polymorphism with age at the onset of AD symptoms among subjects lacking the ε4 allele,7 and association of a haplotype spanning TOMM40 with expression of APOE.8 However, other studies did not find an effect of TOMM40 after adjusting for APOE.9,10 A polymorphism immediately upstream of the APOC1 gene has also been proposed as a possible risk locus for AD.11,12 This polymorphism is in strong linkage disequilibrium (LD) with the APOE risk locus, but this pattern varies substantially by population.13 Studies in mice and humans indicate that APOC1 expression has an effect on memory.14–16 Other studies reported that APOC1 modifies the risk of AD independent of or through interaction with APOE.17,18
The GWASs conducted by several large consortia have identified robust evidence of an association with genes outside the APOE region, including clusterin (CLU [OMIM *185430]),19–21 phosphatidylinositol-binding clathrin assembly protein (PICALM [OMIM *603025]),20,21 complement component (3b/4b) receptor 1 (CR1 [OMIM *120620]),19 bridging integrator 1 (BIN1 [OMIM *601248]),20 CD2-associated protein (CD2AP [OMIM *604241]),22 ephrin type-A receptor 1 (EPHA1 [OMIM *179610]),20,22 the membrane-spanning 4A (MS4A) gene cluster,22,23 myeloid-associated antigen CD33 (CD33 [OMIM *159590]),22 and ATP-binding cassette, subfamily A (ABC1), member 7 (ABCA7 [OMIM *605414]).23 Findings with CLU, PICALM, CR1, and ABCA7 have been replicated.22–24
Because there are population differences in LD and allele frequencies, most association studies have focused on a single population to decrease genetic background noise and reduce the likelihood of false-positive findings due to confounding. Thus, confirmation in other populations is required to determine the generalizability of the contribution of each gene to AD risk and the possibility of population-specific causative variants. Although the effect of APOE has been investigated extensively in multiple populations,4,25,26 few African American cohorts have been included in GWASs for AD.
In the present study, we genotyped more than 1000 African American cases and controls for more than 600 000 SNPs covering the entire genome. Genotypes for 2.5 million SNPs imputed from HapMap reference panels were used to investigate the contribution of genes previously implicated in whites to AD risk and to identify novel AD risk variants in this population. We also analyzed a comparable set of SNPs in 5 white AD GWAS data sets containing more than 9700 subjects to replicate novel findings and for comparison with previously obtained results.
METHODS
Subjects were ascertained from 2 genetic studies of AD focused on African Americans. One subject group includes participants of the Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study, which contains primarily discordant sibling pairs. Enrollment, data collection, and diagnostic procedures in the MIRAGE Study are explained in detail elsewhere.27 A second group of primarily unrelated individuals includes participants of the Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans (GenerAAtions) Study, who were identified through the electronic claims database of the Henry Ford Health System. Community-dwelling African Americans 65 years or older who had at least 1 encounter with the Henry Ford Health System in the 3 years before their recruitment and who had an available proxy informant were eligible for this study. Cases met criteria of the National Institute of Neurological and Communicative Diseases and Stroke–Alzheimer’s Disease and Related Disorders Association for possible or probable AD, determined in a consensus conference that included a behavioral neurologist (R.S.), psychiatrist, neuropsychologist, and a behavioral neurology nurse practitioner.
For comparison, we also examined 5 white AD GWAS data sets containing 3568 cases and 6205 controls, namely, the MIRAGE Study white families, and 4 data sets obtained from a public database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap), including the Alzheimer Disease Neuroimaging Initiative (ADNI),28,29 a Canadian study on genetics of Alzheimer disease associations (GenADA),30 the National Institute on Aging–Late-Onset Alzheimer’s Disease Family Study (NIA-LOAD),31 and the Framingham Heart Study.32–34 The numbers of cases and controls in each data set are shown in Table 1.
Table 1.
Sample Sizes of African American (Discovery) and White (Replication) Data Sets
| Data Sets
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| African American
|
White
|
||||||||
| MIRAGE | GenerAAtions | Total | MIRAGE | ADNI | GenADA | NIA-LOAD | FHS | Total | |
| No. of cases | 267 | 246 | 513 | 609 | 254 | 669 | 1839 | 197 | 3568 |
| No. of controls | 292 | 204 | 496 | 875 | 169 | 713 | 1983 | 2465 | 6205 |
| Total | 559 | 450 | 1009 | 1484 | 423 | 1382 | 3822 | 2662 | 9773 |
Abbreviations: ADNI, Alzheimer Disease Neuroimaging Initiative; FHS, Framingham Heart Study; GenADA, Canadian study on genetics of Alzheimer disease associations; GenerAAtions, Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans; MIRAGE, Multi-Institutional Research on Alzheimer Genetic Epidemiology; NIA-LOAD, National Institute on Aging–Late-onset Alzheimer’s Disease Family Study.
Genotyping methods, procedures for data cleaning and imputation, and statistical methods are described in detail in the supplementary material (http://www.bumc.bu.edu/genetics/results/aa_alzheimer). Briefly, the APOE genotyping method varied by study. Imputation of autosomal SNP genotypes was performed using the Markov Chain Haplotyping (MaCH) software35 based on the HapMap 2 and 3 reference SNP panels (http://hapmap.ncbi.nlm.nih.gov/). Imputed SNPs were tested for association with AD in the family-based data sets using generalized estimating equations (GEE)36,37 to account for nonindependence of family members. Analysis of the case-control data sets was performed using logistic regression models. All tests of association were adjusted for sex and age at examination. Two models were evaluated for each SNP, one with and the other without a term for APOE genotype coded as the number of APOE ε4 alleles. Unless otherwise noted, all results are from the ε4-unadjusted model. An additional analysis of SNPs in the APOE region included an adjustment for APOE genotype classified into one of the following 4 categories: ε2/ε2 and ε2/ε3; ε3/ε3; ε2/ε4 and ε3/ε4; and ε4/ε4. The SNP association results obtained from individual data sets were combined by meta-analysis using the inverse variance method implemented in the software package METAL.38 Nominal (uncorrected for multiple testing) P values are reported throughout. In the genome-wide analysis, a P value of 5×10−8 was used as the threshold for significance, and a threshold of P<10−5 was considered suggestive evidence of association.
RESULTS
APOE REGION
The frequency distributions of APOE genotypes for African American and white cohorts are shown in Table 2. In the African American cohort, the ε4 allele is very significantly associated with AD (P=9.69×10−23). Analysis of individual genotypes showed evidence of a significant protective effect of ε2 (ε2/ε2 and ε2/ε3 genotypes) compared with the ε3/ε3 genotype and an exponential increase in risk associated with the dose of ε4 (Table 3). The odds ratio (OR) estimates and APOE allele frequencies, showing a higher rate of ε4 alleles in African American controls compared with white controls, are in agreement with a previous study of the APOE association in the MIRAGE Study African American cohort.26
Table 2.
APOE Genotype Frequency by Population and Alzheimer Disease Status
| Population | Genotype, No. (%) of Subjectsa |
||||||
|---|---|---|---|---|---|---|---|
| ε2/ε2 | ε2/ε3 | ε2/ε4 | ε3/ε3 | ε3/ε4 | ε4/ε4 | Missing | |
| African American | |||||||
| Cases | 0 | 25 (4.9) | 19 (3.7) | 147 (28.7) | 227 (44.2) | 78 (15.2) | 17 (3.3) |
| Controlsb | 2 (0.4) | 67 (13.3) | 12 (2.4) | 210 (41.7) | 172 (34.1) | 18 (3.6) | 15 (3.0) |
| White | |||||||
| Cases | 9 (0.3) | 112 (3.1) | 93 (2.6) | 1045 (29.3) | 1743 (48.9) | 552 (15.5) | 14 (0.4) |
| Controls | 21 (0.3) | 673 (10.8) | 136 (2.2) | 3562 (57.4) | 1489 (24.0) | 182 (2.9) | 139 (2.2) |
Abbreviation: APOE, apolipoprotein E.
Percentages have been rounded and might not total 100.
The total number of controls sums to 6202 rather than 6205 owing to the presence of 3 rare APOE isoforms observed in the Framingham Heart Study subjects.
Table 3.
Odds of Alzheimer Disease for APOE Genotypes Relative to ε3/ε3
| Genotypes | OR (95% CI) | P Value |
|---|---|---|
| ε2/ε2 or ε2/ε3 | 0.43 (0.26–0.71) | .0094 |
| ε2/ε4 or ε3/ε4 | 2.08 (1.58–2.74) | 1.96 × 10−7 |
| ε4/ε4 | 8.23 (4.78–14.15) | 2.62 × 10−14 |
Abbreviations: APOE, apolipoprotein E; OR, odds ratio.
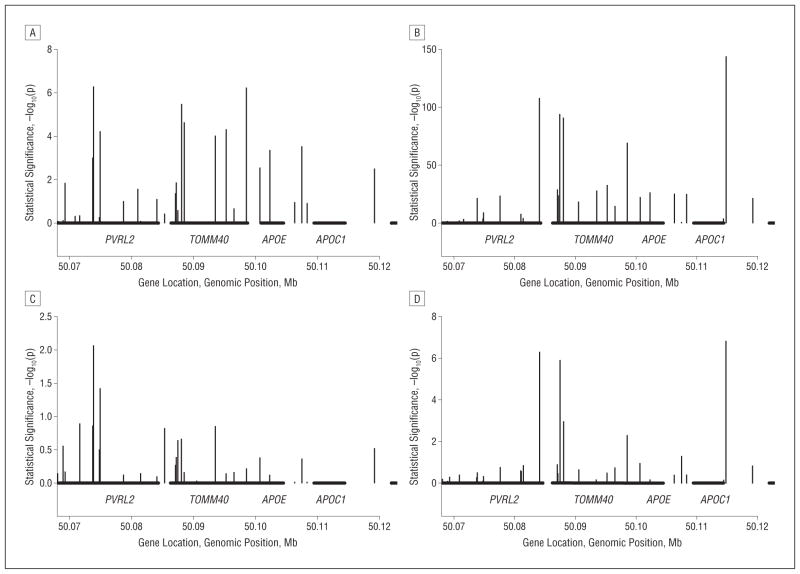
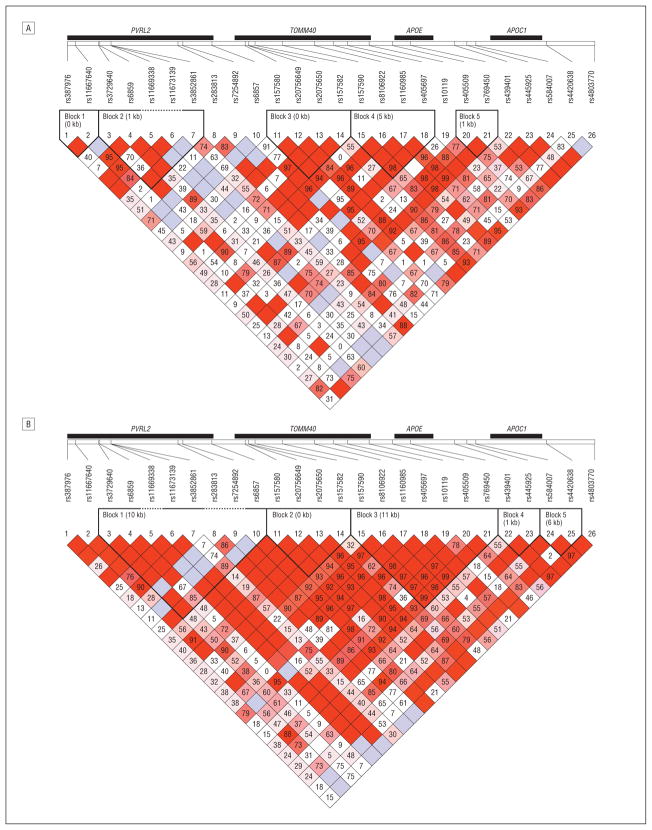
Analyses of the APOE region in the African American data sets revealed a highly significant association with 3 markers within 25 kb of APOE, including PVRL2 SNP rs6859 (P=5.39×10−7) and TOMM4 SNPs rs157582 (P=3.26×10−6) and rs10119 (P=5.95×10−7) (Table 4 lists top SNPs in the region; see eTable 1 in the supplementary material for all nominally significant SNPs). Only rs6859 remained significant after adjustment for APOE genotype (P=.0087). Figure 1 shows the unadjusted and APOE genotype–adjusted results for all SNPs in the region immediately flanking APOE. Figure 2 shows the estimated LD in the region for the African American and MIRAGE Study white data sets. In the white cohorts, ε4 was strongly associated with AD (P=6.80×10−147). In addition, without adjustment for the ε4 allele, 19% of SNPs in this region were very significantly associated with AD (P < 10−5). The top-ranked SNPs in this group are rs4420638 in APOC1 (P=1.07×10−144), rs6857 in PVRL2 (P = 1.49 × 10−108), and rs2075650 in TOMM40 (P=1.70×10−94). After adjustment for APOE genotype, only 7 SNPs remained significant at P<.05, including rs6857 (P=4.98×10−7), rs4420638 (P=1.54×10−7), and rs2075650 in TOMM40 (P=1.25×10−6).
Table 4.
Most Significantly Associated Markers in the APOE Region in African American and White Cohorts
| Marker | Position, dbSNP Build 129. bp | Gene | Effect Allele | Unadjusted for APOE |
APOE Genotype Adjusted
|
||||
|---|---|---|---|---|---|---|---|---|---|
| OR | P Value | Directiona | OR | P Value | Directiona | ||||
| African American | |||||||||
| rs6859 | 50 073 874 | PVRL2 | A | 1.58 | 5.39 × 10−7 | + + | 1.30 | .0087 | + + |
| rs3852861 | 50 074 901 | PVRL2 | T | 0.64 | 5.99 × 10−5 | − − | 0.78 | .038 | − − |
| rs157582 | 50 088 059 | TOMM40 | T | 1.60 | 3.26 × 10−6 | + + | 1.15 | .222 | − + |
| rs157583 | 50 088 513 | TOMM40 | T | 2.08 | 2.48 × 10−5 | + + | 0.92 | .700 | − − |
| rs8106922 | 50 093 506 | TOMM40 | A | 1.48 | 9.73 × 10−5 | + + | 1.18 | .144 | − + |
| rs1160985 | 50 095 252 | TOMM40 | T | 0.65 | 4.92 × 10−5 | − − | 1.04 | .730 | + − |
| rs10119 | 50 098 513 | TOMM40 | A | 1.80 | 5.95 × 10−7 | + + | 1.07 | .614 | − + |
| rs445925 | 50 107 480 | 5′ APOC1 | A | 1.57 | 3.02 × 10−4 | + + | 1.13 | .443 | + + |
| White | |||||||||
| rs6857 | 50 084 094 | PVRL2 | T | 3.23 | 1.49 × 10−108 | + + ?? + | 1.50 | 4.98 × 10−7 | + + ?? + |
| rs157580 | 50 087 106 | TOMM40 | A | 1.70 | 2.77 × 10−29 | + + ?? + | 1.09 | .130 | + + ?? + |
| rs2075650 | 50 087 459 | TOMM40 | A | 0.35 | 1.70 × 10−94 | − − ?? − | 0.71 | 1.25 × 10−6 | − − ?? − |
| rs157582 | 50 088 059 | TOMM40 | T | 2.73 | 2.54 × 10−91 | + + ?? + | 1.30 | .0011 | + − ?? + |
| rs8106922 | 50 093 506 | TOMM40 | A | 1.66 | 2.06 × 10−28 | + + ?? + | 1.02 | .679 | + − ? − |
| rs1160985 | 50 095 252 | TOMM40 | T | 0.58 | 2.78 × 10−33 | − − ?? − | 0.95 | .326 | − + ?? + |
| rs10119 | 50 098 513 | TOMM40 | A | 2.48 | 8.15 × 10−70 | + + ?? + | 1.22 | .0051 | + − ?? + |
| rs4420638 | 50 114 786 | APOC1 | A | 0.23 | 1.07 × 10−144 | − − ? − − | 0.61 | 1.54 × 10−7 | − − ? − − |
Abbreviations: APOC1, apolipoprotein C-I; APOE, apolipoprotein E; bp, base pairs; dbSNP, database single-nucleotide polymorphism; OR, odds ratio; PVRL2, poliovirus receptor-related 2; TOMM40, translocase of outer mitochondrial membrane 40 yeast homologue.
For the African American cohort, symbols indicate increased (+) or decreased (−) risk in the Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans and Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) data sets; for the white cohort, increased (+) or decreased (−) risk in the MIRAGE, Alzheimer Disease Neuroimaging Initiative, Framingham Heart Study, Canadian Study on Genetics of Alzheimer Disease Associations, and National Institute on Aging–Late-Onset Alzheimer’s Disease Family Study data sets.
Figure 1.
Association and linkage disequilibrium in the apolipoprotein E (APOE) region. A and B, Unadjusted findings in African Americans and whites, respectively; C and D, APOE-genotype adjusted findings in African Americans and whites, respectively. APOC1 indicates apolipoprotein C-I; Mb, megabase; PVRL2, poliovirus receptor–related 2; and TOMM40, translocase of outer mitochondrial membrane 40 yeast homologue.
Figure 2.
Linkage disequilibrium in the apolipoprotein E (APOE) region. A, African American cohort data sets. B, White cohort data sets. Other gene names are described in the legend to Figure 1.
PREVIOUSLY IMPLICATED REGIONS
Results for African Americans in the regions of AD associations from the white GWAS are summarized in Table 5. There was no evidence of association in African Americans with 2 of these 3 CLU SNPs, including rs11136000, which was consistently significant across multiple studies in white samples. A nominally significant association (P=.034) was observed with rs2279590 that had been previously reported in whites. However, the minor allele (T) was associated with increased AD risk in this African American sample (OR, 1.41), whereas the T allele is protective in the white sample (OR, 0.87). Two additional nominally significant SNPs were observed in CLU, the most significant of which was rs9331926 (P=.020); complete results including all nominally significant SNPs in previously implicated regions are summarized in eTable 2 in the supplementary material.
Table 5.
Association of Alzheimer Disease With Genome-wide Significant Regions in White GWASs
| Gene, SNP | Effect Allele | AFCEU | Previously Reported Results in White Cohort
|
Results in African American Cohort
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | AF | P Value | OR (95% CI) | AF | P Value | OR (95% CI) | ||||
| CLU | ||||||||||
| rs2279590 | T | 0.31 | Lambert et al,19 2009 | NR | 9.2 × 10−8 | 0.87 (0.83–0.92) | 0.12 | .034 | 1.41a (1.03–1.95) | |
| rs11136000 | T | 0.35 | Lambert et al,19 2009 | NR | 2.0 × 10−8 | 0.86 (0.82–0.91) | 0.56 | .951 | 1.01 (0.84–1.20) | |
| Seshadri et al,20 2010 | 0.39 | 1.62 × 10−16 | 0.85 (0.82–0.88) | |||||||
| Harold et al,21 2009 | 0.40 | 8.5 × 10−10 | 0.86 (0.82–0.90) | |||||||
| rs9331888 | G | 0.32 | Lambert et al,19 2009 | NR | 1.4 × 10−7 | 1.15 (1.09–1.22) | 0.20 | >.99 | 1.00 (0.76–1.32) | |
| rs9331926b | G | 0.07 | . . . | . . . | . . . | . . . | 0.04 | .020 | 1.96 (1.11–3.48) | |
| PICALM | ||||||||||
| rs3851179 | T | 0.41 | Seshadri et al,20 2010 | 0.37 | 3.16 × 10−12 | 0.87 (0.84–0.91) | 0.16 | .157 | 0.85 (0.68–1.07) | |
| Harold et al,21 2009 | 0.37 | 1.3 × 10−9 | 0.86 (0.82–0.90) | |||||||
| rs17148827c | C | 0.00 | . . . | . . . | . . . | . . . | 0.04 | .0089 | 2.01 (1.19–3.40) | |
| rs12795381d | C | 0.14 | . . . | . . . | . . . | . . . | 0.04 | .0086 | 0.49 (0.29–0.84) | |
| CR1 | ||||||||||
| rs6656401 | A | 0.24 | Lambert et al,19 2009 | NR | 7.9 × 10−9 | 1.20 (1.13–1.28) | 0.07 | .227 | 0.79 (0.54–1.16) | |
| rs3818361 | A | 0.26 | Lambert et al,19 2009 | NR | 1.4 × 10−7 | 1.18 (1.11–1.25) | 0.43 | .466 | 0.94 (0.79–1.11) | |
| BIN1 | ||||||||||
| rs744373 | G | 0.31 | Seshadri et al,20 2010 | 0.29 | 1.59 × 10−11 | 1.15 (1.11–1.20) | 0.48 | .999 | 1.00 (0.84–1.20) | |
| rs11685593e | T | 0.21 | . . . | . . . | . . . | . . . | 0.06 | .0098 | 1.66 (1.13–2.45) | |
| rs11691237f | T | 0.27 | . . . | . . . | . . . | . . . | 0.10 | .0098 | 1.52 (1.11–2.09) | |
| rs7585314g | T | 0.85 | . . . | . . . | . . . | . . . | 0.33 | .0030 | 0.75 (0.62–0.91) | |
| CD2AP | ||||||||||
| rs9349407 | C | 0.28 | Naj et al,22 2011 | 0.27 | 8.6 × 10−9 | 1.11 (1.07–1.15) | 0.22 | .854 | 0.98 (0.78–1.22) | |
| EPHA1 | ||||||||||
| rs11767557 | C | 0.20 | Naj et al,22 2011 | 0.19 | 6.0 × 10−10 | 0.90 (0.86–0.93) | 0.18 | .586 | 1.06 (0.86–1.31) | |
| rs11762262h | T | 0.20 | . . . | . . . | . . . | . . . | 0.23 | .034 | 1.27 (1.02–1.59) | |
| rs4595035i | T | 0.37 | . . . | . . . | . . . | . . . | 0.43 | .0094 | 1.25 (1.06–1.47) | |
| MS4A | ||||||||||
| rs4938933 | C | 0.50 | Naj et al,22 2011 | 0.39 | 1.7 × 10−9 | 0.88 (0.85–0.92) | 0.30 | .493 | 1.06 (0.88–1.29) | |
| rs610932 | T | 0.54 | Hollingworth et al,23 2011 | . . . | 1.8 × 10−14 | 0.90 (0.87–0.92) | 0.49 | .299 | 0.91 (0.77–1.08) | |
| rs10792258j | T | 0.20 | . . . | . . . | . . . | . . . | 0.37 | .010 | 0.79 (0.66–0.95) | |
| ABCA7 | ||||||||||
| rs3752246 | G | 0.19 | Naj et al,22 2011 | 0.19 | 5.8 × 10−7 | 1.15 (1.09–1.21) | 0.04 | .375 | 0.82 (0.53–1.27) | |
| rs3764650 | G | 0.11 | Hollingworth et al,23 2011 | . . . | 4.5 × 10−17 | 1.23 (1.18–1.30) | 0.28 | .019 | 1.27 (1.04–1.55) | |
| rs3764647k | G | 0.04 | . . . | . . . | . . . | . . . | 0.25 | .0087 | 1.32 (1.07–1.63) | |
| CD33 | ||||||||||
| rs3865444 | A | 0.32 | Naj et al,22 2011 | 0.30 | 1.6 × 10−9 | 0.91 (0.88–0.93) | 0.09 | .732 | 0.95 (0.70–1.29) | |
| rs10419982l | A | 0.45 | . . . | . . . | . . . | . . . | 0.40 | .00054 | 1.38 (1.15–1.65) | |
Abbreviations: ABCA7, ATP-binding cassette, subfamily A (ABC1), member 7; AF, effect allele frequency; BIN1, bridging integrator 1; CD2AP, CD2-associated protein; CD33, myeloid-associated antigen CD33; CEU, HapMap’s CEPH (Utah residents with ancestry from Northern and Western Europe) Population; CLU, clusterin; CR1, complement component (3b/4b) receptor 1; ellipses, missing information; EPHA1, ephrin type-A receptor 1; GWAS, genome-wide association study; kb, kilobase; MS4A, the membrane-spanning 4A gene cluster; NPR, not previously reported as associated with Alzheimer disease in whites; NR, not reported; OR, odds ratio; PICALM, phosphatidylinositol binding clathrin assembly protein; SNP, single-nucleotide polymorphism.
Effect estimate directions differ between whites and African Americans.
This SNP was not previously reported as associated with Alzheimer disease in whites (NPR). The lowest P value is in CLU.
This SNP was NPR and is located 15 kb from rs3851179.
This SNP was NPR. This represents the lowest P value observed in PICALM.
This SNP was NPR and flanks rs744375, 6.5 kb away from rs744373.
This SNP was NPR. This represents the most significant P value observed in BIN1.
This SNP was NPR. This represents the lowest P value observed in the BIN1 region.
This SNP was NPR and flanks rs11767557.
This SNP was NPR. This represents the smallest P value observed in the region of EPHA1.
This SNP was NPR. This represents the smallest P value observed in the MS4A region.
This SNP was NPR. This represents the smallest P value observed in ABCA7.
This SNP was NPR and is 63 kb from rs3865444. This represents the lowest P value observed in the CD33 region.
Harold et al21 found genome-wide significant evidence of association with rs3851179, located 88 kb upstream from PICALM. This SNP was not associated with AD in our African American sample (P=.16), although the estimated OR (0.85) is nearly identical to the OR reported in the white sample (0.87). However, we observed nominally significant association with 8 of 287 other SNPs tested in the region, including rs12795381 (P=.0086) and rs17148827 (P=.0089), which is monomorphic in whites. The rs12795381 finding is consistent with modest evidence of association with multiple SNPs in the PICALM coding region.20 We also evaluated the interaction of PICALM SNPs with APOE as reported in a large white sample.24 Stratified analysis revealed evidence of association with rs12795381 in subjects with the APOE ε4 allele (P=.04) but not in those without it (P=.61). However, we were unable to perform a formal test of interaction owing to the relatively small sample size and low minor allele frequency.
None of the 88 tested CR1 SNPs (including the 2 reported as significant by Lambert et al19) and none of the 112 tested CD2AP SNPs (including rs9349407, which was reported as significant by Naj et al22) were associated with AD in African Americans. The most promising result among these SNPs was obtained with rs12734030 in CR1 (P=.09).
Seshadri et al20 proposed BIN1 as a candidate gene for AD on the basis of a genome-wide significant P value observed for rs744373 located approximately 30 kb from the BIN1 coding region. Although this result was not replicated in our African American sample (P>.99), several adjacent SNPs were nominally significant, including rs11685593 (P=.0098). Association was also observed with multiple SNPs within the BIN1 coding region, the most significant of which was rs11691237 (P=.0098). The most significant association in the region was observed with rs7585314 (P=.0030), which is 68 kb from rs744373 in CYP27C1.
Located approximately 6 kb from EPHA1, rs11767557 is the only genome-wide significant result in this region reported by Naj et al22 and was not associated with AD in African Americans (P=.59). However, rs11762262, which is approximately 1260 bp closer to EPHA1 than rs11767557, was nominally significant (P=.034). We observed multiple nominally significant SNPs spread throughout the EPHA1 region in the African American sample. The strongest evidence for association in this region was obtained with rs4595035 (P=.0094), which is 32 kb upstream from rs11767557.
Naj et al22 also observed genome-wide significant association with many SNPs in the MS4A cluster. We evaluated association with all SNPs across this cluster, which spans about 500 kb. The most significant finding in the MS4A region was observed with rs10792258 (P=.010). This SNP is 394 bp distal from rs1582763 and 253 bp proximal to rs1562990, both of which were strongly associated with AD (P=5.92×10−11 and P=2.47×10−9, respectively) in the meta-analysis of large white data sets by Naj et al.22 A similar level of significance was observed with rs3802957 (P=.011) in the 3′ untranslated region of MS4A1, 203 kb from rs10792258.
We did not see association (P=.38) with an SNP in ABCA7 (rs3752246), which approached genome-wide significance in the study by Naj et al.22 We did, however, observe nominally significant association with rs3764650 (P=.019), which was reported as genome-wide significant in the study by Hollingworth et al.23 The effect of this SNP on risk of AD in our study (OR, 1.27) was very similar to that observed previously (OR, 1.23). Several other nominally significant SNPs were observed in the region, of which the most significant, synonymous coding SNP rs376647 (P=.0087), is located 11 kb from rs3752246.
We also did not observe an association with CD33 SNP rs3865444 (P=.73), which was found to have genome-wide significance in the GWAS by Naj et al.22 The most significant result in the CD33 region in the African American sample was obtained with rs10419982 (P=.0005), 69 kb away from rs3865444. This SNP almost survives correction for the 200 SNPs examined in the region. However, given the great distance of this SNP from CD33, there is not adequate evidence to consider this result a replication.
NOVEL GENE DISCOVERY
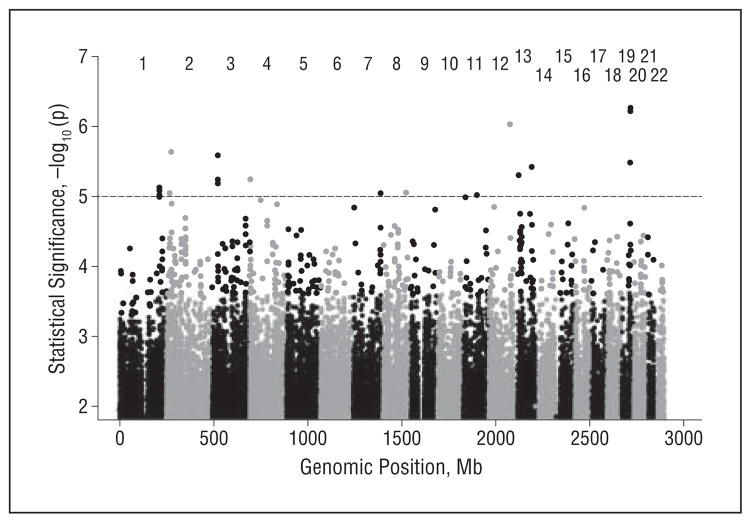
Genotypes were evaluated for 2 505 093 imputed SNPs that passed minor allele frequency criteria and imputation quality thresholds. A Manhattan plot of the results of the genome screen is presented in Figure 3. No SNP achieved a genome-wide level of significance. Eleven SNPs achieved suggestive levels of association (P<10−5) (Table 6). The direction of effect for these SNPs was consistent across the African American cohorts. Four of these SNPs (rs11889338, rs2221154, rs956225, and rs10850408) are more than 50 kb from the nearest characterized gene. Strong evidence of association was obtained with rs340849 (P=7.52×10−6), located 34 kb from PROX1, and with rs3888908 (P=9.52×10−6), located 19 kb upstream from P4HA3. The most significant finding was obtained for rs10850408 (P=9.25×10−7), a chromosome 12 SNP more than 250 kb from the nearest gene. The SNPs in ZC3H3, TMTC1, and ENOX1 showed suggestive evidence of an association in analyses adjusting for APOEε4 (see eTable 3 in the supplementary material for all SNPs with P<10−5). None of these findings were replicated in the white meta-analysis (details are provided in the eAppendix in the supplementary material).
Figure 3.
Manhattan plot of genome-wide association study results for the meta-analysis of the African American cohorts. The dotted line indicates suggestive evidence of association (P<10−5).
Table 6.
Top-Ranked Genetic Association Findings From Genome-wide Survey in African Americans
| CHR | Position, dbSNP Build 129, bp | Gene | SNP | P Value | Effect Allele | AF | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| 1 | 212 184 713a | – | rs340849a | 7.52 × 10−6 | A | 0.20 | 0.59 (0.47–0.75) |
| 2 | 17 291 066 | – | rs11889338 | 8.94 × 10−6 | A | 0.26 | 1.55 (1.28–1.88) |
| 2 | 27 760 977 | SLC4A1AP | rs17006206 | 2.30 × 10−6 | G | 0.10 | 2.05 (1.52–2.76) |
| 3 | 28 903 864 | – | rs2221154 | 2.58 × 10−6 | T | 0.19 | 0.57 (0.45–0.72) |
| 4 | 2 072 894 | POLN | rs1923775 | 5.61 × 10−6 | T | 0.25 | 1.60 (1.30–1.95) |
| 7 | 146 528 336 | CNTNAP2 | rs10273775 | 8.94 × 10−6 | G | 0.42 | 1.52 (1.27–1.84) |
| 8 | 122 978 868 | – | rs956225 | 8.71 × 10−6 | G | 0.03 | 0.30 (0.18–0.51) |
| 11 | 73 710 714 | – | rs3888908 | 9.52 × 10−6 | A | 0.15 | 1.72 (1.36–2.20) |
| 12 | 113 864 776 | – | rs10850408 | 9.25 × 10−7 | T | 0.34 | 0.63 (0.52–0.76) |
| 13 | 25 622 328 | – | rs17511627 | 5.01 × 10−6 | C | 0.17 | 1.75 (1.37–2.22) |
| 13 | 97 929 295 | STK24 | rs912330 | 3.79 × 10−6 | T | 0.14 | 0.54 (0.41–0.70) |
| Other SNPs of Interest From APOE ε4 Adjusted Analysis | |||||||
| 8 | 144 692 178 | ZC3H3 | rs3750208 | 7.28 × 10−6 | A | 0.04 | 0.37 (0.24–.057) |
| 12 | 29 812 934 | TMTC1 | rs302318 | 1.97 × 10−6 | C | 0.26 | 0.59 (0.48–0.74) |
| 13 | 43 064 019 | ENOX1 | rs17460623 | 9.37 × 10−6 | C | 0.10 | 0.49 (0.36–0.67) |
Abbreviations: AF, effect allele frequency; bp, base pairs; CHR, chromosome; CNTNAP2, contactin-associated protein-like 2; dbSNP, database single-nucleotide polymorphism; ENOX1, ecto-NOX disulfidethiol exchanger 1; OR, odds ratio; POLN, polymerase (DNA directed) nu; SLC4A1AP, solute carrier family 4 (anion exchanger), member 1, adaptor protein; SNP, single-nucleotide polymorphism; STK24, serine/threonine kinase 24; TMTC1, transmembrane and tetratricopeptide repeat containing 1; ZC3H3, zinc finger CCCH-type containing 3; –, indicates that the SNP is more than 50 kilobases from the nearest characterized gene.
Does not include multiple SNPs on CHR 1 that were redundant owing to strong linkage disequilibrium (pairwise R2 > 0.8) and CHR 3 (R2 > 0.7) and 1 SNP that did not meet minor allele frequency criteria in subjects from the Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans Study.
COMMENT
This is, to our knowledge, the first comprehensive genetic association study of AD in African Americans. This study is timely and important for several reasons. African Americans are about twice as likely as non-Hispanic whites to have AD.39 Although differences in AD etiology across populations have been widely studied, they are still poorly understood. The occurrence of multiple demented individuals in African American families is significantly higher than in white families, although the genetic risk of AD is similar in these 2 populations.27 The increased familial risk in African Americans is likely a result of higher rates of risk factors, such as poor education, diabetes mellitus, and smoking.39 However, comparisons of risk in African American and white cohorts are complicated by differences in assessment of cognitive decline across studies and by population differences in willingness to participate in medical research.40–42
We obtained incontrovertible evidence of an association with the APOE ε4 allele, thus confirming findings from several smaller genetic studies of African Americans.4,26 In non-Hispanic whites, homozygosity for ε4 is associated with a 13- to 15-fold increased odds of developing AD compared with those with the most common genotype, ε3/ε3.4 We showed previously in a set of 308 African American AD cases and 409 ethnically matched controls that persons with the ε3/ε4 and ε4/ε4 genotypes had 2.6- and 10.5-fold increased odds of AD, respectively, compared with persons with the ε3/ε3 genotype.26 These risks decreased substantially after 68 years of age. The risk of AD was lower among individuals with the ε2/ε3 genotype. We observed similar risks in the present study. Approximately one-third of the African American sample in this study overlaps with the sample in our earlier report.
Our present study and previous studies in white populations identified highly significant evidence of an association with genes adjacent to APOE, most notably TOMM40 and APOC1 (reviewed by Ertekin-Taner5). Arguably, the distinction of such findings from confounding with APOE is intractable because of the tight LD spanning the genes in this region.9,10 However, we identified highly significant evidence of an association with several SNPs in the APOE region in African Americans and whites after adjustment for APOE genotype. This finding is consistent with an AD risk locus distinct from APOE. The observation that different SNPs in this region are significant in African Americans and whites after adjustment for APOE may reflect differences in LD structure in this region (Figure 2). The residual association in these other genes may also represent unmeasured effects of variants in regulatory regions of APOE.43–45 Additional studies in larger African American samples are needed to determine which of these explanations is more likely.
Among the African Americans in the present study, a subset of 180 cases and 200 controls from the MIRAGE Study and 221 cases and 186 controls from the GenerAAtions Study was included in another recent study24 that evaluated the association of AD with PICALM, CLU, and CR1 SNPs highlighted in the original studies reports.19,21 The authors did not find evidence of an association with any of the SNPs examined in the African American data sets. We did not replicate the genome-wide significant associations with these loci in a larger set of African American cases and controls, even at a nominal significance level. However, we observed an association in African Americans with other previously unreported variants in each of these regions and in the most recently reported regions of genome-wide significant association,22,23 except CR1 and CD2AP. Only one previously reported genome-wide significant association (rs3764650 in ABCA7) was confirmed in our African American sample.23 Discordance in the association patterns between whites and African Americans could be related to population differences in allele frequencies or LD patterns. This explanation is consistent with our findings of association in the African Americans between SNPs, which have very low frequency in whites (eg, rs17148827 in PICALM), and one of the previously reported AD-associated CLU SNPs (rs2279590), but with an opposite pattern of effect. Alternatively, the AD risk variants in these genes may differ across populations (ie, allelic heterogeneity), as we observed previously in SORL1.46 However, lack of replication might also be a result of small sample size when compared with recent consortium-based GWASs.19–23 In most instances, the confidence intervals for the effect estimates in African Americans included the point estimates in whites.
Analysis of the entire autosomal genome revealed evidence suggestive of an association with several novel candidate genes that may play a role in AD pathogenesis. PROX1 (OMIM *601546) is a prospero-related transcription factor that plays a critical role in the development of various organs, including the mammalian lymphatic and central nervous systems.47,48 This transcription factor has recently been shown to play a key role in adult neurogenesis, suggesting it may be involved in memory development.49 The contactin-associated protein-like 2 gene (CNTNAP2 [OMIM *604569]) is involved in brain development and has been implicated in susceptibility to autism and language disorders.50–53 In 2009, Harold et al21 reported a SNP in the contactin gene, CNTN5 (OMIM *607219), to have a GWAS P value of 2×10−5. Subsequently, the same SNP was shown to be associated with a variety of magnetic resonance imaging measures in the Alzheimer Disease Neuroimaging Initiative cohort.54 Serine/threonine kinase 24 (STK24 [OMIM *604984]) is expressed in the brain.55,56 An isoform of STK24 has been shown to be a regulator of axon growth and axon regeneration after injury.57,58 However, we did not observe association in these regions in a meta-analysis of a replication sample of 5 white AD data sets containing 3568 cases and 6205 controls. A study of a larger independent sample of African American and possibly white samples will be needed to determine whether these associations are spurious or reflect population-specific variants or variable LD patterns among populations.
This study represents an important step in elucidating the genetic basis of AD in African Americans. Our results suggest that African Americans share some but not all AD genetic risk factors with whites. Further research would not only lead to a more accurate understanding of the genetic risk factors that could be incorporated in diagnostic and predictive testing protocols specific for African Americans but may also yield new gene discovery and clues for subsequent interventions useful to all populations at risk for AD.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by grants R01-AG09029, R01-AG025259, R01-HG02213, R01-HG005092, 5R01AG020688, K24-AG027841, P30-AG13846, P30-AG10129, and K01 MH076100 from the National Institutes of Health.
Group Members: Members of the MIRAGE Study Group include Drs Farrer, Green, Baldwin, Cupples, Lunetta, and Logue (Boston University); Dr Griffith, Abimbola Akomolafe, MD, MPH, Angela Ashley, MD, Lorin Freedman, MD, and Elizabeth Ofili, MD (Morehouse School of Medicine); Helena Chui, MD (University of Southern California, Los Angeles); Ranjan Duara, MD (Mt Sinai Medical Center, Miami, Florida); Tatiana Foroud, PhD, and Martin Farlow, MD (Indiana University School of Medicine, Indianapolis); Robert Friedland, MD (University of Louisville, Louisville, Kentucky); Dr Go and Lindy Harrell, MD, PhD (University of Alabama–Birmingham); Alexander Kurz, MD (Technical University, Munich, Germany); Dr Obisesan (Howard University); Helen Petrovitch, MD, and Lon White, MD (Pacific Health Research Institute, Honolulu, Hawaii); Marwan Sabbagh, MD (Sun Health Research Institute, Sun City, Arizona); Dessa Sadovnick, PhD (University of British Columbia, Vancouver); and Magda Tsolaki, MD (University of Aristotle, Thessaloniki, Greece).
Footnotes
Author Contributions: Study concept and design: Logue, Griffith, Obisesan, Shatz, Borenstein, Cupples, Lunetta, Fallin, Baldwin, and Farrer. Acquisition of data: Green, Go, Obisesan, Shatz, Borenstein, Fallin, Baldwin, and Farrer. Analysis and interpretation of data: Logue, Schu, Vardarajan, Buros, Green, Go, Cupples, Lunetta, and Farrer. Drafting of the manuscript: Logue, Schu, Buros, Green, Baldwin, and Farrer. Critical revision of the manuscript for important intellectual content: Logue, Vardarajan, Go, Griffith, Obisesan, Shatz, Borenstein, Cupples, Lunetta, Fallin, Baldwin, and Farrer. Statistical analysis: Logue, Schu, Vardarajan, Buros, Green, Cupples, Lunetta, Fallin, and Farrer. Obtained funding: Go, Shatz, Borenstein, and Farrer. Administrative, technical, and material support: Schu, Buros, Green, Obisesan, Borenstein, Baldwin, and Farrer. Study supervision: Logue, Obisesan, Borenstein, Lunetta, Fallin, Baldwin, and Farrer.
Financial Disclosure: None reported.
Online-Only Material: The eTables, eMethods, and eAppendix are available at http://www.bumc.bu.edu/genetics/results/aa_alzheimer.
Additional Contributions: The GenerAAtions Study Group provided input in design, implementation, and consensus diagnosis efforts, including Hugh Hendrie, MD, and Fred Unverzagt, PhD (Indiana University), and Susan Bassett, PhD (Johns Hopkins). Michael Wake, MPH, MSW, provided project coordination.
References
- 1.Bienvenu OJ, Davydow DS, Kendler KS. Psychiatric “diseases” versus behavioral disorders and degree of genetic influence. Psychol Med. 2011;41(1):33–40. doi: 10.1017/S003329171000084X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrer LA, Myers RH, Connor L, Cupples LA, Growdon JH. Segregation analysis reveals evidence of a major gene for Alzheimer disease. Am J Hum Genet. 1991;48(6):1026–1033. [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43(8):1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 4.Farrer LA, Cupples LA, Haines JL, et al. APOE Alzheimer Disease Meta Analysis Consortium. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- 5.Ertekin-Taner N. Genetics of Alzheimer disease in the pre- and post-GWAS era. Alzheimers Res Ther. 2010;2(1):3. doi: 10.1186/alzrt26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swerdlow RH, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis: an update. Exp Neurol. 2009;218(2):308–315. doi: 10.1016/j.expneurol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz MW, Crenshaw DG, Saunders AM, Roses AD. Genetic variation at a single locus and age of onset for Alzheimer’s disease. Alzheimers Dement. 2010;6(2):125–131. doi: 10.1016/j.jalz.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bekris LM, Galloway NM, Montine TJ, Schellenberg GD, Yu CE. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):409–417. doi: 10.1002/ajmg.b.30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu CE, Seltman H, Peskind ER, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer’s disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89(6):655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takei N, Miyashita A, Tsukie T, et al. Japanese Genetic Study Consortium for Alzheimer Disease. Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93(5):441–448. doi: 10.1016/j.ygeno.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Poduslo SE, Neal M, Schwankhaus J. A closely linked gene to apolipoprotein E may serve as an additional risk factor for Alzheimer’s disease. Neurosci Lett. 1995;201(1):81–83. doi: 10.1016/0304-3940(95)12158-z. [DOI] [PubMed] [Google Scholar]
- 12.Poduslo SE, Neal M, Herring K, Shelly J. The apolipoprotein CI A allele as a risk factor for Alzheimer’s disease. Neurochem Res. 1998;23(3):361–367. doi: 10.1023/a:1022409617539. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Berglund L, Ramakrishnan R, et al. A common Hpa I RFLP of apolipoprotein C-I increases gene transcription and exhibits an ethnically distinct pattern of linkage disequilibrium with the alleles of apolipoprotein E. J Lipid Res. 1999;40(1):50–58. [PubMed] [Google Scholar]
- 14.Bartrés-Faz D, Junqué C, Clemente IC, et al. MRI and genetic correlates of cognitive function in elders with memory impairment. Neurobiol Aging. 2001;22 (3):449–459. doi: 10.1016/s0197-4580(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 15.Abildayeva K, Berbée JF, Blokland A, et al. Human apolipoprotein C-I expression in mice impairs learning and memory functions. J Lipid Res. 2008;49(4):856–869. doi: 10.1194/jlr.M700518-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Ki CS, Na DL, Kim DK, Kim HJ, Kim JW. Genetic association of an apolipoprotein C-I (APOC1) gene polymorphism with late-onset Alzheimer’s disease. Neurosci Lett. 2002;319(2):75–78. doi: 10.1016/s0304-3940(01)02559-9. [DOI] [PubMed] [Google Scholar]
- 17.Scacchi R, Gambina G, Ruggeri M, et al. Plasma levels of apolipoprotein E and genetic markers in elderly patients with Alzheimer’s disease. Neurosci Lett. 1999;259(1):33–36. doi: 10.1016/s0304-3940(98)00889-1. [DOI] [PubMed] [Google Scholar]
- 18.Drigalenko E, Poduslo S, Elston R. Interaction of the apolipoprotein E and CI loci in predisposing to late-onset Alzheimer’s disease. Neurology. 1998;51(1):131–135. doi: 10.1212/wnl.51.1.131. [DOI] [PubMed] [Google Scholar]
- 19.Lambert JC, Heath S, Even G, et al. European Alzheimer’s Disease Initiative Investigators. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 20.Seshadri S, Fitzpatrick AL, Ikram MA, et al. CHARGE Consortium; GERAD1 Consortium; EADI1 Consortium. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun G, Naj AC, Beecham GW, et al. Alzheimer’s Disease Genetics Consortium. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67(12):1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang MX, Stern Y, Marder K, et al. The APOE-ε4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279 (10):751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 26.Graff-Radford NR, Green RC, Go RC, et al. Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch Neurol. 2002;59(4):594–600. doi: 10.1001/archneur.59.4.594. [DOI] [PubMed] [Google Scholar]
- 27.Green RC, Cupples LA, Go R, et al. MIRAGE Study Group. Risk of dementia among white and African American relatives of patients with Alzheimer disease. JAMA. 2002;287(3):329–336. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- 28.Saykin AJ, Shen L, Foroud TM, et al. Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Wetten S, Li L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65 (1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Cheng R, Graff-Radford N, Foroud T, Mayeux R National Institute on Aging Late-Onset Alzheimer’s Disease Family Study Group. Analyses of the National Institute on Aging Late-Onset Alzheimer’s Disease Family Study: implication of additional loci. Arch Neurol. 2008;65(11):1518–1526. doi: 10.1001/archneur.65.11.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawber TR, Kannel WB. The Framingham Study: an epidemiological approach to coronary heart disease. Circulation. 1966;34(4):553–555. doi: 10.1161/01.cir.34.4.553. [DOI] [PubMed] [Google Scholar]
- 33.Splansky GL, Corey D, Yang Q, et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165(11):1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 34.Cobb JL, Wolf PA, Au R, White R, D’Agostino RB. The effect of education on the incidence of dementia and Alzheimer’s disease in the Framingham Study. Neurology. 1995;45(9):1707–1712. doi: 10.1212/wnl.45.9.1707. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Ding J, Abecasis GR. MaCH 1.0: rapid haplotype reconstruction and missing genotype inference [abstract 2290] Am J Hum Genet. 2006;79(suppl):416. [Google Scholar]
- 36.Liang KY, Zeger SL. Longitudinal data analysis using linear models. Biometrika. 1986;73(1):13–22. doi: 10.1093/biomet/73.1.13. [DOI] [Google Scholar]
- 37.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42(1):121–130. [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alzheimer’s Association. 2010 Alzheimer’s disease facts and figures. Alzheimers Dement. 2010;6(2):158–194. doi: 10.1016/j.jalz.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Dilworth-Anderson P, Hendrie HC, Manly JJ, Khachaturian AS, Fazio S Social, Behavioral and Diversity Research Workgroup of the Alzheimer’s Association. Diagnosis and assessment of Alzheimer’s disease in diverse populations. Alzheimers Dement. 2008;4(4):305–309. doi: 10.1016/j.jalz.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Connell CM, Scott Roberts J, McLaughlin SJ, Akinleye D. Racial differences in knowledge and beliefs about Alzheimer disease. Alzheimer Dis Assoc Disord. 2009;23(2):110–116. doi: 10.1097/WAD.0b013e318192e94d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danner DD, Smith CD, Jessa P, Hudson J. African Americans with memory loss: findings from a community clinic in Lexington, Kentucky. Nurs Clin North Am. 2008;43(3):437–447. ix–x. doi: 10.1016/j.cnur.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roks G, Cruts M, Bullido MJ, et al. The –491 A/T polymorphism in the regulatory region of the apolipoprotein E gene and early-onset Alzheimer’s disease. Neurosci Lett. 1998;258(2):65–68. doi: 10.1016/s0304-3940(98)00857-x. [DOI] [PubMed] [Google Scholar]
- 44.Town T, Paris D, Fallin D, et al. The –491A/T apolipoprotein E promoter polymorphism association with Alzheimer’s disease: independent risk and linkage disequilibrium with the known APOE polymorphism. Neurosci Lett. 1998;252 (2):95–98. doi: 10.1016/s0304-3940(98)00567-9. [DOI] [PubMed] [Google Scholar]
- 45.Artiga MJ, Bullido MJ, Sastre I, et al. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998;421(2):105–108. doi: 10.1016/s0014-5793(97)01543-3. [DOI] [PubMed] [Google Scholar]
- 46.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galeeva A, Treuter E, Tomarev S, Pelto-Huikko M. A prospero-related homeo-box gene Prox-1 is expressed during postnatal brain development as well as in the adult rodent brain. Neuroscience. 2007;146(2):604–616. doi: 10.1016/j.neuroscience.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Lavado A, Oliver G. Prox1 expression patterns in the developing and adult murine brain. Dev Dyn. 2007;236(2):518–524. doi: 10.1002/dvdy.21024. [DOI] [PubMed] [Google Scholar]
- 49.Lavado A, Lagutin OV, Chow LM, Baker SJ, Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010;8(8):e1000460. doi: 10.1371/journal.pbio.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alarcón M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arking DE, Cutler DJ, Brune CW, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakkaloglu B, O’Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and re-sequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vernes SC, Newbury DF, Abrahams BS, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biffi A, Anderson CD, Desikan RS, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI) Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67(6):677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou TH, Ling K, Guo J, et al. Identification of a human brain-specific isoform of mammalian STE20-like kinase 3 that is regulated by cAMP-dependent protein kinase. J Biol Chem. 2000;275(4):2513–2519. doi: 10.1074/jbc.275.4.2513. [DOI] [PubMed] [Google Scholar]
- 56.Schinkmann K, Blenis J. Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J Biol Chem. 1997;272(45):28695–28703. doi: 10.1074/jbc.272.45.28695. [DOI] [PubMed] [Google Scholar]
- 57.Irwin N, Li YM, O’Toole JE, Benowitz LI. Mst3b, a purine-sensitive Ste20-like protein kinase, regulates axon outgrowth. Proc Natl Acad Sci U S A. 2006;103 (48):18320–18325. doi: 10.1073/pnas.0605135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorber B, Howe ML, Benowitz LI, Irwin N. Mst3b, an Ste20-like kinase, regulates axon regeneration in mature CNS and PNS pathways. Nat Neurosci. 2009;12(11):1407–1414. doi: 10.1038/nn.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.