Abstract
Sulfatides are one of the major sphingoglycolipids in mammalian serum and are synthesized and secreted mainly from the liver as a component of lipoproteins. Recent studies revealed a protective role for serum sulfatides against arteriosclerosis and hypercoagulation. Although peroxisome proliferator-activated receptor (PPAR) α has important functions in hepatic lipoprotein metabolism, its association with sulfatides has not been investigated. In this study, sulfatide levels and the expression of enzymes related to sulfatide metabolism were examined using wild-type (+/+), Ppara-heterozygous (+/−), and Ppara-null (−/−) mice given a control diet or one containing 0.1% fenofibrate, a clinically used hypolipidemic drug and PPARα activator. Fenofibrate treatment increased serum and hepatic sulfatides in Ppara (+/+) and (+/−) mice through a marked induction of hepatic cerebroside sulfotransferase (CST), a key enzyme in sulfatide synthesis, in a PPARα-dependent manner. Furthermore, increases in CST mRNA levels were correlated with mRNA elevations of several known PPARα target genes, and such changes were not observed for other sulfatide-metabolism enzymes in the liver. These results suggest that PPARα activation enhances hepatic sulfatide synthesis via CST induction and implicate CST as a novel PPARα target gene.
1. Introduction
Sulfatides are sphingoglycolipids composed of sphingoid, fatty acid, galactose, and sulfate [1] that are distributed in various tissues such as the central nervous system, kidney, liver, and gastrointestinal tract [1–4]. Glycolipids are also present in the serum as one of the major components of lipoproteins [1]. Several studies have revealed a protective role for serum sulfatides against arteriosclerosis and hypercoagulation [5]. Serum levels of sulfatides are markedly decreased in humans with end-stage renal failure [6] but normalize after renal transplantation [7]. However, the precise mechanism regulating serum sulfatide concentrations in humans remains unclear. Previously studies demonstrated that serum sulfatide levels were strongly correlated with hepatic, but not renal, sulfatide levels in mice with protein overload nephropathy, and that decreased serum sulfatide levels were also associated with the downregulation of hepatic expression of cerebroside sulfotransferase (CST), a key enzyme in sulfatide synthesis [8]. These and previous findings suggest the possible participation of hepatic peroxisome proliferator-activated receptor (PPAR) in the regulation of serum and liver sulfatide metabolisms. To examine this possibility, serum and liver sulfatide concentrations and hepatic expression of a series of sulfatide-metabolizing enzymes were analyzed using Ppara-homozygous (+/+), Ppara-heterozygous (+/−), and Ppara-null (−/−) mice fed a control diet or one containing fenofibrate, a typical PPARα activator.
2. Materials and Methods
2.1. Mice and Treatment
All animal experiments were conducted in accordance with animal study protocols approved by the Shinshu University School of Medicine. Wild-type (+/+), Ppara (+/−), and Ppara (−/−) mice on a 129/Sv genetic background were generated as described previously [9–11]. These mice were maintained in a pathogen-free environment under controlled conditions (25°C; 12 h light/dark cycle) with tap water ad libitum and a standard rodent diet. Twelve-week-old male wild-type (+/+), Ppara (+/−), and Ppara (−/−) mice weighing 25–30 g were used for the following experiments. Mice of each genotype were randomly divided into two groups (n = 6 in each group of the same genotype). One mouse group was treated with a regular diet containing 0.1% fenofibrate (Wako Pure Chemical Industries, Osaka, Japan), and the other group was continued on a regular diet as a control. In an additional experiment, Ppara (+/+), Ppara (+/−), and Ppara (−/−) mice were randomly divided into two groups (n = 6 in each group of the same genotype) and were treated with a regular diet with or without 0.5% clofibrate (Wako Pure Chemical Industries). Seven days after commencing treatment, the mice were sacrificed under anesthesia for collection of blood and tissues.
2.2. Extraction and Measurement of Lipids
Total lipids in the serum and liver were extracted using the hexane/isopropanol method [12], and serum/liver sulfatides were determined as forms of lysosulfatides (LS; sulfatides without fatty acids) by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) as previously described [13]. Sulfatides levels were calculated as the sum of the levels of seven LS molecular species: LS-sphingadienine (LS-d18 : 2), LS-(4E)-sphingenine (LS-d18 : 1), LS-sphinganine (LS-d18 : 0), LS-4D-hydroxysphinganine (LS-t18 : 0), LS-(4E)-icosasphingenine (LS-d20 : 1), LS-icosasphinganine (LS-d20 : 0), and LS-4D-hydroxyicosasphinganine (LS-t20 : 0). Triglyceride (TG) levels in the serum and liver were measured using a Triglyceride E-test kit (Wako Pure Chemical Industries).
2.3. Immunoblot Analysis
Liver nuclear and cytosolic fractions were prepared from each mouse using NE-PER Nuclear and Cytoplasmic Extraction Regents (Thermo Fisher Scientific, Rockford, IL, USA) [14], and their protein concentrations were determined with a BCA protein assay kit (Thermo Fisher Scientific) [15]. Nuclear fractions (10 μg protein) were used for immunoblot analysis of PPARs and TATA box-binding protein (TBP). For detection of other proteins, cytosolic fractions (5 μg protein) were employed. Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After blocking, the membranes were incubated with primary antibodies followed by alkaline phosphatase-conjugated secondary antibodies [16–18]. Primary antibodies against long-chain acyl-CoA synthase (LACS), liver fatty acid-binding protein (L-FABP), and medium-chain acyl-CoA dehydrogenase (MCAD) were prepared as described previously [19–21]. Antibodies against other proteins were purchased commercially: cerebroside sulfotransferase (CST) from Abnova (Taipei, Taiwan), arylsulfatase A (ARSA) from Everest Biotech (Oxfordshire, UK), TBP from Abcam (Cambridge, UK), and ceramide galactosyltransferase (CGT), galactosylceramidase (GALC), microsomal transfer protein (MTP), PPARα, PPARβ/δ, PPARγ, and actin from Santa Cruz Biotechnology (Santa Cruz, CA, USA). TBP and actin were used as loading controls for nuclear and cytosolic protein extracts, respectively. Band intensities were measured densitometrically, normalized to those of TBP or actin, and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet.
2.4. Analysis of mRNA
Total liver RNA was extracted using an RNeasy Mini Kit (QIAGEN, Hilden, Germany), and samples of 2 μg of RNA were reverse-transcribed using oligo-dT primers and SuperScript II reverse transcriptase (Invitrogen Corporation, Carlsbad, CA, USA). Levels of mRNA were quantified by real-time polymerase chain reaction (PCR) using an SYBR Premix Ex Taq II (Takara Bio, Otsu, Japan) on a Thermal Cycler Dice TP800 system (Takara Bio) [10, 16]. Specific primers were designed by Primer Express software (Applied Biosystems, Foster City, CA, USA) as shown in Table 1. The mRNA levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as an internal control. Measurements of mRNA levels were normalized to those of GAPDH and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet.
Table 1.
Primer pairs used for the RT-PCR.
| Gene | GeneBank accession number | Primer sequence | |
|---|---|---|---|
| ARSA | NM_009713 | F | 5′-ACCACCCCTAACCTGGATCAGT-3′ |
| R | 5′-ATGGCGTGCACAGAGACACA-3′ | ||
| CGT | NM_011674 | F | 5′-TGGGTCCAGCCTATGGATGT-3′ |
| R | 5′-GCAGCGTTGGTCTTGGAAAC-3′ | ||
| CST | NM_016922 | F | 5′-ATGGCCTTCACGACCTCAGA-3′ |
| R | 5′-CGGTCTTGTGCGTCTTCATG-3′ | ||
| GALC | NM_008079 | F | 5′-GAGTGAGAATCATAGCGAGCGATA-3′ |
| R | 5′-AGTTCCTGGTCCAGCAGCAA-3′ | ||
| GAPDH | M32599 | F | 5′-TGCACCACCAACTGCTTAG-3′ |
| R | 5′-GGATGCAGGGATGATGTTCTG-3′ | ||
| LACS | NM_007981 | F | 5′-TCCTACGGCAGTGATCTGGTG-3′ |
| R | 5′-GGTTGCCTGTAGTTCCACTTGTG-3′ | ||
| L-FABP | NM_017399 | F | 5′-GCAGAGCCAGGAGAACTTTGAG-3′ |
| R | 5′-TTTGATTTTCTTCCCTTCATGCA-3′ | ||
| MCAD | NM_007382 | F | 5′-TGCTTTTGATAGAACCAGACCTACAGT-3′ |
| R | 5′-CTTGGTGCTCCACTAGCAGCTT-3′ | ||
| MTP | NM_008642 | F | 5′-GAGCGGTCTGGATTTACAACG-3′ |
| R | 5′-GTAGGTAGTGACAGATGTGGCTTTTG-3′ | ||
| PPARα | NM_011144 | F | 5′-CCTCAGGGTACCACTACGGAGT-3′ |
| R | 5′-GCCGAATAGTTCGCCGAA-3′ | ||
| PPARβ/δ | XM_128500 | F | 5′-TCAACATGGAATGTCGGGTT-3′ |
| R | 5′-ATACTCGAGCTTCATGCGGATT-3′ | ||
| PPARγ | NM_011146 | F | 5-TTCCACTATGGAGTTCATGCTTGT-3′ |
| R | 5′-TCCGGCAGTTAAGATCACACCTA-3′ |
F: forward sequence; R: reverse sequence.
2.5. Assays for DNA-Binding Activity of PPARs
The DNA-binding activity of nuclear PPARαPPARβ/δ, and PPARγ was determined using PPARα, PPARβ/δ, and PPARγ Transcription Factor Assay kits (Cayman Chemical, Ann Arbor, MI, USA) [22–24], respectively. These assays are based on an enzyme-linked immunosorbent assay using PPAR response element (PPRE) immobilized microplates and specific PPAR antibodies, thus offering similar results to those from the conventional radioactive electrophoretic mobility shift assay. DNA-binding assays were carried out according to the manufacturer's instructions using nuclear extracts (50 μg protein) prepared as described previously. Results are expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet.
2.6. Statistical Analysis
All data are expressed as mean ± standard deviation (SD). Statistical analysis was performed using one-way ANOVA with Bonferroni correction (SPSS Statistics 17.0; SPSS Inc, Chicago, IL, USA). Correlation coefficients were calculated using Spearman's rank correlation analysis. A probability value of less than 0.05 was considered to be statistically significant.
3. Results
3.1. Fenofibrate Increased Serum/Liver Sulfatides in a PPARα-Dependent Manner
Fenofibrate treatment increased serum, and more notably liver, sulfatide concentrations in Ppara (+/+) and (+/−) mice only (Figure 1(a)). However, the increases in serum/liver sulfatides were not detected in Ppara (−/−) mice with fenofibrate treatment. These results demonstrate that fenofibrate increases serum/liver sulfatide levels in a PPARα-dependent manner. The treatment did not affect the composition of sulfatides (Table 2). Fenofibrate also decreased serum/liver TG levels in a PPARα-dependent manner (Figure 1(b)), which was in agreement with previous reports [25, 26].
Figure 1.
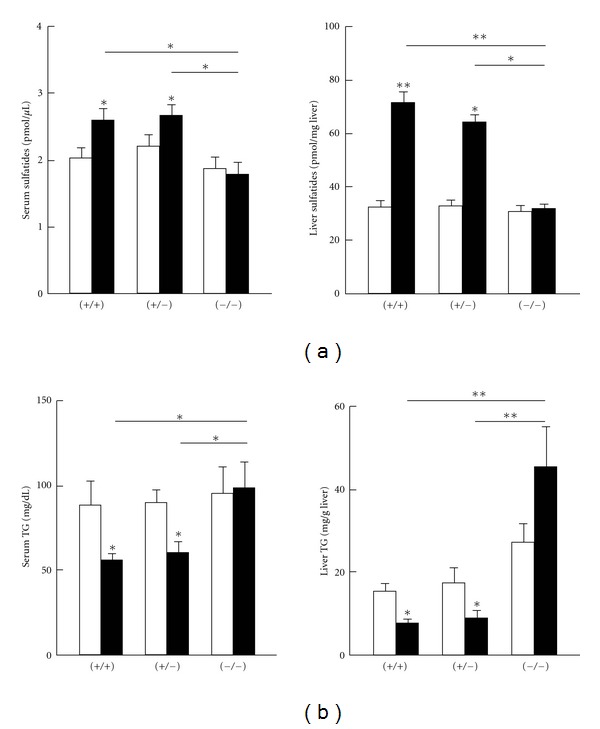
Changes in serum and hepatic levels of sulfatides (a) and TG (b). Ppara (+/+), (+/−), and (−/−) mice were treated without (open bars) or with (closed bars) 0.1% fenofibrate for 7 days. Results are expressed as mean ± SD (n = 6/group). *P < 0.05; **P < 0.01.
Table 2.
Composition of serum and liver sulfatides.
| Serum | Liver | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (+/+) | (+/−) | (−/−) | (+/+) | (+/−) | (−/−) | |||||||
| (−) | (+) | (−) | (+) | (−) | (+) | (−) | (+) | (−) | (+) | (−) | (+) | |
| d18 : 2 | 7 | 9 | 8 | 7 | 8 | 7 | 12 | 11 | 12 | 13 | 11 | 12 |
| d18 : 1 | 34 | 31 | 33 | 36 | 33 | 35 | 29 | 30 | 30 | 28 | 30 | 31 |
| d18 : 0 | 11 | 11 | 12 | 10 | 11 | 10 | 11 | 10 | 12 | 11 | 10 | 12 |
| t18 : 0 | 7 | 9 | 8 | 7 | 8 | 7 | 6 | 6 | 6 | 5 | 7 | 6 |
| d20 : 1 | 8 | 11 | 9 | 8 | 9 | 8 | 12 | 11 | 10 | 12 | 10 | 10 |
| d20 : 0 | 5 | 7 | 6 | 6 | 6 | 6 | 10 | 9 | 9 | 10 | 9 | 8 |
| t20 : 0 | 28 | 22 | 24 | 26 | 25 | 27 | 20 | 23 | 21 | 21 | 23 | 21 |
(−): mice treated with a control diet; (+): mice treated with fenofibrate; d18 : 2: sphingadienine; d18 : 1: (4E)-sphingenine; d18 : 0: sphinganine; t18 : 0: 4D-hydroxysphinganine; d20 : 1: (4E)-icosasphingenine; d20 : 0: icosasphinganine; t20 : 0: 4D-hydroxyicosasphinganine.
Data are expressed as percentages.
3.2. Fenofibrate Upregulated Hepatic CST in a PPARα-Dependent Manner
We assessed several major hepatic sulfatide-metabolizing enzymes to determine the mechanistic basis of the changes observed in sulfatide concentrations. CST and ARSA, respectively, catalyze the forward and reverse reactions from galactosylceramides to sulfatides, and a similar relationship exists for CGT and GALC in the synthesis of galactosylceramides from ceramides [8]. Fenofibrate treatment significantly increased levels of mRNA encoding CST in Ppara (+/+) and (+/−) mice (Figure 2(a)), with the extent of induction higher in the Ppara (+/+) group. Upregulation of CST expression by fenofibrate was not observed in Ppara (−/−) mice. PPARα-dependent increases in CST mRNA corresponded to increases in CST protein levels (Figure 2(b)). Fenofibrate treatment did not affect expression of the other sulfatide-metabolizing enzymes, ARSA, CGT, and GALC, at either the mRNA or the protein level (Figure 2). Since hepatic CST mRNA levels were strongly correlated with sulfatide levels in the serum (r = 0.886, P = 0.019) and liver (r = 0.943, P = 0.005), the increased serum/liver sulfatide levels found after treatment were viewed as mainly due to the significant induction of hepatic CST.
Figure 2.
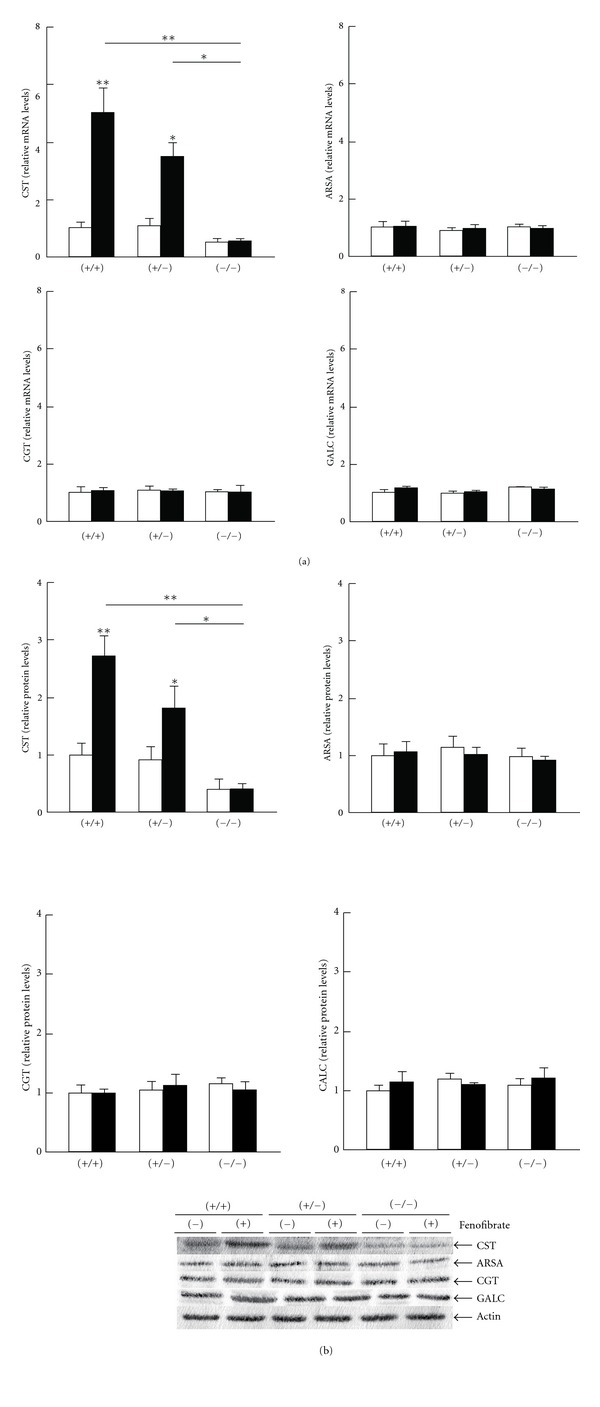
Changes in hepatic expression of sulfatide-metabolizing enzymes by fenofibrate treatment. Open and closed bars indicate mice treated without or with 0.1% fenofibrate, respectively. Data are expressed as mean ± SD (n = 6/group). *P < 0.05; **P < 0.01. (a) The mRNA levels of CST, ARSA, CGT, and GALC. Hepatic mRNA levels were normalized to those of GAPDH and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet. (b) Immunoblot analysis. Actin was used as the loading control. Band intensities were measured densitometrically, normalized to those of actin, and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet.
3.3. Hepatic CST Was Induced by PPARα Activation
As expected, fenofibrate treatment significantly enhanced hepatic expression of PPARα and several representative PPARα target genes, including LACS, MCAD, L-FABP, and MTP (Figures 3 and 4) [27–29]. The DNA binding activity levels of PPARα were also elevated by fenofibrate (Figure 3(b)). The treatment did not influence the expression and activity of PPARβ/δ or PPARγ (Figure 3), nor did it affect levels of CST mRNA or protein in the livers of Ppara (−/−) mice (Figure 2). The mRNA levels of CST were strongly correlated with those of PPARα target gene products (r = 0.886, P = 0.019 for LACS; r = 0.928, P = 0.008 for MCAD; r = 0.943, P = 0.005 for L-FABP; and r = 0.943, P = 0.005 for MTP). PPARα-dependent induction of CST mRNA levels was also observed in mice treated with clofibrate, another typical PPARα activator (Figure 5). These results indicated that the induction of hepatic CST was closely associated with PPARα activation in mice.
Figure 3.

Changes in hepatic expression of PPARs by fenofibrate treatment. Open and closed bars indicate mice treated without or with 0.1% fenofibrate, respectively. Data are expressed as mean ± SD (n = 6/group). *P < 0.05; **P < 0.01. (a) The mRNA levels of PPARs. Hepatic mRNA levels were normalized to those of GAPDH and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet. (b) PPAR-binding activity based on an enzyme-linked immunosorbent assay. Detailed protocols are described in Section 2. Results are expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet. (c) Immunoblot analysis. TBP was used as the loading control. Band intensities were measured densitometrically, normalized to those of TBP, and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet.
Figure 4.
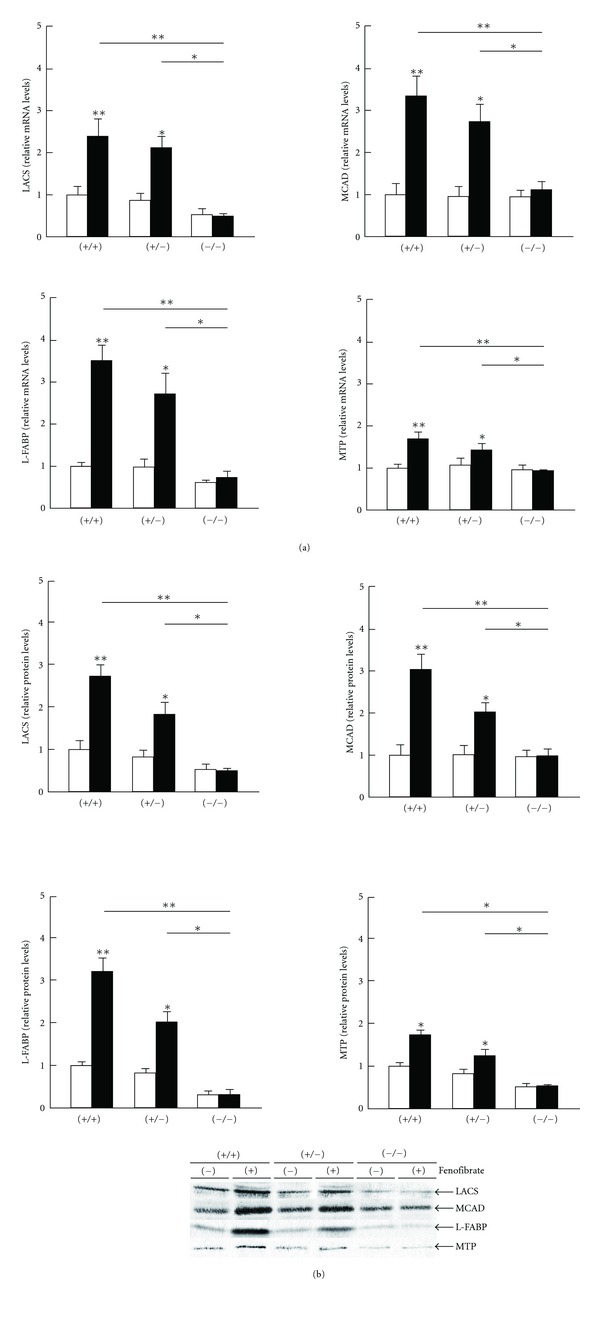
Changes in hepatic expression of conventional PPARα target genes by fenofibrate treatment. Open and closed bars indicate mice treated without or with fenofibrate, respectively. Data are expressed as mean ± SD (n = 6/group). *P < 0.05; **P < 0.01. (a) Analysis of mRNA. Hepatic mRNA levels were normalized to those of GAPDH and then expressed as fold changes relative to levels of Ppara (+/+) mice treated with a control diet. (b) Immunoblot analysis. Band intensities were measured densitometrically, normalized to those of actin, and then expressed as fold changes relative to those of Ppara (+/+) mice treated with a control diet.
Figure 5.
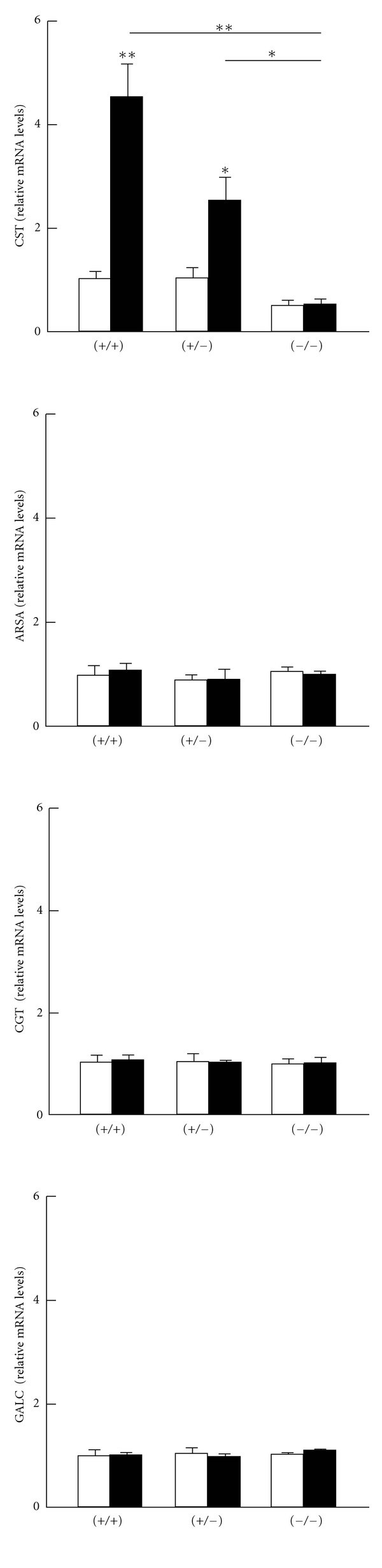
PPARα-dependent induction of CST mRNA levels by clofibrate treatment. Open and closed bars indicate mice treated without or with 0.5% clofibrate, respectively. Data are expressed as mean ± SD (n = 6/group). *P < 0.05; **P < 0.01.
4. Discussion
The present study revealed that fenofibrate treatment increased serum/liver sulfatide levels and the expression of hepatic CST mRNA and protein through PPARα activation. As CST mRNA levels were closely correlated with those of four known PPARα target genes, these findings suggest that CST may be a novel PPARα target gene candidate.
While CST is a key enzyme in sulfatide metabolism, little is known about its transcriptional regulation. We recently reported that an increase in hepatic oxidative stress downregulated CST expression in mice [8], although the mechanism remains unclear. A search for putative PPRE regions in the mouse CST gene [30, 31] revealed several candidates: −1,434/−1,422 (AGGTCTAAGGGCA), −1,202/−1,190 (TGGACTTTGCCCT), and −896/−884 (AGGACAAAGAGCA) from exon 1a; −1,499/−1,487 (AGGCTACAGTTCA) from exon 1e; and −1,569/−1,557 (AGGTCAGAGCACA) and −302/−290 (AGGACAGAGCCCA) from exon 1f. These regions may be useful for analysis in future in vitro experiments.
The degree of increases in serum sulfatides was lower than that in hepatic sulfatides by fenofibrate treatment (1.27-fold in the serum versus 2.20-fold in liver in Ppara (+/+) mice and 1.22-fold in the serum versus 1.95-fold in the liver of Ppara (+/−) mice). Sulfatides synthesized in the liver are secreted into the blood together with TG as a component of very-low-density lipoprotein (VLDL) [32]. Thus, hepatic TG content was reduced by fenofibrate treatment probably due to the enhanced of mitochondrial β-oxidation ability resulting in a reduction of hepatic VLDL synthesis as seen in other experiments using cultured hepatocytes [33]. Further studies are required to determine sulfatide metabolism in the serum and liver since they are significantly influenced by numerous pathophysiological events and treatments, including acute kidney injury [8, 34], clofibrate pretreatment [35], chronic kidney disease [6], and kidney transplantation [7].
The role of PPARα has been clarified in several liver diseases. For instance, PPARα is downregulated in alcoholic liver disease [11, 36] as well as after liver transplantation[37]. Persistent activation of PPARα ameliorates hepatic steatosis and inflammation in mice but may also induce hepatocarcinogenesis [10]. The association between liver disease and sulfatide metabolism may be of interest for further research.
Lastly, several animal studies have uncovered a protective role for serum sulfatides against arteriosclerosis and hypercoagulation [5]. We also reported a close relationship between lower serum sulfatide concentrations and higher incidences of cardiovascular disease in patients with end-stage renal failure [6], in whom sulfatide levels returned to normal following kidney transplantation [7]. Accordingly, increasing or maintaining serum sulfatide levels using fibrates may be useful in reducing the risk of cardiovascular events, which is consistent with the known beneficial effect of fibrates seen in randomized controlled studies [38]. Furthermore, these findings show a need to examine sulfatide metabolism in cardiomyocytes, endothelial cells, and vascular smooth cells to disclose any novel protective roles of PPARα in cardiovascular inflammation and atherosclerosis, particularly in relation to CST upregulation.
Conflicts of Interests
The authors have declared that no conflict of interests exists.
Acknowledgment
The authors thank Trevor Ralph for his English editorial assistance.
Abbreviations
- ARSA:
Arylsulfatase A
- CGT:
Ceramide galactosyltransferase
- CST:
Cerebroside sulfotransferase
- GALC:
Galactosylceramidase
- GAPDH:
Glyceraldehyde-3-phosphate dehydrogenase
- LACS:
Long-chain acyl-CoA synthase
- L-FABP:
Liver fatty acid-binding protein
- LS:
Lysosulfatides
- MCAD:
Medium-chain acyl-CoA dehydrogenase
- MTP:
Microsomal triglyceride transfer protein
- PCR:
Polymerase chain reaction
- PPAR:
Peroxisome proliferator-activated receptor
- PPRE:
PPAR response element
- TG:
Triglycerides.
References
- 1.Ishizuka I. Chemistry and functional distribution of sulfoglycolipids. Progress in Lipid Research. 1997;36(4):245–319. doi: 10.1016/s0163-7827(97)00011-8. [DOI] [PubMed] [Google Scholar]
- 2.Honke K, Zhang Y, Cheng X, Kotani N, Taniguchi N. Biological roles of sulfoglycolipids and pathophysiology of their deficiency. Glycoconjugate Journal. 2004;21(1-2):59–62. doi: 10.1023/B:GLYC.0000043749.06556.3d. [DOI] [PubMed] [Google Scholar]
- 3.Nagai KI, Tadano-Aritomi K, Niimura Y, Ishizuka I. Higher expression of renal sulfoglycolipids in marine mammals. Glycoconjugate Journal. 2008;25(8):723–726. doi: 10.1007/s10719-008-9132-x. [DOI] [PubMed] [Google Scholar]
- 4.Eckhardt M. The role and metabolism of sulfatide in the nervous system. Molecular Neurobiology. 2008;37(2-3):93–103. doi: 10.1007/s12035-008-8022-3. [DOI] [PubMed] [Google Scholar]
- 5.Kyogashima M. The role of sulfatide in thrombogenesis and haemostasis. Archives of Biochemistry and Biophysics. 2004;426(2):157–162. doi: 10.1016/j.abb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Hu R, Li G, Kamijo Y, et al. Serum sulfatides as a novel biomarker for cardiovascular disease in patients with end-stage renal failure. Glycoconjugate Journal. 2007;24(9):565–571. doi: 10.1007/s10719-007-9053-0. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Kamijo Y, Matsumoto A, et al. Kidney transplantation recovers the reduction level of serum sulfatide in ESRD patients via processes correlated to oxidative stress and platelet count. Glycoconjugate Journal. 2011;28(3-4):125–135. doi: 10.1007/s10719-011-9329-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Nakajima T, Kamijo Y, et al. Acute kidney injury induced by protein-overload nephropathy down-regulates gene expression of hepatic cerebroside sulfotransferase in mice, resulting in reduction of liver and serum sulfatides. Biochemical and Biophysical Research Communications. 2009;390(4):1382–1388. doi: 10.1016/j.bbrc.2009.10.164. [DOI] [PubMed] [Google Scholar]
- 9.Lee SST, Pineau T, Drago J, et al. Targeted disruption of the α isoform of the peroxisome proliferator- activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Molecular and Cellular Biology. 1995;15(6):3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPARα activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. Journal of Clinical Investigation. 2008;118(2):683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajima T, Kamijo Y, Tanaka N, et al. Peroxisome proliferator-activated receptor α protects against alcohol-induced liver damage. Hepatology. 2004;40(4):972–980. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- 12.Hara A, Radin NS. Lipid extraction of tissues with a low toxicity solvent. Analytical Biochemistry. 1978;90(1):420–426. doi: 10.1016/0003-2697(78)90046-5. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Hu R, Kamijo Y, et al. Establishment of a quantitative, qualitative, and high-throughput analysis of sulfatides from small amounts of sera by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Analytical Biochemistry. 2007;362(1):1–7. doi: 10.1016/j.ab.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 14.Smirnova IV, Bittel DC, Ravindra R, Jiang H, Andrews GK. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. Journal of Biological Chemistry. 2000;275(13):9377–9384. doi: 10.1074/jbc.275.13.9377. [DOI] [PubMed] [Google Scholar]
- 15.Aoyama T, Yamano S, Waxman DJ, et al. Cytochrome P-450 hPCN3, a novel cytochrome P-450 IIIA gene product that is differentially expressed in adult human liver. cDNA and deduced amino acid sequence and distinct specificities of cDNA-expressed hPCN1 and hPCN3 for the metabolism of steroid hormones and cyclosporine. Journal of Biological Chemistry. 1989;264(18):10388–10395. [PubMed] [Google Scholar]
- 16.Kamijo Y, Hora K, Tanaka N, et al. Identification of functions of peroxisome proliferator-activated receptor α in proximal tubules. Journal of the American Society of Nephrology. 2002;13(7):1691–1702. doi: 10.1097/01.asn.0000018403.61042.56. [DOI] [PubMed] [Google Scholar]
- 17.Aoyama T, Ueno I, Kamijo T, Hashimoto T. Rat very-long-chain acyl-CoA dehydrogenase, a novel mitochondrial acyl- CoA dehydrogenase gene product, is a rate-limiting enzyme in long-chain fatty acid β-oxidation system. cDNA and deduced amino acid sequence and distinct specificities of the cDNA-expressed protein. Journal of Biological Chemistry. 1994;269(29):19088–19094. [PubMed] [Google Scholar]
- 18.Aoyama T, Uchida Y, Kelley RI, et al. A novel disease with deficiency of mitochondrial very-long-chain acyl-CoA dehydrogenase. Biochemical and Biophysical Research Communications. 1993;191(3):1369–1372. doi: 10.1006/bbrc.1993.1368. [DOI] [PubMed] [Google Scholar]
- 19.Shindo Y, Hashimoto T. Acyl-coenzyme A synthetase and fatty acid oxidation in rat liver peroxisomes. Journal of Biochemistry. 1978;84(5):1177–1181. doi: 10.1093/oxfordjournals.jbchem.a132234. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka N, Zhang X, Sugiyama E, et al. Eicosapentaenoic acid improves hepatic steatosis independent of PPARα activation through inhibition of SREBP-1 maturation in mice. Biochemical Pharmacology. 2010;80(10):1601–1612. doi: 10.1016/j.bcp.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuta S, Mayazawa S, Hashimoto T. Purification and properties of rat liver Acyl-CoA dehydrogenases and electron transfer flavoprotein. Journal of Biochemistry. 1981;90(6):1739–1750. doi: 10.1093/oxfordjournals.jbchem.a133651. [DOI] [PubMed] [Google Scholar]
- 22.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocrine Reviews. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 23.Duplus E, Glorian M, Forest C. Fatty acid regulation of gene transcription. Journal of Biological Chemistry. 2000;275(40):30749–30752. doi: 10.1074/jbc.R000015200. [DOI] [PubMed] [Google Scholar]
- 24.Gervois P, Torra IP, Fruchart JC, Staels B. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clinical Chemistry and Laboratory Medicine. 2000;38(1):3–11. doi: 10.1515/CCLM.2000.002. [DOI] [PubMed] [Google Scholar]
- 25.Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation. 1998;98(19):2088–2093. doi: 10.1161/01.cir.98.19.2088. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Tanaka N, Nakajima T, Kamijo Y, Gonzalez FJ, Aoyama T. Peroxisome proliferator-activated receptor α-independent peroxisome proliferation. Biochemical and Biophysical Research Communications. 2006;346(4):1307–1311. doi: 10.1016/j.bbrc.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 27.Mandard S, Müller M, Kersten S. Peroxisome proliferator-activated receptor α target genes. Cellular and Molecular Life Sciences. 2004;61(4):393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoyama T, Peters JM, Iritani N, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα) Journal of Biological Chemistry. 1998;273(10):5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 29.Améen C, Edvardsson U, Ljungberg A, et al. Activation of peroxisome proliferator-activated receptor α increases the expression and activity of microsomal triglyceride transfer protein in the liver. Journal of Biological Chemistry. 2005;280(2):1224–1229. doi: 10.1074/jbc.M412107200. [DOI] [PubMed] [Google Scholar]
- 30.Podvinec M, Kaufmann MR, Handschin C, Meyer UA. NUBIScan, an in Silico approach for prediction of nuclear receptor response elements. Molecular Endocrinology. 2002;16(6):1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- 31.Hirahara Y, Tsuda M, Wada Y, Honke K. cDNA cloning, genomic cloning, and tissue-specific regulation of mouse cerebroside sulfotransferase. European Journal of Biochemistry. 2000;267(7):1909–1916. doi: 10.1046/j.1432-1327.2000.01139.x. [DOI] [PubMed] [Google Scholar]
- 32.Hara A, Taketomi T. Occurrence of sulfatide as a major glycosphingolipid in WHHL rabbit serum lipoproteins. Journal of Biochemistry. 1987;102(1):83–92. doi: 10.1093/oxfordjournals.jbchem.a122044. [DOI] [PubMed] [Google Scholar]
- 33.Hahn SE, Goldberg DM. Modulation of lipoprotein production in HEP G2 cells by fenofibrate and clofibrate. Biochemical Pharmacology. 1992;43(3):625–633. doi: 10.1016/0006-2952(92)90586-8. [DOI] [PubMed] [Google Scholar]
- 34.Kamijo Y, Hora K, Kono K, et al. PPARα protects proximal tubular cells from acute fatty acid toxicity. Journal of the American Society of Nephrology. 2007;18(12):3089–3100. doi: 10.1681/ASN.2007020238. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Kamijo Y, Hora K, et al. Pretreatment by low-dose fibrates protects against acute free fatty acid-induced renal tubule toxicity by counteracting PPARα deterioration. Toxicology and Applied Pharmacology. 2011;252(3):237–249. doi: 10.1016/j.taap.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okiyama W, Tanaka N, Nakajima T, et al. Polyenephosphatidylcholine prevents alcoholic liver disease in PPARα-null mice through attenuation of increases in oxidative stress. Journal of Hepatology. 2009;50(6):1236–1246. doi: 10.1016/j.jhep.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa K, Tanaka N, Morita M, et al. PPARα is down-regulated following liver transplantation in mice. Journal of Hepatology. 2012;56(3):586–594. doi: 10.1016/j.jhep.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keech A, Simes A, Barter P, et al. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366(9500):1849–1861. doi: 10.1016/S0140-6736(05)67667-2. [DOI] [PubMed] [Google Scholar]


