Abstract
Many membrane associated enzymes including those of the phospholipase C (PLC) superfamily are regulated by specific interactions with lipids. Previously we have shown that the C2 domain of PLC δ1 is required for phosphatidyl serine (PS) dependent enzyme activation, and that activation requires the presence of Ca2+. To identify the site of interaction and the role of Ca2+ in the activation mechanism, we mutagenized three highly conserved Ca2+ binding residues (Asp 653, Asp-706 and Asp-708) to Gly in the C2 domain of PLC δ1. The PS-dependent Ca2+ binding affinities of the mutant enzymes D653G, D706G and D708G were reduced by an order of magnitude and the maximal Ca2+ binding was reduced to half of that of the native enzyme. The Ca2+ dependent PS binding was also reduced in the mutant enzymes. Under basal conditions, the Ca2+ dependence and maximal hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) was not altered in the mutants. However, the Ca2+ dependent PS stimulation was severely defective. PS reduces the Km of the native enzyme almost 20 fold, but far less for the mutants. Replacing Asp-653, Asp-706 and Asp-708 simultaneously to glycine in the C2 domain of PLC δ1, leads to a complete and selective loss of the stimulation and binding by PS. These results show that D653, 706 and 708 are required for Ca2+ binding in the C2 domain and demonstrate a mechanism by which C2 domains can mediate regulation of enzyme activity by specific lipid ligands.
INTRODUCTION
The family of phospholipase C enzymes consist of six subfamilies (δ, β, γ, ε, ζ and η) of structurally and functionally distinct isozymes. A common functional feature is the ability of all members to catalyze the hydrolysis of polyphosphoinositides such as phoshatidylinositol 4,5-bisphosphate (PIP2) to generate the second messengers inositol trisphosphate (IP3) and diacylglycerol (DAG). This common phosphodiesterase activity is often the result of direct or indirect activation by receptor-coupled signaling pathways at the surface of eucaryotic cells. The mechanisms by which each family is regulated is different In general, the β isoforms are regulated by the Gq family of heterotrimeric G proteins, the γ isoforms by receptor and non-receptor tyrosine kinases, the ε family by G12 and G13 as well as by small G proteins such as Ras and Rap2B, the ζ family by calcium, and the δ family by calcium and lipid ligands such as phosphatidylserine (PS) and PIP2 as well as Ral and Gh (1, 2), though in recent years there have been a number of reported cases that do not fall into the traditional regulation paradigm according to each PLC subtype (3). While there is generally little conservation of primary structure among the six families, three major motifs are found in all eukaryotic isoforms, the catalytic X and Y domains, the pleckstrin homology (PH) domain, and the C2 domain.
PLC δ1 is highly responsive to lipid stimulations. For example, the activity of PLC δ1 is enhanced by PIP2 through the PH domain (4, 5). In cells this PIP2 activation is observed both in the plasma membrane and in the nucleus (6–8). Also PLC δ1 can be activated by other anionic lipids such as PS (9) and free fatty acids (10, 11).
The C2 domain is a conserved motif of approximately 130 amino acids that is predicted to exist in 295 distinct proteins by the human genome project (12). The structural motif of the C2 domain is an anti-parallel β-sandwich consisting of eight β-strands. Many of these C2 domain-containing proteins are involved in intracellular signaling or membrane trafficking. The function of the C2 domain has been examined in several of these molecules including protein kinase C, cytoplasmic phospholipase A2, phospholipase C and synaptotagmin. PKC α C2 is known to bind both calcium and PS (13). As for PLC δ family, the C2 domain is also considered important in membrane targeting. The isolated C2 domains from δ1 and δ3 translocate specifically towards the plasma membrane whereas the C2 domain of δ4 is localized to various cellular membranes including the perinuclear membranes and plasma membrane upon stimulation with ionomycin, a calucium-mobilizing ionophore, which is presumably resulted from a difference in their lipid specificity, especially towards PS (14). In addition, the C2 domains of δ1 and δ3 exhibit a strong calcium-dependent membrane affinity, whereas the C2 domain of δ4 has significant calcium-independent membrane affinity. Most studies have suggested a role for the C2 domain in membrane targeting, though a few studies have also suggested a role for calcium induced conformational changes (15).
Previously, we demonstrated the ability of free calcium and PS to bind to the C2 domain of PLC δ1 and activate phosphodiesterase activity 10–20 fold (9). In this work we identify the structural determinants for calcium and PS binding. Three aspartic acid residues in the loop region of the C2 domain are identified at position 653, 706 and 708 to each contribute to calcium and lipid binding. These studies identify the C2 domain as an allosteric modulatory domain, and demonstrate the role of C2 domains in mediating the effects of calcium and phospholipid on protein function.
EXPERIMENTAL PROCEDURES
Materials
The expression vector pRSETA was from Invitrogen. To express PLC δ1 under the control of the T7 promoter, the coding sequence for PLC δ1 was cloned into pRSETA. The resulting expression construct (pRSETAplc) was transformed into the Escherichia coli strain BL21(DE3)pLys (Novagen), and the protein was isolated and purified as described previously (16). Phosphatidylethanolamine, PA, PC, and PS were obtained from Avanti Polar Lipids Inc. PIP2 and dodecyl maltoside was obtained from Calbiochem.
Phospholipid Binding Assay
Phospholipid vesicles composed of PS/PC or PA/PC mixtures were prepared as described by Mueller et al. (17) with slight modifications (16, 18). A dry phospholipid film was formed by slowly blowing 0.25 ml of chloroform/methanol (2:1 v/v) containing mixed lipids (300 nmol or the indicated concentration of each of the indicated phospholipids) under a stream of nitrogen followed by freeze-drying under vacuum for 4 h. The phospholipid film was hydrated under nitrogen with 0.5 ml of nitrogen-aerated 0.18 M sucrose for 18 h at 4 °C followed by mixing with an equal volume of distilled H2O. Vesicles were isolated from the pellet by centrifuging the hydrated phospholipids at 1200 × g for 20 min. The phospholipid vesicles were washed once with 1 ml of 50 mM HEPES, pH 7.0, 100 mM KCl, 2 mM EGTA (binding buffer) and resuspended in 0.5 ml of the same buffer.
Centrifugation Binding Assay
The binding of PLC δ1 to phospholipid vesicles was estimated by a centrifugation assay (16, 19). The free Ca2+ concentration was calculated according to Fabiato and Fabiato (20). To perform the assay, 1 μg of enzyme was incubated with 200 μl of 50 mM HEPES, pH 7.0, 100 mM KCl, 2 mM EGTA, 150 μM phospholipid vesicle, and various concentrations of CaCl2 to yield the indicated concentration of free calcium. The reaction was carried out at 30 °C for 15 min. The free and bound PLC δ1 were separated by sedimentation at 50,000 × g for 30 min. An equal proportion of the supernatant and pellet fractions were resolved by 12% SDS-polyacrylamide gel electrophoresis, and the amount of PLC δ1 in each fraction was estimated by Western blotting analysis.
Calcium Binding Measurements
Calcium binding was determined by a nitrocellulose membrane binding assay similar to that described by Nakamura (21) and Kawasaki et al. (22). 150 μM PS/PC or PA/PC phospholipid vesicles containing the indicated mole fractions of lipids were incubated with PLC δ1 (final concentration, 0.15 μM) in 100 μl of 50 mM HEPES, pH 7.0, 100 mM KCl, 10 μM EGTA containing 5–120 μM of 45CaCl2 (total cpm, 1.5 × 107) to yield the indicated concentration of free Ca2+. Millipore polyvinylidene difluoride Immobilon-P transfer membrane was immersed in methanol and washed three times with 10 ml of 50 mM HEPES, pH 7.0, 100 mM KCl, and 10 mM EGTA. The membrane was mounted on a 96-well Bio-Dot filtration apparatus according to the manufacturer’s instructions (Bio-Rad). After incubating at 30 °C for 30 min, the reaction was filtered through the membrane at a constant flow rate of 0.6–1 ml/min. Each well was washed six times with 100 μl of ice-cold 50 mM HEPES, pH 7.0, 100 mM KCl, 10 mM EGTA. The membrane was cut into sections corresponding to each sample, and the retention of 45Ca2+ on the membrane was determined by liquid scintillation counting. The amount of 45Ca2+ bound to the enzyme was determined in the presence or absence of vesicles containing the indicated composition of phospholipids. Calcium binding to phospholipid vesicles in the absence of enzyme was determined under the same experimental conditions. The amount of 45Ca2+ binding directly to phospholipid vesicles was relatively low but consistent with published apparent Kd values for Ca2+-PS interactions (23, 24). The calcium bound to membranes or to phospholipid vesicles was considered nonspecific binding and was subtracted from the amount of Ca2+ bound by samples containing protein plus phospholipid.
PIP2 Hydrolysis Assay
PIP2 hydrolysis in dodecyl maltoside/PIP2 mixed micelles was performed in a manner similar to that described by Cifuentes et al. (4) with slight modifications. In brief, the indicated amount of PIP2/[3H]PIP2 (4 ×105 cpm) in chloroform/methanol (19:1) in the presence or absence of the indicated phospholipids was dried under a stream of N2 and lyophilized for 30 min. Lipids were solubilized by probe sonication in 0.95 ml of 50 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM EGTA plus the indicated concentration of dodecyl maltoside. Bovine serum albumin in the same buffer was added to 500 μg/ml. PLC activity was determined as a function of the concentration of substrate by keeping the total concentration of nonsubstrate phospholipid plus dodecyl maltoside at 500 μM and varying the mole fraction of PIP2. The reaction at 30 °C was initiated by adding various concentration of CaCl2 to yield the indicated concentration of free calcium. The reaction was continued for 1–5 min and was stopped by adding 0.34 ml of 10% ice-cold trichloroacetic acid and 0.17 ml of bovine serum albumin (10 mg/ml). After incubation on ice for 15 min, the unhydrolyzed [3H]PIP2 (pellet) was separated from [3H]IP3 (supernatant) by centrifugation at 2000 × g for 10 min at 4 °C.
Analysis of Kinetic Data
Surface dilution kinetics were employed to study PIP2 hydrolysis catalyzed by PLC δ1. Case III conditions previously described for phospholipase A2 (25) and PLC (26) were employed. In Case III conditions, the total concentration of diluent detergent (dodecyl maltoside plus nonsubstrate phospholipid) was fixed, and the PLC activity was measured with increasing concentrations of substrate. A dual phospholipid binding model of catalysis was used to analyze the kinetic data as previously described (9).
RESULTS
Asp653, 706 and 708 are essential for PS stimulation of PLC δ1
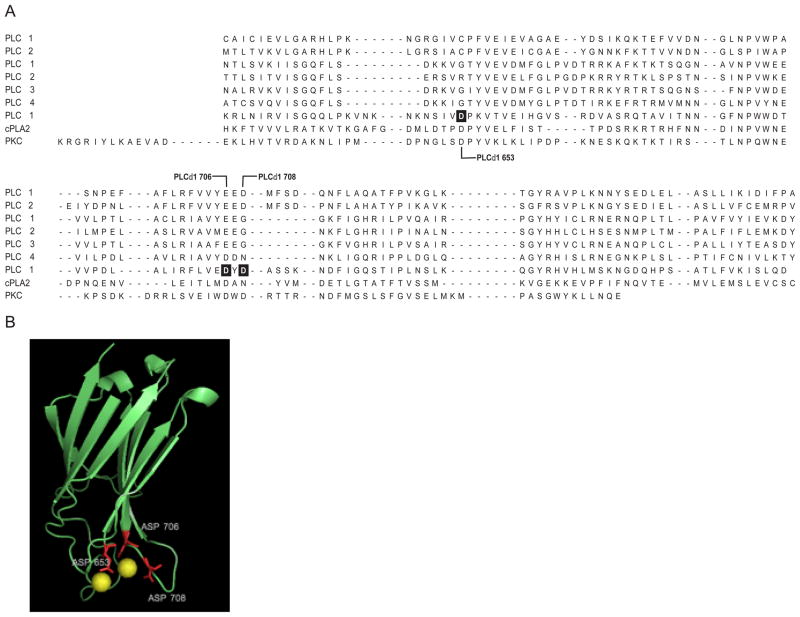
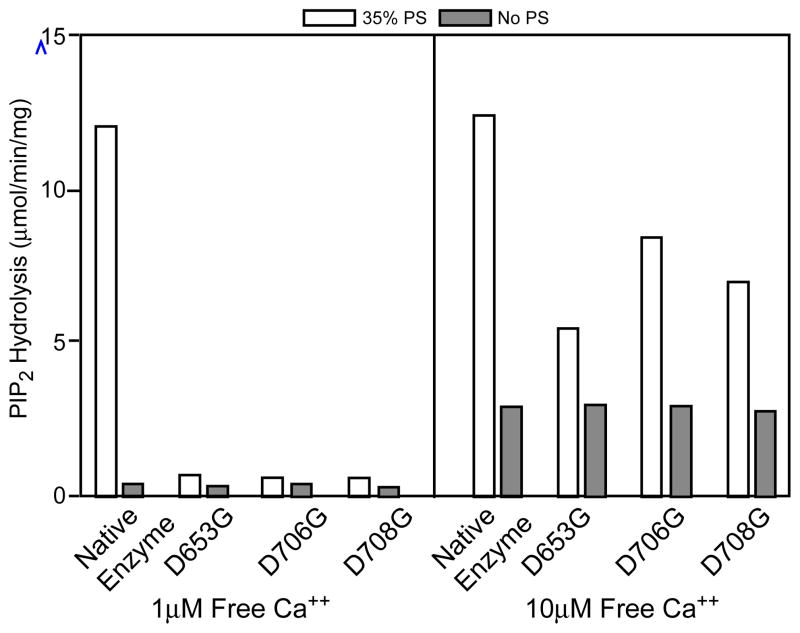
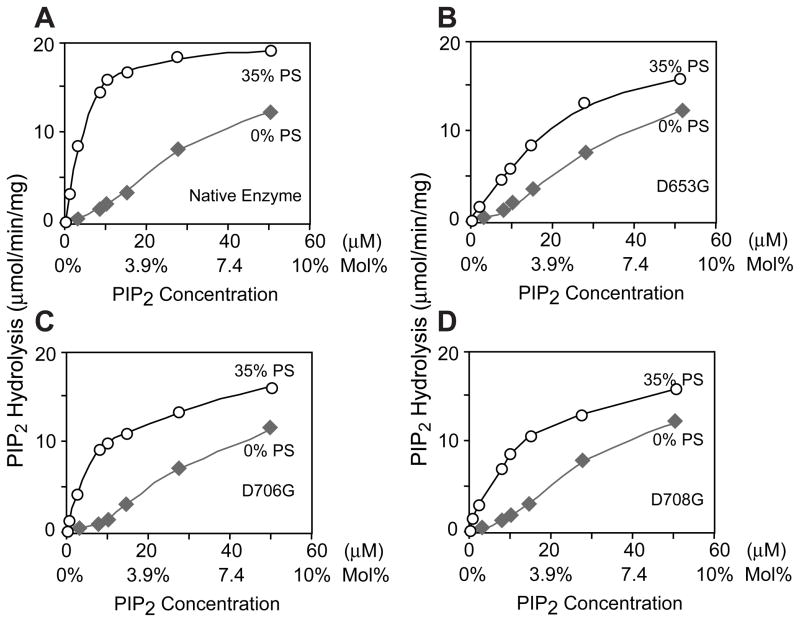
Three highly conserved negatively charged amino acid residues (Asp-653, Asp-706, and Asp-708) were identified in the δ isoforms of PLC by aligning the primary sequences of the C2 domains from PLA2, PKCα and all eucaryotic PI-PLCs (Figure 1A & 1B). Crystalographic studies have implicated these three residues in coordinating calcium binding (27). To investigate the role of these residues in Ca2+ binding and enzyme function, all three Asp were individually mutated by site-directed mutagenesis to Gly. These residues do not participate in the Ca2+-dependent basal activity of PLC δ1, but are required for the Ca2+-dependent PS stimulation of phosphodiesterase activity. As shown in Fig. 2, the rate of hydrolysis of PIP2 was very similar for the native and mutant enzymes in PS-free dodecyl maltoside mixed micelles at the two free Ca2+ concentrations tested (the hydrolysis rates were ~0.5 and ~3.3 μmol PIP2 hydrolyzed/min/mg at 1 μM and 10 μM free Ca2+, respectively). However, when 35% PS was included in the mixed micelles, significantly higher activity was seen for the native enzyme than for the mutant enzymes at both of the two free Ca2+ concentrations (Fig. 2). At 10 μM free Ca2+, PS stimulated the activities of the native, D653G, D706G and D708G PLC δ1 to 12.7, 5.5, 9.24 and 7.6 μmol PIP2 hydrolyzed/min/mg, respectively. The fold increase in activity for the native enzyme, D653G, D706G and D708G was 3.5, 1.7, 2.8 and 2.3, respectively. More remarkable stimulation by PS on the native enzyme was seen at lower free Ca2+ (1 μM), the native enzyme was stimulated by a factor of 20, while the activities of all three mutant enzymes remained unchanged. The activities of the mutant enzymes in the absence of PS were comparable to those of the native enzyme, irrespective of the concentration of free Ca2+ used.
Fig. 1.
(A) Amino acid alignment of C2 domains. Asp residues mutated in PLC δ1 are highlighted. (B) The crystal structure of the C2 domain of PLC δ1 (PDB: 1DJI). The residues colored in red are D653, D706 and D708.
Fig. 2.
Phosphatidylserine stimulation of the native, D653G, D706G and D708G mutant PLC δ1. Hydrolysis of 5 μM (1 mol%) PIP2 in PIP2/dodecyl maltoside mixed micelles in the presence (open bar) or absence of 35% PS (solid bar). The catalytic reaction was carried out in either 1 or 10 μM of free Ca2+. The total concentration of dodecyl maltoside plus nonsubstrate phospholipid was constant at 495 μM. The catalytic reaction was reaction was carried out in 50 μl of 50 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM EGTA plus 500 μg/ml BSA and CaCl2 to yield 1 or 10 μM of free Ca2+. The free Ca2+ concentration in an EGTA-CaCl2 buffer was calculated according to Fabiato and Fabiato (20). The reaction was carried at 30°C for 1–5 min, stopped and quantitated as described in Experimental Procedures.
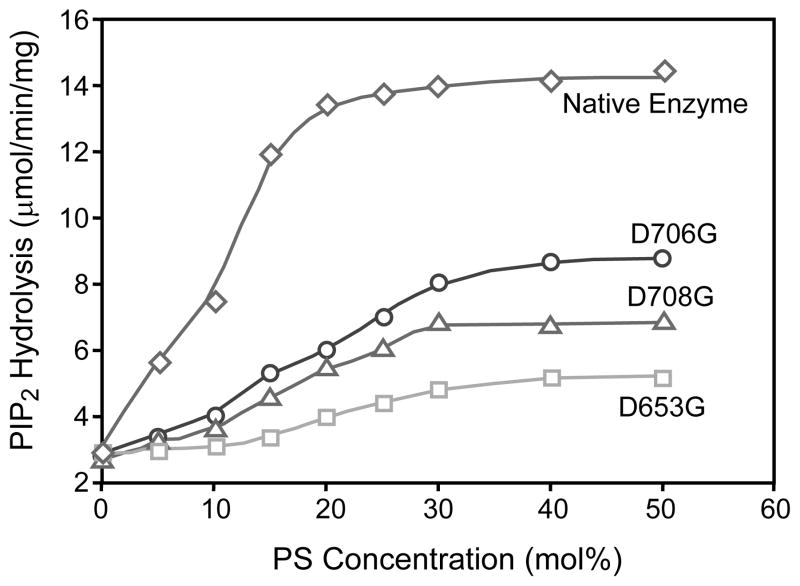
These results demonstrate that D653, D706 and D708 are not required for basal phosphodiesterase activity of PLC δ1, but are essential for PS mediated stimulation of activity. This is more clearly illustrated in Fig. 3. The PS dose response curves show that the rate of PIP2 hydrolysis by native and mutant enzymes is dependent on the mole fraction of PS. The potency and maximal effect of PS is markedly reduced for the mutant enzymes.
Fig. 3.
Dependence of PS concentration on PIP2 hydrolysis by the native and mutant PLC δ1. Catalytic hydrolysis of 5 μM of PIP2 in dodecyl maltoside mixed micelles containing increasing concentration of PS (from 0 to 50 mol%) by the native (diamond), D653G (square), D706G (circle) and D708G (triangle) mutant enzyme. The total concentration of dodecyl maltoside plus nonsubstrate phospholipid was constant at 495 μM. The catalytic reaction was carried out as described in Fig. 1 and Experimental Procedures.
Asp653, 706 and 708 mediate Ca2+ regulation of PLC δ1
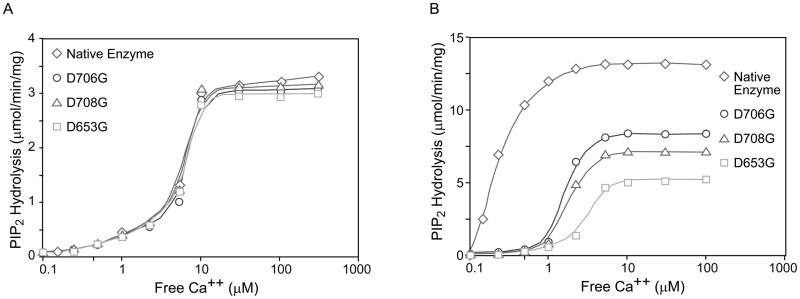
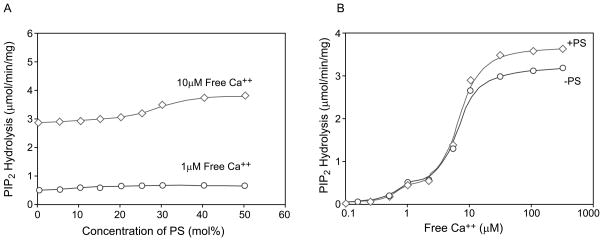
We have previously shown that PS stimulates PLC δ1 by virtue of its ability to form a Ca2+/PS/PLC δ1 complex via the C2 domain. PS and Ca2+ are interdependent; neither one can interact with nor regulate PLC δ1 without the other. Therefore, replacing aspartate 653, 706 and 708 with glycine may reduce the ability of PS to increase the potency of Ca2+ for stimulating catalysis. This is clearly illustrated in Fig. 4. In the absence of PS, the calcium dose response curves for the mutant and native enzymes were identical to each other (Fig. 4A). The maximal catalytic activity of the native enzyme, D653G, D706G and D708G were estimated to be 3.3 μmol PIP2 hydrolyzed/min/mg, and the free Ca2+ concentration required for half-maximal activity of each enzyme (EC50) was approximately 6 μM. Including 35 mol% PS in the mixed micelles shifted the Ca2+ dose response curves up and to the left for native and mutant enzymes, though the shift was much more pronounced for the native enzyme (Fig. 4B). PS increased both the potency of Ca2+ and the maximal activity. The maximal activity of the native enzyme was increased from 3.2 to 13 μmol/min/mg (Fig. 4A and 4B). The free Ca2+ concentration for half maximal activity (EC50) of the native enzyme was reduced by a factor of at least 20 (from 6 to 0.25 μM). The maximal activity of the mutants was 40 to 70% that of the native PLC δ1 in the presence of PS. The maximal PS stimulated activity of D653G, D706G and D708G PLC δ1 was 4.9, 8.2 and 6.9 μmol of PIP2/min/mg, respectively, while that of the native enzyme was 13 μmol/min/mg. With 35% PS, Ca2+ is at least 5 to 10 fold less potent in stimulating PIP2 hydrolysis by D653G, D706G and D708G PLC δ1 than for the native enzyme. As shown in Fig. 4B, the free Ca2+ concentration required for half-maximal hydrolysis of PIP2 in mixed micelles containing 35 mol % PS by D653G, D706G and D708G PLC δ1 was estimated to be 2.5, 2.0 and 1.8 μM, respectively. This reduction in potency of Ca2+ for activation of the mutant enzymes may explain why we were not able to observe the stimulatory effect of PS on the mutants when assays were carried out at 1 μM free Ca2+. In the absence of PS, the calcium dose response curves for native and mutant enzymes are identical. However, in the presence of PS calcium has a reduced potency and reduced maximal effect on the mutant enzymes as compared to the PS effect on the native enzyme. The mutant enzymes have a selective deficiency in PS activation.
Fig. 4.
Effect of PS on the Ca2+ dependence of the native and the mutant PLC δ1 catalytic activities. Calcium concentration dependence of hydrolysis of 5 μM PIP2 (corresponding to 1 mol% in the mixed micelles) by native (diamond), D653G (square), D706G (circle) and D708G (triangle) mutant PLC δ1 in PIP2/dodecyl maltoside mixed micelles containing 0 (A) or 35 (B) mol% PS. The reaction was carried out in a buffer of 50 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM EGTA plus 500 μg/ml BSA and various CaCl2 to yield indicated concentration of free Ca2+. PLC δ1 catalyzed hydrolysis of PIP2 in the mixed micelles was measured as described (Fig. 1 and Experimental Procedures).
D653G, D706G and D708G have lower affinity for substrate than the native enzyme
We have previously shown that the stimulation of PIP2 hydrolysis by PS was primarily due to an increase in the affinity for the substrate at the catalytic site. (9) To understand the enzymatic mechanism underlying the impaired phenotypes of the D653G, D706G and D708G mutant enzymes, the effect of PS on the substrate concentration dependence of PLC δ1 catalysis was examined. When the micellar concentration of PIP2 was increased from 0.1 to 9%, the hydrolysis of PIP2 by the native enzyme in mixed micelles containing 35 mol % PS sharply increased from 3.1 to 19 μmol/min/mg and was saturated as the PIP2 concentration approached 4 mol%. PIP2 hydrolysis catalyzed by the native enzyme in PS-free mixed micelles increased slowly and did not reach a maximum even at 9 mol% PIP2. Comparing the substrate dependence of PIP2 hydrolysis in the presence and absence of PS, demonstrated that the most dramatic stimulatory effect of PS on the hydrolysis of PIP2 was at low substrate concentrations. As shown in Fig. 5A, when the PIP2 concentration was less than 2 mol %, PIP2 hydrolysis in mixed micelles containing 35 mol % PS was at least 10-fold higher than in PS-free mixed micelles. The stimulatory effect of PS diminished as the concentration of PIP2 increased. The substrate dependence for the D653G, D706G and D708G mutant enzymes was very comparable to that of the native enzyme, when the hydrolysis of PIP2 was carried out in PS free mixed micelles. The rates of PIP2 hydrolysis by the D653G, D706G and D708G mutant enzymes were much lower than that of native enzyme if the reactions were carried out in the presence of 35 mol% PS. Specifically, the stimulatory effect of PS on the mutant enzymes was much less than that of the native enzyme at low substrate concentrations. This observation reflects a decrease in substrate affinity for the mutant enzymes. The kinetic parameters of the native and the mutant enzymes revealed that the mutant enzymes were partially impaired in PS stimulated- reduction in the interfacial Michaelis constant (Km), which reflects the affinity for the substrate. Km of the native enzyme was reduced by a factor 20 in the presence of 35% of PS, while those of D653G, D706G and D708G was reduced by a factor of 3, 6 and 4.5, respectively. PS had little effect on the maximal rate of catalysis (Vmax) or on the affinity for the interface (Ks) for all enzymes.
Fig. 5.
Influence of PS on the substrate dependence of catalytic hydrolysis of PIP2 by the native and the mutant PLC δ1. Hydrolysis of increasing concentrations of PIP2 by the native (A), D653G (B), D706G (C) and D708G (D) PLC δ1 in PIP2/dodecyl maltoside mixed micelles containing 0 (diamond) and 35 (circle) mol% PS. The mol% of PIP2 was increased while the concentration of PS was fixed at 0 and 35 mol%, and the combined total concentration of PS plus dodecyl maltoside was maintained at 495 μM, see Experimental Procedures. The reaction was carried out in the presence of 5 μM free Ca2+, stopped and the [3H]IP3 was separated and quantitated as described in Experimental Procedures.
D653G, D706G and D708G are defective in Ca2+ binding
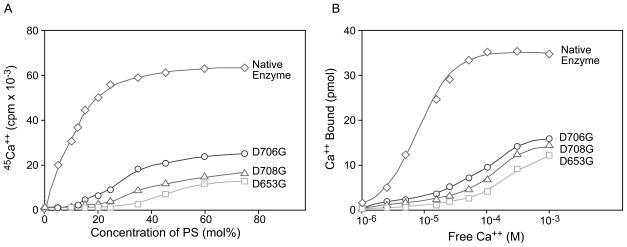
PS activation of PLC δ1 requires the simultaneous binding of Ca2+ and PS to the C2 domain of the enzyme. To test whether D653, D706 and D708 are involved in the lipid dependent Ca2+ binding to the C2 domain, we measured the binding of 45Ca2+ to PLC δ1 as a function of the PS concentration and Ca2+ concentration. As illustrated in Fig. 6A, the binding of Ca2+ by the native enzyme is highly dependent on the concentration of PS; the binding was enhanced by as low as 5 mol % PS and plateaued as PS reached 30 mol%. Replacing D653, D706 or D708 to glycine reduced the total Ca2+ binding to PLC δ1 at saturating concentrations of PS and also required higher concentrations of PS to reach saturation (Fig. 6A).
Fig. 6.
Dependence of PS and free Ca2+ on the binding of Ca2+ to the native and the mutant PLC δ1. PS dependence of the binding of Ca2+ to the native (diamond), D653G (square), D706G (circle) and D708G (triangle) mutant PLC δ1. Binding of 0.15 μM enzyme with 100 μM free 45Ca2+ (total 1.5 × 107 cpm) in 0.1 ml buffer containing 50 mM Hepes, pH 7.0, 100 mM KCl, and 10 μM EGTA and 150 μM PS/PC vesicles containg 0–75 mol% of PS, the determination of bound Ca2+ were carried as described in Experimental Procedures (A). Binding of 0.15 μM native (diamond), D653G (square), D706G (circle) and D708G (triangle) mutant PLC δ1 to increasing concentrations of free 45Ca2+ (total 1.5 × 107 cpm) in the presence of 150 μM PS/PC vesicles composed of 50 mol% of PS was carried out as described in Experimental Procedures (B).
Since little Ca2+ would bind to PLC δ1 in the absence of PS/PC vesicles, the Ca2+ saturation binding isotherm was carried out in the presence of PS/PC vesicles containing 40 mol% PS. In the presence of PS/PC vesicles, 45Ca2+ bound to the native and the mutant PLC δ1 in a dose dependent and saturable manner Fig. 6B. Ca2+ bound to the native enzyme when the free Ca2+ was as low as 2 μM and the binding increased sharply until the free concentration reached 100 μM. The binding of Ca2+ to mutant enzymes was much lower than that of the native PLC δ1, and increased slowly as the free Ca2+ concentration increased from 10 to 1000 μM. Saturable binding was still not obtained as the free Ca2+ concentration reached 1 μM. As shown in Fig. 6B, the maximal binding of Ca2+ to 0.15 μM protein was estimated to be 35 pmole, corresponding to approximately 2.3 pmole Ca2+/pmole protein. The binding of Ca2+ to D653G, D706G and D708G mutant enzymes at 1 mM free Ca2+ was 12.8, 16.5 and 15 pmole corresponding to 0.84, 1.08 and 0.98 pmole Ca2+/pmole protein, respectively. The concentration of free Ca2+ required for half-maximal binding to the native enzyme was estimated to be 7 μM, while that for D653G, D706G and D708G mutants it was 128, 70 and 90 μM, respectively. The Ca2+ binding isotherm revealed that the affinity for Ca2+ binding to PLC δ1 was reduced by a factor of 10 in D653G, D706G and D708G (Fig. 6B). Therefore, D653, D706 and D708 are required for high affinity PS-dependent Ca2+ binding and mutation of these resides leads to a 10-fold decrease in affinity for Ca2+ and a decrease in maximal Ca2+ binding.
D653, D706 and D708 are required for PS binding to PLC δ1
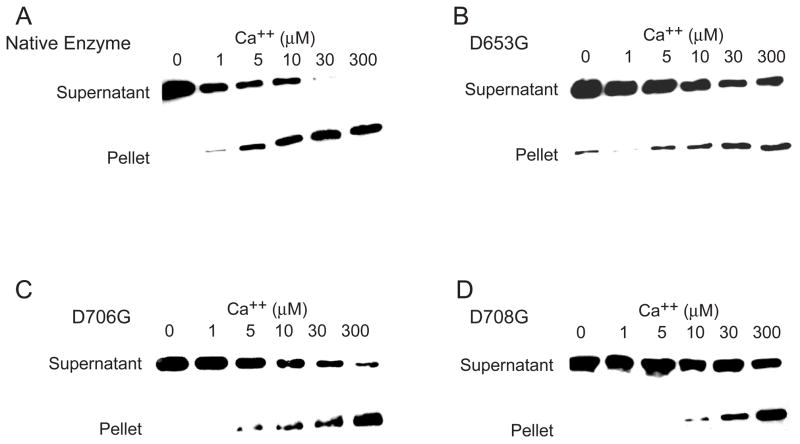
Since D653G, D706G and D708G are defective in Ca2+ binding and PS stimulation, these mutants may be also defective in PS binding. To test this possibility, the binding of native and mutant PLC δ1 to sucrose loaded PS/PC vesicles was determined in the presence of increasing concentrations of free Ca2+. The native enzyme bound to PS in a Ca2+-dependent manner and saturable binding was obtained when the free Ca2+ reached 30 μM. As shown in Fig. 7, increasing amounts of PLC δ1 accumulated in the pellet fraction as the Ca2+ increased from 0 to 300 μM, a consequence of direct binding of PLC δ1 to the sucrose-loaded PS/PC vesicles. More than 90% of the native enzyme was bound to PS/PC vesicles at 30 μM Ca2+. The Ca2+-dependent binding of PS to D653G, D706G and D708G mutant enzymes was impaired. Less than 50% of the mutant protein was bound to PS at 30 μM free Ca2+, and binding is not saturated even if the free Ca2+ was increased to 300 μM (Fig. 7).
Fig. 7.
Ca2+ -dependent phospholipid binding to the native and the mutant PLC δ1. Centrifugation binding assay of PLC δ1 and sucrose loaded PS/PC vesicles in the presence of the indicated concentration of free Ca2+ ion. 1μg of the native (A), D653G (B), D706G (C) and D708G (D) mutant protein was incubated with 150 μM PS/PC vesicles (molar ratio = 1:1) in 0.2 ml of 50 mM Hepes, pH 7.0, 100 mM KCl, 2 mM EGTA and various concentrations of CaCl2 to yield the indicated concentration of free Ca2+. The reaction was incubated at 30°C for 15 min. The bound enzyme (pellet fraction) and the free enzyme (supernatant fraction) were separated and quantitated as described under Experimental Procedures.
The results thus far show that residues D653, D706 and D708 are essential for Ca2+-dependent PS stimulation of PLC δ1. However, each single residue mutant D653G, D706G and D708G still displayed some residual binding and regulation by both PS and Ca2+. This indicates that D653, 706 and 708 may independently contribute to the interaction of PS and Ca2+ with PLC δ1. To test this hypothesis, we mutated D653, 706, and 708 to glycine simultaneously. The ability of PS to stimulate the triple residue mutant enzyme and its dependence on free Ca2+ was examined. Analysis of the PS dose response for the D653, 706, 708G enzyme reveals that this mutant enzyme is barely stimulated by PS even at very high concentrations of PS and Ca2+ (10 μM Ca2+ and 50 mol % PS Fig. 8A). When the reactions were carried out at 10 μM free Ca2+, the activity of the D653/D706/D708G mutant slightly increased from 2.9 to 3.9 μmol PIP2 hydrolyzed/min/mg when the PS in the mixed micelles increased from 0 to 50 mol%.
Fig. 8.
Ca2+ and PS dependence of PIP2 hydrolysis by the D653/D706/D708G mutant PLC δ1. PS concentration dependence of the D653/D706/D708G mutant PLC δ1catalyzed hydrolysis of 5 μM PIP2 (corresponding to 1 mol % in the mixed micelles) in the presence of 1 (circle) or 10 (diamond) μM free Ca2+ (A). Ca2+ dependence of hydrolysis of 5 μM of PIP2 (corresponding to 1 mol % in the mixed micelles) by the D653/D706/D708G mutant PLC δ1 in mixed micelles containing 0 (circle) or 50 (diamond) mol % of PS is shown. The catalytic reaction was carried as described (Fig. 2 and 3, and Experimental Procedures).
As shown in Fig. 8B, PIP2 hydrolysis by D653/D706/D708G PLC δ1 in the presence of increasing concentrations of free Ca2+ revealed that the activity of D653/D706/D708G mutant PLC δ1 was stimulated as free Ca2+ increased from 1 μM and the activity reached a plateau when the free Ca2+ approached 10 μM. This pattern of Ca2+ dependence is very similar to that of the native enzyme under basal conditions. In contrast to the native enzyme, the activity in the presence of PS was only slight higher than that in the absence of PS. The estimated free Ca2+ (5.5 μM) required for half maximal stimulation in the absence of PS was comparable to that (6 μM) in the presence of PS. These results indicate that the Ca2+ affinity for the D653/D706/D708G mutant PLC δ1 was not affected by the presence of PS. This observation is quite in contrast to the native enzyme, and the single residue mutants D653G, D706G and D708G, where the Ca2+ affinity was increased 20, 2.3, 3.3 and 3 -fold respectively, by the presence of PS.
The D653/D706/D708G PLC δ1 triple residue mutant does not bind PS nor Ca2+, nor does PS effect the affinity for substrate
The physical binding of Ca2+ and PS to PLC δ1 was eliminated by simultaneous mutation of D653, D706 and D708. As shown in Fig. 9A, Ca2+ barely binds to the triple mutant enzyme even in the presence of 35 mol% of PS. Centrifugation phospholipid vesicle binding assays also revealed that the triple mutant binds very little to PS/PC vesicles when the free Ca2+ concentration ranged from 0 to 300 μM (Fig. 9B). Although the triple mutant displays Ca2+-dependence during basal catalysis similar to that of native enzyme, the mutant enzyme is severely defective in Ca2+ and PS binding. Lack of this physical interaction with the mutant enzyme leads to a severe loss of activation by PS. Analysis of the substrate concentration dependence of the triple mutant revealed that the mutant enzyme is barely stimulated by PS (Fig. 10). The substrate affinity for the triple mutant was not affected by PS. These results demonstrate that each of the three aspartate residues at position 653, 706, and 708 mediate the effects of PS and Ca2+ on the activation of PLC δ1.
Fig. 9.
Binding of Ca2+ and PS to the D653, 706, 708G mutant PLC δ1. Binding of 0.15 μM the D653/D706/D708G mutant enzyme with indicated concentration of free 45Ca2+ (total 1.5 × 107 cpm) in 0.1 ml buffer containing 50 mM Hepes, pH 7.0, 100 mM KCl, and 10 μM EGTA and 150 μM PS/PC vesicles containing 0 (diamond) and 35 (circle) mol% of PS, the determination of bound Ca2+ were carried as described in Experimental Procedures (A). Centrifugation binding assay of the D653/D706/D708G mutant enzyme and sucrose loaded PS/PC vesicles in the presence of the indicated concentration of free Ca2+ ion. Binding of 1μg protein to150 μM PS/PC vesicles (molar ratio = 1:1) in the presence of indicated concentration of free Ca2+ was carried as described (Fig. 6 and Experimental Procedures) (B).
Fig. 10.
Effect of PS on the substrate dependence of the PIP2 hydrolysis catalyzed by the D653/D706/D708G mutant PLC δ1. Hydrolysis of increasing concentrations of PIP2 by the D653/D706/D708G mutant PLC δ1 in PIP2/dodecyl maltoside mixed micelles containing 0 (diamond) and 50 (circle) mol% PS. The reaction was carried out in the presence of 20 μM free Ca2+, stopped and the [3H]IP3 was separated and quantitated as described in Fig. 4 and Experimental Procedures.
DISCUSSION
This extensive structure-function analysis is a follow-up to our previous paper that described the ability of free calcium and phosphatidylserine (PS) to potentiate the phosphodiesterase activity of PLC δ1 through a C-terminal C2 domain (9). C2 domains mediate the regulatory effects of the second messenger calcium for many proteins that act at an interface between cytosol and plasma membrane, nuclear membrane, and endoplasmic reticulum (15, 28). They have a very conserved tertiary structure consisting of an eight-stranded antiparallel β-sandwich. Despite this conservation of tertiary structure, C2 domains vary substantially in their primary structure, which likely explains their differential ability to bind ligands such as calcium (a minority of C2 domains do not bind nor are regulated by calcium), phospholipids, and proteins. Much of the diversity in primary structure resides in the loops that connect the β-strands at the bottom and top of the β-sandwich. Specific residues from these loops have been implicated in calcium and phospholipid binding (29, 30, 49 and 50).
The aspartic acid residues at positions 653, 706 and 708 of the loop regions of the C2 domain of PLC δ1 are highly conserved in the C2 domains from a number of signaling molecules that display Ca2+-dependent activation. Subsets of C2 domains that are calcium-independent are missing most if not all of these conserved residues. The solution of a crystal structure of PLC δ1 complexed with the calcium analog lanthanum revealed that the negatively charged carboxyl groups of residues Asp 653, 706 and 708 participated in coordinating the binding of three lanthanum ions to the C2 domain of PLC δ1 (27). To investigate the functional role of calcium binding to the C2 domain of PLC δ1, we eliminated the negative charges in the holo protein at these three positions by mutating each Asp residue to Gly, individually and all together to create a triple mutant.
Replacement of all three Asp residues with Gly (D653/D706/D708G) ablates the ability of PLC δ1 to bind Ca2+ in the absence of substrate and also ablates PS binding. Since substrate is necessary for Ca2+ binding to the catalytic domain but not to the C2 domain, non-catalytic Ca2+ binding sites are assessed by excluding substrate from the assay mixture. Ca2+ can still bind to the catalytic site of the triple mutant, since Ca2+ binding to the catalytic domain is absolutely necessary for hydrolysis and the basal activity of this enzyme is unchanged. In fact, the activity of D653/D706/D708G PLC δ1 is still responsive to increasing Ca2+ concentrations, its Ca2+ dose response curve is similar to that of the native enzyme in the absence of PS. Because the binding of Ca2+ and PS is interdependent, D653/D706/D708G PLC δ1 does not bind PS. This finding is also in agreement with our previous results that showed the simultaneous presence of PS, Ca2+ and PLC δ1 is required for the formation of the ternary Ca2+-PS-enzyme activation complex(9). Because D653/D706/D708G PLC δ1 cannot bind PS, the enzyme is not regulated at all by PS, unlike the native enzyme which is stimulated 20 fold by μM concentrations of PS and calcium. The lack of PS stimulation and binding to D653/D706/D708G PLC δ1 definitively identifies the C2 domain as the site of Ca2+ and PS interaction, and defines a role for the C2 domain in enzyme activation. Data from Ananthanarayanan et al. support this model. They have demonstrated Ca2+-dependent translocation of the C2 domain of PLC δ1 to the inner plasma membrane which is considered rich in PS content (14). It is concluded that Ca2+ binding to the C2 domain is essential for the enzyme to interact with and be regulated by PS. Work by Corbin et al. also demonstrate that PKC alpha and cPLA2-alpha C2 associate to the membrane via calcium drive interactions with specific lipids (49).
We have determined the stochiometry of Ca2+ binding to PLC δ1 to be 2.3, a number in agreement with the co-crystal structures (27). From analysis of the single point mutants, D653G, D706G, and D708G, it is concluded that all three residues play a role in Ca2+ and PS binding. Each of the point mutants has an intermediate phenotype between wild type and triple mutant D653/D706/D708G PLC δ1 enzyme. For example, each of the single point mutants could be regulated by PS and Ca2+, however, the potency of PS and Ca2+ was substantially reduced as was the maximal efficacy of these compounds. The reduced potency and maximal efficacy was a result of the reduced affinity of Ca2+ and PS for these mutant enzymes. While it is still not known how many Ca2+ ions actually bind to the C2 domain of PLC δ1, it is at least two and perhaps as many as three. It is clear, however, that all three Asp acid residues (D653, D706, and D708G) in the C2 domain of PLC δ1 play an equal role in binding to these Ca2+ ions, and that the binding of multiple Ca2+ ions is cooperative. Grobler and Hurley reported binding of a total of four Ca2+ ions, one to the catalytic pocket and three to the C2 domain, estimated from the Hill coefficients of calorimetric titration (31). On the other hand, Cifuentes et al. approximated the number of bound Ca2+ ions to be two for both the intact and active proteolytic fragments of PLC δ1 (4). It may be possible that the method we employed here was able to detect only strong interactions between the metal cation and the protein, however, the obtained number of bound Ca2+ is in good agreement with the co-crystal structures.
Ca2+ can have multiple roles on regulating C2 domain function. Calcium can alter the electrostatic potential of the surface of the C2 domain, can act as a bridge between the C2 domain and a ligand, can induce conformational changes, and/or promote protein-protein interactions with adaptor proteins (30, 32). Several reports suggest that the role of the C2 domain of PLC δ1 is to correctly orient the catalytic domain to the interface (14, 33). This may indeed be a role for the C2 domain, however, it’s dramatic effect on enzyme catalysis is likely to involve more complex mechanisms. Kinetic analysis revealed that PS stimulates PLC δ1 by virtue of its ability to increase substrate affinity. The native enzyme’s affinity for PIP2 was increased 20-fold in the presence of PS, while the affinity between PIP2 and the triple mutant enzyme remained unchanged. It appears that the C2 domain of PLC δ1 acts primarily as an allosteric regulator, since the Km for substrate is reduced dramatically after Ca2+ and PS binding. Kinetic analyses comparing wildtype and C2 domain mutant enzymes using a soluble substrate such as cIP, should be able to discriminate true allosteric effects from those mediated by interactions with the interface.
Our approach examines the C2 domain as it exists in nature, as a part of a macromolecule. This approach not only has the potential to be more informative, but is also a more conservative approach than studies that seek to examine the function of isolated C2 domains. It is especially important to determine the functional role(s) of the C2 domain of PLC δ1 as part of the holo protein, since the C2 domain is very closely apposed to the catalytic domain in this phospholipase (33). The extensive contact between the C2 and catalytic domain is likely necessary for proper folding of both domains, and allows for the transfer of allosteric effects. The crystal structure of PLC δ1 reveals that there is an extensive and rigid interface between C2 domain and core structure of catalytic domain (33). Additional evidence suggesting that the C2 domain is specifically important for enzymatic activity, was provided by Kanematsu et al. In their study the replacement of the C2 domain with that of its related molecule, p130, completely abolished the activity of the enzyme (34). The C2 domain of the PLC family is considered to have evolved together with two other domains, the catalytic and EF-hand domains (35). More recently, a fragment of PLC β3 comprising the most of the enzyme including the C2 domain, was co-crystallized with Gαq and the structure of the complex was examined (36). The interface between the two proteins included the region between the C2 and catalytic domains of PLC β3, whose mutatations markedly reduced the PLC activity in the presence of Gαq. It is possible that PS/Ca2+-binding to the C2 domain of PLC δ1 could subsequently cause a structural change in the C2 domain itself as well as the region between the C2 and catalytic domains similar to the one in PLC β3 complexed with Gαq, which ultimately affects the enzymatic activity.
C terminal deletion mutagenesis of PLC δ1 resulted in production of an inactive protein (37, 38), furthermore, the Ca2+ binding loops in the C2 domain are located in the same face as the cleavage site. These findings led to the proposal that membrane binding to the C2 domain of PLC δ1 was essential to correctly orient the catalytic site to the substrate containing interface (33). However, the proposal did not distinguish between direct effects of the C2 domain on activity, on protein stability or both. The isolated C2 domain of PLC δ1 or of the C2 domain truncation enzyme is expressed mainly in the insoluble fraction of bacterial extracts, suggesting that correct folding of the catalytic core and of the C2 domain of PLC δ1 is interdependent. The present biochemical studies of mutated PLC δ1 demonstrate that Ca2+ binding to the C2 domain is not essential for catalysis of substrate but has a direct affect on the catalytic center. The affinity for the substrate PIP2 at the active site is increased by 20-fold as the enzyme binds to Ca2+ and membrane containing PS via its C2 domain. The modification of Km is a common mechanism by which enzymes are regulated.
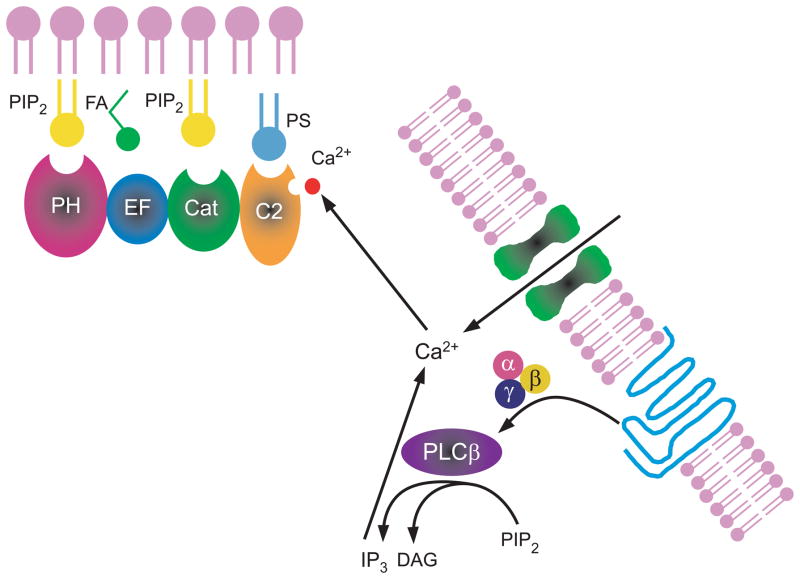
Why does deletion of the Ca2+ binding residues in the C2 domain lead to a reduction in the PS mediated increase in affinity for substrate? The simplest explanation is that the mutant enzymes are not able to interact with PS, since this interaction requires the binding of Ca2+ to the C2 domain. Disruption of the Ca2+ binding leads to defective PS-PLC δ1 interaction thus PS is not able to stimulate PLC δ1. Because PS binding to the C2 domain is highly sensitive to the free Ca2+ concentration, we postulate that Ca2+ is the primary regulator of PLC δ1, since Ca2+ concentrations clearly can vary several magnitudes while the concentration of PS likely varies little in the cell (Figure 11). This mechanism may be particularly important for the regulation of PLC δ1 in the cell. The affinity of the enzyme for PIP2 has been determined by kinetic analysis from several laboratories. The Km values ranged from 2 to 20 mol% of membrane lipids. (4) Since the concentration of PIP2 in the plasma membrane is below 1%, it is highly unlikely that PLC δ1 can catalyze the hydrolysis of PIP2 in the cell unless there is a mechanism to increase its affinity for substrate. The present results provide a mechanism whereby Ca2+ may function as an activator of PLC δ1 in the cell. Actually, several observations reveal that activation of the PLC δ1 isozyme might occur as an event secondary to receptor-mediated activation of other PLC isozymes or Ca2+ channels (39, 40). In addition, to directly participating in the catalytic reaction, the present results shows that Ca2+ may increase the affinity for substrate by promoting PS binding via the C2 domain.
Fig 11.
Schematic depicting proposed model for PLC δ1 activation.
Although D653, D706, and D708 are conserved in PLC δ1, primary sequence analysis shows that these Ca2+ binding residues are not conserved in the C2 domains of the β and γ families of PLC. Asp-653 is replaced with Gly and Cys in PLC β and γ, respectively. Asp708 is replaced with a Asn or Gly in PLC β. The activity of the δ class is much more sensitive to the concentration of free Ca2+ than that of the β, γ or ε enzymes. Only the newly identified ζ isozyme whose expression is limited to sperm appears to have similar or even greater sensitivity to free Ca2+ (41). Since elimination of any one of these three conserved aspartates in PLC δ1 leads to a substantial reduction in Ca2+ dependent enzyme activation, it is predicted that the β and γ isoenzymes should be much less sensitive to the concentration of free calcium. This prediction is consistent with both in vitro and in vivo findings of the Ca2+ dependence of PLC isoforms (39, 42). PLC δ1 but not PLC β1 nor PLC γ1 is stimulated by physiologic concentrations of Ca2+ to hydrolyze cellular inositol lipids (40).
Many studies in excitable tissues show that a rise in intracellular Ca2+ activates PLC activity (40, 43–47). Indeed, it has been demonstrated that PLC δ1 is activated by capacitative calcium entry that follows PLC β activation upon bradykinin stimulation (39). We hypothesize that this calcium induced PLC activity may be primarily due to activation of PLC δ1 and demonstrate a potential mechanism for Ca2+ to regulate PLC δ1 through the C2 domain. Thus activation of a receptor coupled to PLC β leads to increased intracellular free Ca2+, which in turn can further activate a Ca2+-sensitive form of PLC (like PLC δ1) forming a positive feedback loop of Ca2+ and inositol phospholipid signaling (Figure 11). Because receptor mediated PLC activation is usually transient and rapidly desensitized (48), the positive Ca2+-inositol phospholipid loop could prolong the action of PLC and may operate in slower or more prolonged synaptic or cellular responses. We propose that PLC δ1 is in fact responsible for these prolonged effects, such as those seen in hypertension and cardiac hypertrophy.
These studies identify the C2 domain as an allosteric modulatory domain, and demonstrate the role of C2 domains in mediating the effects of calcium and phospholipid on protein function. Since many other molecules contain C2 domains, these studies suggest a mechanism by which these other molecules could be regulated.
Table 1.
Effect of PS on the kinetic properties of the native and the mutant PLC δ1
| PLC δ1 PS concentration (mol %) | Vmax (μmol/min/mg) | Km (mol fraction) | Ks (μM) | |
|---|---|---|---|---|
| Native PLC δ1 | 0 | 17±3 | 0.045±0.001 | 70±2 |
| 35 | 19±4 | 0.0025±0.0002 | 45±3 | |
| D653G PLC δ1 | 0 | 14±2 | 0.045±0.001 | 71±3 |
| 35 | 17±3 | 0.01±0.01 | 47±2 | |
| D706G PLC δ1 | 0 | 15±2 | 0.051±0.003 | 69±2 |
| 35 | 18±3 | 0.008±0.002 | 44±2 | |
| D708G PLC δ1 | 0 | 14±3 | 0.048±0.004 | 73±4 |
| 35 | 18±4 | 0.009±0.001 | 46±2 |
Hydrolysis of increasing concentration of PIP2 at 5 μM free Ca2+ by PLC δ1 was measured in PIP2/dodecyl maltoside mixed micelles containing 35 and 0 mol % PS (see Experimental Procedures). Vmax, Km and Ks corresponding to the constants defined in the surface dilution model; values of Vmax, Km and Ks were calculated by fitting the data to Equation 1 in ref (26).
Acknowledgments
This work was supported by grants from the National Institutes of Health HL 03961 and HL 55591 (to JWL), and National Sciences Council of the Republic of China, the Institute of Biomedical Sciences and Academia Sinica Intramural Funding (to K.K.)
The abbreviations used are
- PIP2
phosphatidylinositol 4,5-bisphosphate
- IP3
inositol 1,4,5-trisphosphate
- PI
phosphatidylinositol
- PC
hosphatidylcholine
- PA
phosphatidic acid
- PS
phosphatidyl serine
- PLC
phosphoinositde-specific phospholipase C
References
- 1.Murthy SN, Lomasney JW, Mak EC, Lorand L. Interactions of G(h)/transglutaminase with phospholipase Cdelta1 and with GTP. Proc Natl Acad Sci U S A. 1999;96:11815–11819. doi: 10.1073/pnas.96.21.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sidhu RS, Clough RR, Bhullar RP. Regulation of phospholipase C-delta1 through direct interactions with the small GTPase Ral and calmodulin. J Biol Chem. 2005;280:21933–21941. doi: 10.1074/jbc.M412966200. [DOI] [PubMed] [Google Scholar]
- 3.Katan M. New insights into the families of PLC enzymes: looking back and going forward. Biochem J. 2005;391:e7–e9. doi: 10.1042/BJ20051506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cifuentes ME, Honkanen L, Rebecchi MJ. Proteolytic fragments of phosphoinositide-specific phospholipase C-delta 1. Catalytic and membrane binding properties. Journal of Biological Chemistry. 1993;268:11586–11593. [PubMed] [Google Scholar]
- 5.Lomasney JW, Cheng HF, Wang LP, Kuan YS, Liu SM, Fesik SW, King K. Phosphatidylinositol 4,5-bisphosphate binding to the pleckstrin homology domain of phospholipase C-delta 1 enhances enzyme activity. J Biol Chem. 1996;271(41):25316–25326. doi: 10.1074/jbc.271.41.25316. [DOI] [PubMed] [Google Scholar]
- 6.Yagisawa H, Sakuma K, Paterson HF, Cheung R, Allen V, Hirata H, Watanabe Y, Hirata M, Williams RL, Katan M. Replacements of Single Basic Amino Acids in the Pleckstrin Homology Domain of Phospholipase C-Delta-1 Alter the Ligand Binding, Phospholipase Activity, and Interaction With the Plasma Membrane. Journal of Biological Chemistry. 1998;273:417–424. doi: 10.1074/jbc.273.1.417. [DOI] [PubMed] [Google Scholar]
- 7.Stallings JD, Tall EG, Pentyala S, Rebecchi MJ. Nuclear translocation of phospholipase C-delta1 is linked to the cell cycle and nuclear phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2005;280:22060–22069. doi: 10.1074/jbc.M413813200. [DOI] [PubMed] [Google Scholar]
- 8.Stallings JD, Zeng YX, Narvaez F, Rebecchi MJ. Phospholipase C-delta1 expression is linked to proliferation, DNA synthesis, and cyclin E levels. J Biol Chem. 2008;283:13992–14001. doi: 10.1074/jbc.M800752200. [DOI] [PubMed] [Google Scholar]
- 9.Lomasney JW, Cheng HF, Roffler SR, King K. Activation of Phospholipase C delta1 through C2 Domain by a Ca(2+)- Enzyme-Phosphatidylserine Ternary Complex. J Biol Chem. 1999;274:21995–22001. doi: 10.1074/jbc.274.31.21995. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi M, Gryczynski Z, Lukomska J, Feng J, Roberts MF, Lakowicz JR, Lomasney JW. Spectroscopic characterization of the EF-hand domain of phospholipase C delta1: identification of a lipid interacting domain. Arch Biochem Biophys. 2005;440:191–203. doi: 10.1016/j.abb.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi M, Mutharasan RK, Feng J, Roberts MF, Lomasney JW. Identification of Hydrophobic Interactions between Proteins and Lipids: Free Fatty Acids Activate Phospholipase C delta1 via Allosterism. Biochemistry. 2004;43:7522–7533. doi: 10.1021/bi035966c. [DOI] [PubMed] [Google Scholar]
- 12.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 13.Ochoa WF, Corbalan-Garcia S, Eritja R, Rodriguez-Alfaro JA, Gomez-Fernandez JC, Fita I, Verdaguer N. Additional binding sites for anionic phospholipids and calcium ions in the crystal structures of complexes of the C2 domain of protein kinase calpha. J Mol Biol. 2002;320:277–291. doi: 10.1016/S0022-2836(02)00464-3. [DOI] [PubMed] [Google Scholar]
- 14.Ananthanarayanan B, Das S, Rhee SG, Murray D, Cho W. Membrane targeting of C2 domains of phospholipase C-delta isoforms. J Biol Chem. 2002;277:3568–3575. doi: 10.1074/jbc.M109705200. [DOI] [PubMed] [Google Scholar]
- 15.Cho W. Membrane targeting by C1 and C2 domains. J Biol Chem. 2001;276:32407–32410. doi: 10.1074/jbc.R100007200. [DOI] [PubMed] [Google Scholar]
- 16.Cheng HF, Jiang MJ, Chen CL, Liu SM, Wong LP, Lomasney JW, King K. Cloning and identification of amino acid residues of human phospholipase C delta 1 essential for catalysis. Journal of Biological Chemistry. 1995;270:5495–5505. doi: 10.1074/jbc.270.10.5495. [DOI] [PubMed] [Google Scholar]
- 17.Mueller P, Chien TF, Rudy B. Formation and properties of cell-size lipid bilayer vesicles. Biophysical Journal. 1983;44:375–381. doi: 10.1016/S0006-3495(83)84311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang LP, Lim C, Kuan YS, Chen CL, Chen HF, King K. Positive charge at position 549 is essential for phosphatidylinositol 4,5-bisphosphate-hydrolyzing but not phosphatidylinositol-hydrolyzing activities of human phospholipase C delta 1. J Biol Chem. 1996;271(40):24505–24516. doi: 10.1074/jbc.271.40.24505. [DOI] [PubMed] [Google Scholar]
- 19.Rebecchi M, Peterson A, McLaughlin S. Phosphoinositide-specific phospholipase C-delta 1 binds with high affinity to phospholipid vesicles containing phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1992;31:12742–12747. doi: 10.1021/bi00166a005. [DOI] [PubMed] [Google Scholar]
- 20.Fabiato A, Fabiato F. Calculator programs for computing the composition of the solutions containing multiple metals and ligands used for experiments in skinned muscle cells. Journal de Physiologie. 1979;75:463–505. [PubMed] [Google Scholar]
- 21.Nakamura J. Calcium-dependent non-equivalent characteristics of calcium binding sites of the sarcoplasmic reticulum Ca2+-ATPase. Biochim Biophys Acta. 1986;870:495–501. doi: 10.1016/0167-4838(86)90258-x. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki H, Kasai H, Okuyama T. Protein analyses and reagents: microscale assay of calcium-binding activity of proteins and peptides using a nitrocellulose membrane. Anal Biochem. 1985;148:297–302. doi: 10.1016/0003-2697(85)90232-5. [DOI] [PubMed] [Google Scholar]
- 23.Portis A, Newton C, Pangborn W, Papahadjopoulos D. Studies on the mechanism of membrane fusion: evidence for an intermembrane Ca2+-phospholipid complex, synergism with Mg2+, and inhibition by spectrin. Biochemistry. 1979;18:780–790. doi: 10.1021/bi00572a007. [DOI] [PubMed] [Google Scholar]
- 24.Nelsestuen GL, Lim TK. Equilibria involved in prothrombin- and blood-clotting factor X- membrane binding. Biochemistry. 1977;16:4164–4171. doi: 10.1021/bi00638a005. [DOI] [PubMed] [Google Scholar]
- 25.Hendrickson HS, Dennis EA. Kinetic analysis of the dual phospholipid model for phospholipase A2 action. Journal of Biological Chemistry. 1984;259:5734–5739. [PubMed] [Google Scholar]
- 26.James SR, Paterson A, Harden TK, Downes CP. Kinetic analysis of phospholipase C beta isoforms using phospholipid-detergent mixed micelles - Evidence for interfacial catalysis involving distinct micelle binding and catalytic steps. J Biol Chem. 1995;270(20):11872–11881. doi: 10.1074/jbc.270.20.11872. [DOI] [PubMed] [Google Scholar]
- 27.Essen LO, Perisic O, Lynch DE, Katan M, Williams RL. A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1. Biochemistry. 1997;36:2753–2762. doi: 10.1021/bi962466t. [DOI] [PubMed] [Google Scholar]
- 28.Hurley JH, Misra S. Signaling and subcellular targeting by membrane-binding domains. Annu Rev Biophys Biomol Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bittova L, Sumandea M, Cho W. A structure-function study of the C2 domain of cytosolic phospholipase A2. Identification of essential calcium ligands and hydrophobic membrane binding residues. J Biol Chem. 1999;274:9665–9672. doi: 10.1074/jbc.274.14.9665. [DOI] [PubMed] [Google Scholar]
- 30.Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 31.Grobler JA, Hurley JH. Catalysis By Phospholipase C Delta(1) Requires That Ca2+ Bind to the Catalytic Domain, But Not the C2 Domain. Biochemistry. 1998;37:5020–5028. doi: 10.1021/bi972952w. [DOI] [PubMed] [Google Scholar]
- 32.Stahelin RV, Cho W. Roles of calcium ions in the membrane binding of C2 domains. Biochem J. 2001;359:679–685. doi: 10.1042/0264-6021:3590679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 34.Kanematsu T, Yoshimura K, Hidaka K, Takeuchi H, Katan M, Hirata M. Domain organization of p130, PLC-related catalytically inactive protein, and structural basis for the lack of enzyme activity. Eur J Biochem. 2000;267:2731–2737. doi: 10.1046/j.1432-1327.2000.01291.x. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez JL, Smith GR, Contreras-Moreira B, Sgouros JG, Meunier FA, Bates PA, Schiavo G. Functional recycling of C2 domains throughout evolution: a comparative study of synaptotagmin, protein kinase C and phospholipase C by sequence, structural and modelling approaches. J Mol Biol. 2003;333:621–639. doi: 10.1016/j.jmb.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 36.Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, Wang X, Montell C, Kozasa T, Sondek J, Harden TK. Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science. 330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis MV, Carne A, Katan M. Structural requirements of phosphatidylinositol-specific phospholipase C delta 1 for enzyme activity. European Journal of Biochemistry. 1993;213:339–347. doi: 10.1111/j.1432-1033.1993.tb17767.x. [DOI] [PubMed] [Google Scholar]
- 38.Yagisawa H, Hirata M, Kanematsu T, Watanabe Y, Ozaki S, Sakuma K, Tanaka H, Yabuta N, Kamata H, Hirata H, Nojima H. Expression and characterization of an inositol 1,4,5- trisphosphate binding domain of phosphatidylinositol-specific phospholipase C-delta(1) J Biol Chem. 1994;269(31):20179–20188. [PubMed] [Google Scholar]
- 39.Kim YH, Park TJ, Lee YH, Baek KJ, Suh PG, Ryu SH, Kim KT. Phospholipase C-delta1 is activated by capacitative calcium entry that follows phospholipase C-beta activation upon bradykinin stimulation. J Biol Chem. 1999;274:26127–26134. doi: 10.1074/jbc.274.37.26127. [DOI] [PubMed] [Google Scholar]
- 40.Allen V, Swigart P, Cheung R, Cockcroft S, Katan M. Regulation of inositol lipid-specific phospholipase cdelta by changes in Ca2+ ion concentrations. Biochem J. 1997;327:545–552. doi: 10.1042/bj3270545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouchi Z, Fukami K, Shikano T, Oda S, Nakamura Y, Takenawa T, Miyazaki S. Recombinant phospholipase Czeta has high Ca2+ sensitivity and induces Ca2+ oscillations in mouse eggs. J Biol Chem. 2004;279:10408–10412. doi: 10.1074/jbc.M313801200. [DOI] [PubMed] [Google Scholar]
- 42.Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 43.Hepler JR, Kozasa T, Smrcka AV, Simon MI, Rhee SG, Sternweis PC. Purification from Sf9 cells and characterization of recombinant Gq alpha and G11 alpha. Activation of purified phospholipase C isozymes by G alpha subunits. Journal of Biological Chemistry. 1993;268:14367–14375. [PubMed] [Google Scholar]
- 44.Smart D, Smith G, Lambert DG. Mu-opioids activate phospholipase C in SH-SY5Y human neuroblastoma cells via calcium-channel opening. Biochem J. 1995;305(Pt 2):577–581. doi: 10.1042/bj3050577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smart D, Wandless A, Lambert DG. Activation of phospholipase C in SH-SY5Y neuroblastoma cells by potassium-induced calcium entry. Br J Pharmacol. 1995;116:1797–1800. doi: 10.1111/j.1476-5381.1995.tb16665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberhard DA, Holz RW. Intracellular Ca2+ activates phospholipase C. Trends Neurosci. 1988;11:517–520. doi: 10.1016/0166-2236(88)90174-9. [DOI] [PubMed] [Google Scholar]
- 47.Myles ME, Fain JN. Carbachol, but not norepinephrine, NMDA, ionomycin, ouabain, or phorbol myristate acetate, increases inositol 1,3,4,5-tetrakisphosphate accumulation in rat brain cortical slices. J Neurochem. 1994;62:2333–2339. doi: 10.1046/j.1471-4159.1994.62062333.x. [DOI] [PubMed] [Google Scholar]
- 48.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 49.Corbin JA, Evans JH, Landgraf KE, Falke JJ. Mechanism of specific membrane targeting by C2 domains: localized pools of target lipids enhance Ca2+ affinity. Biochemistry. 2007;46:4322–4336. doi: 10.1021/bi062140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans JH, Murray D, Leslie CC, Falke JJ. Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Molecular biology of the cell. 2006;17:56–66. doi: 10.1091/mbc.E05-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]