Abstract
A single-nucleotide polymorphism (C/A) located within an E-box at the −20 position of the human angiotensinogen (AGT) promoter may regulate transcriptional activation through differential recruitment of the transcription factors Upstream Stimulatory Factor (USF) 1 and 2. To study the contribution of USF1 on AGT gene expression, mice carrying a (−20C) human AGT transgene were bred with mice harboring a USF1 gene trap allele designed to knock down USF1 expression. USF1 mRNA was reduced relative to controls in liver (9±1%), perigenital adipose (16±3%), kidney (17±1%), and brain (34±2%) in double-transgenic mice. This decrease was confirmed by electrophoretic mobility shift assay. Chromatin immunoprecipitation (ChIP) analyses revealed a decrease in USF1, with retention of USF2 binding at the human AGT promoter in the liver of male mice. Human AGT expression was reduced in the liver and other tissues of female but not male mice. The decrease in endogenous AGT expression was insufficient to alter systolic blood pressure at baseline, but caused reduced systolic blood pressure in female USF1 gene trap mice fed a high fat diet. Treatment of USF1 knockdown males with intravenous adenoviral shRNA targeting USF2 resulted in reduced expression of USF1, USF2 and human AGT protein. Our data from ChIP assays suggests that this decrease in human AGT is due to decreased USF2 binding to the human AGT promoter. In conclusion, both USF1 and USF2 are essential for AGT transcriptional regulation, and distinct gender-specific and tissue-specific mechanisms are involved in the activities of these transcription factors in vivo.
Keywords: Polymorphism, Transcription, Adenovirus, shRNA, Hypertension
Introduction
The angiotensinogen (AGT) gene has long been studied as a candidate gene in hypertension and many studies have illustrated a direct correlation between AGT levels and blood pressure (BP). Increasing the copy number of the AGT gene increases BP by approximately 7 mmHg per copy, delivery of anti-AGT antibodies into the blood reduces BP, and injection of AGT causes increased BP.1–3 However, the precise mechanisms by which genetic polymorphisms in the AGT gene or its promoter region could influence BP remain unclear.
It is widely accepted that hypertension is heritable and consequently has a genetic component. Single Nucleotide Polymorphisms (SNPs) in the human AGT (hAGT) promoter have been implicated in the pathogenesis of high BP, including those at nucleotides −6, −20, −217, −517, and −792 (reviewed in4). Some of the polymorphisms in the hAGT promoter have been reported to affect transcriptional regulation of the gene, thus providing a potential mechanism for dysregulation.5 For example, we reported that one genetic variant in the hAGT promoter (G-6A) might have physiological relevance under high salt diet conditions.6 In that study we examined BP in 4-triple transgenic mouse models expressing either the −6G or −6A haplotypes of hAGT, combined with 2 different human renin transgenes (one well regulated, the other poorly regulated), all on a endogenous murine AGT null genetic background. We found increased salt sensitivity of BP in mice carrying the −6A hAGT haplotype when on a genetic background of poorly controlled human renin expression.
The A-20C variant in hAGT is independently associated with increased BP, cardiac hypertrophy, cerebral small vessel disease, and a blunted aldosterone response to Ang II.7–10 We demonstrated that the −20 position lies within an “E-box (CANNTG)” sequence, which binds the transcription factors Upstream Stimulatory Factor (USF) 1 and USF2, and differential binding of USF1 and USF2 to the −20C or −20A alleles of the AGT promoter may lead to differential expression of AGT in AGT-expressing cell lines.11 Moreover, clinical studies show that AGT expression is higher in human subjects carrying the −20C SNP compared to those carrying the −20A SNP.12 However, the molecular mechanism regulating hAGT expression by transcription factors that bind to physiologically relevant promoter sequences is unclear in vivo.
USF1 and USF2 are ubiquitously expressed transcription factors controlling several critical genes in lipid and glucose metabolism.13 They are recruited to the transcriptional complex to modulate the state of chromatin.14,15USF1-null mice were viable and fertile with elevated levels of USF2, which may compensate for the absence of USF116,17. In contrast, USF2-null mice exhibited reduced levels of USF1 and displayed an obvious growth defect.16 While USF1 or USF2 single knockouts survive, the double-null is lethal, further supporting a functional redundancy in USF1 and USF2. Herein, we determined the in vivo importance of the −20C allele in the hAGT promoter and its possible interaction with USF1 and USF2 by examining transgenic mice carrying the −20C SNP in hAGT in which USF1 expression was knocked down.
Materials and Methods
Please see the online-only Data Supplement for a more detailed Materials and Methods (please see http://hyper.ahajournals.org).
Animals
We employed a USF1 gene trap (USF1GT) allele (USF1Gt (XG830) Byg) derived from ES cells obtained from the Mutant Mouse Regional Resource Center. We also used hAGT transgenic mice are reported.18 All mice were fed standard mouse chow and had access to water ad libitum. All procedures were approved by the University Animal Care and Use Committee at the University of Iowa and conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Molecular Analysis
Gene expression, Western blotting, gel shift and super-shift assays (EMSA), and chromatin immunoprecipitation (ChIP) were performed as detailed in the Supplemental Methods.
Intravenous Adenoviral Delivery in vivo
AdshUSF2.3 expressing an shRNA targeting Usf2 was prepared and administered as described.11
Blood Pressure Measurements
Systolic BP (SBP) was measured using the Visitech Systems BP-2000 tail-cuff BP monitoring system as previously described.19
Statistical analysis
All comparisons were performed between USF1GT and USF1WTlittermates expressing the hAGT transgene. Data were analyzed by t-test or ANOVA as appropriate, and were expressed as mean±SEM. Statistical significance was defined at a value of P<0.05.
Results
Mice carrying −20C SNP of hAGT transgene and USF1GT
To identify the role of USF1 on hAGT mRNA expression in vivo, we bred transgenic mice carrying a high expressing allele (−20C) of hAGT onto the USF1GT genetic background, and then examined human and mouse AGT expression in hAGT/USF1WT and hAGT/USF1GT mice. Body weights between hAGT/USF1WT and hAGT/USF1GT littermates at 3 months of age were not different (26.9±0.7 vs. 25.9±0.9 in male and 19.8±0.4 vs.19.5±0.7 g in female, n≥7). No differences in the weights of liver, kidney, and reproductive adipose tissue, nor glucose levels were found between hAGT/USF1WT and hAGT/USF1GT mice.
Regulation of USF1 and USF2 mRNA
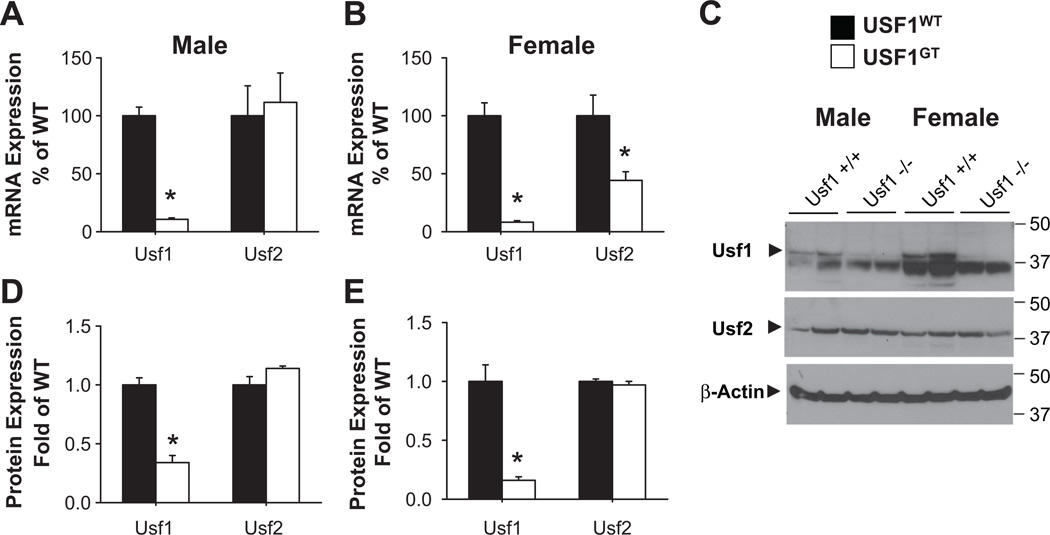
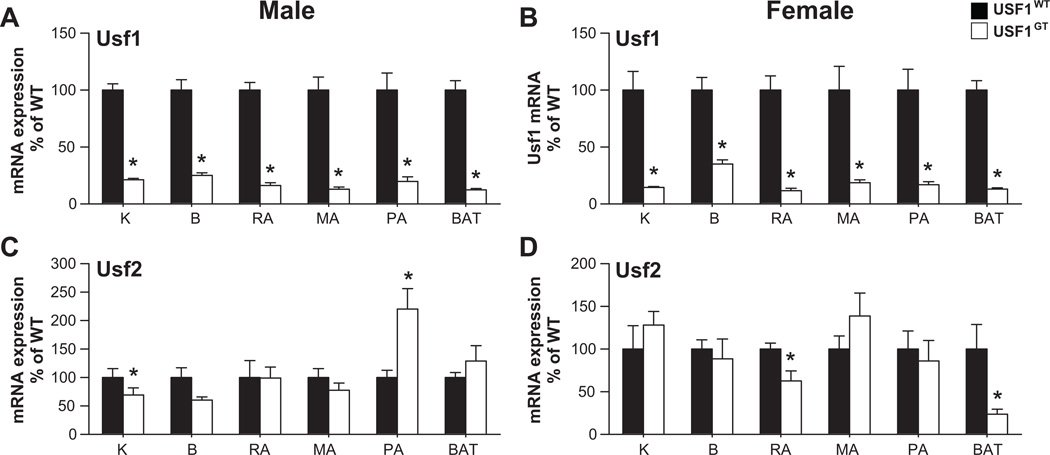
Expression of Usf1 mRNA in USF1GT mice was significantly decreased in male and female liver (11±1 and 8±1% of WT, respectively) (Figure 1A–B). Variable decreases relative to WT were observed in other tissues including kidney and brain, and reproductive, mesenteric, perirenal and brown adipose tissues indicating that the gene trap is not a complete null but results in an effective knockdown of Usf1 mRNA (Figure 2A–B). Although the expression of Usf2 mRNA which possess 2 potential E-Box motifs16,20 was not changed in USF1GT male liver (Figure 1A), it was significantly decreased in USF1GT female liver (Figure 1B). Modest reductions of Usf2 mRNA were found in USF1GT female reproductive and brown adipose tissues (Figure 2D). The levels of USF1, but not USF2 protein in USF1GT male and female liver were reduced significantly (Figure 1C–E).
Figure 1. USF1 and USF2 expression in USF1GTmouse liver.
A–B) Expression of Usf1 and Usf2 mRNA expression in male (A, n=7, 8) and female (B, n=8, 8). C) Western blots showing USF1, USF2, and β-actin in both genders. D-E) Quantitative protein expression in male (D, n=6, 7) and female (E, n=6, 7). *<0.05 vs. USF1WT. All data presented as mean±SEM.
Figure 2. Usf1 and Usf2 mRNA expression in tissues.
Expression of Usf1 (A–B) and Usf2 (C–D) in tissues from male (A, C) and female (B, D) USF1WT and USF1GT mice. Kidney (K), brain (B), and adipose tissues (reproductive adipose; RA, mesenteric adipose; MA, perirenal adipose; PA and brown adipose; BAT) of mice. Each n≥7, *<0.05 vs. USF1WT. All data presented as mean±SEM.
Regulation of hAGT as a USF target gene in vivo
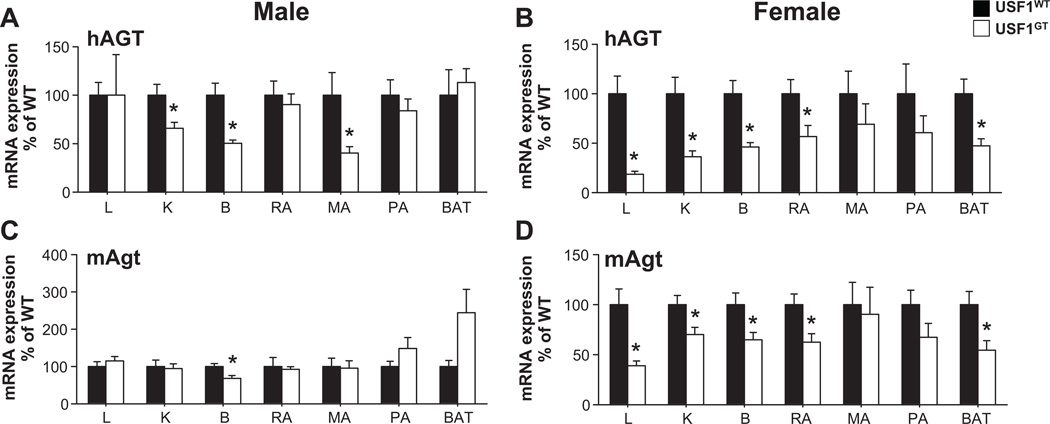
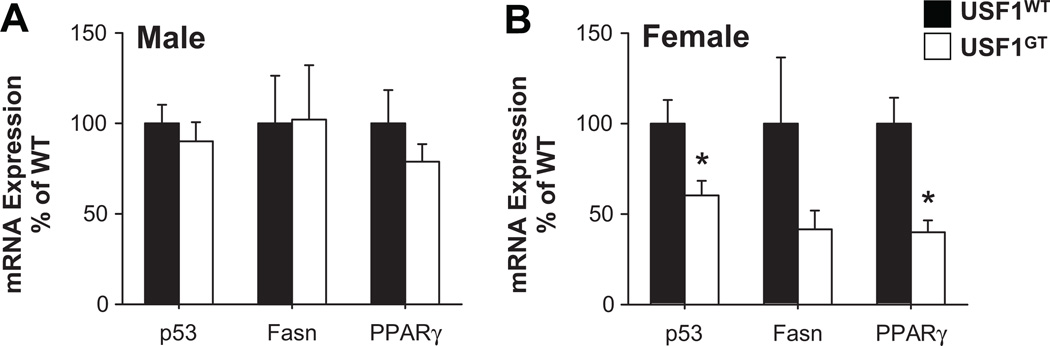
No changes in expression of hAGT mRNA were found in male USF1GT liver and reproductive adipose, which are known as major sites of AGT production, whereas hAGT mRNA expression in male USF1GT kidney, brain and mesenteric adipose tissue were decreased significantly (Figure 3A). In female, the expression of hAGT in most tissues including liver, kidney, brain and reproductive adipose tissue was decreased significantly (Figure 3B). The level of hAGT protein in USF1GT male plasma was unchanged (1.13±0.30 fold vs. WT), but was decreased significantly in plasma from female USF1GT mice (0.65±0.04 fold vs. WT). In males, the expression of endogenous mouse AGT (mAgt) in USF1GT was not changed in liver, kidney and reproductive adipose tissue, but a significant reduction of mAgt mRNA in brain from USF1GT mice was found (Figure 3C). In female USF1GT mice, mAgt mRNA expression was significantly decreased in liver, kidney, brain, reproductive and brown adipose tissue, consistent with hAGT expression patterns in USF1GT female (Figure 3D). The expression of p53, Fasn, PPARγ mRNA, known USF target genes, were not altered in liver from USF1GT males (Figure 4A), but were significantly lower in liver from USF1GT females (Figure 4B).
Figure 3. hAGT and mAgt mRNA expression in USF1GT liver.
Expression of hAGT (A–B) and mAgt (C–D) in tissues from male (A, C) and female (B, D) USF1WT and USF1GT mice. Liver (L), Kidney (K), brain (B), and adipose tissues (reproductive adipose; RA, mesenteric adipose; MA, perirenal adipose; PA and brown adipose; BAT) of mice. Each n≥7, *<0.05 vs. USF1WT. All data presented as mean±SEM.
Figure 4. USF target gene expression in USF1GT liver.
Expression of p53, Fasn, PPARγ in male (A) and female (B) USF1WT and USF1GT mice. Each n≥7, *<0.05 vs. USF1WT. All data presented as mean±SEM.
Blood Pressure in USF1GT mice
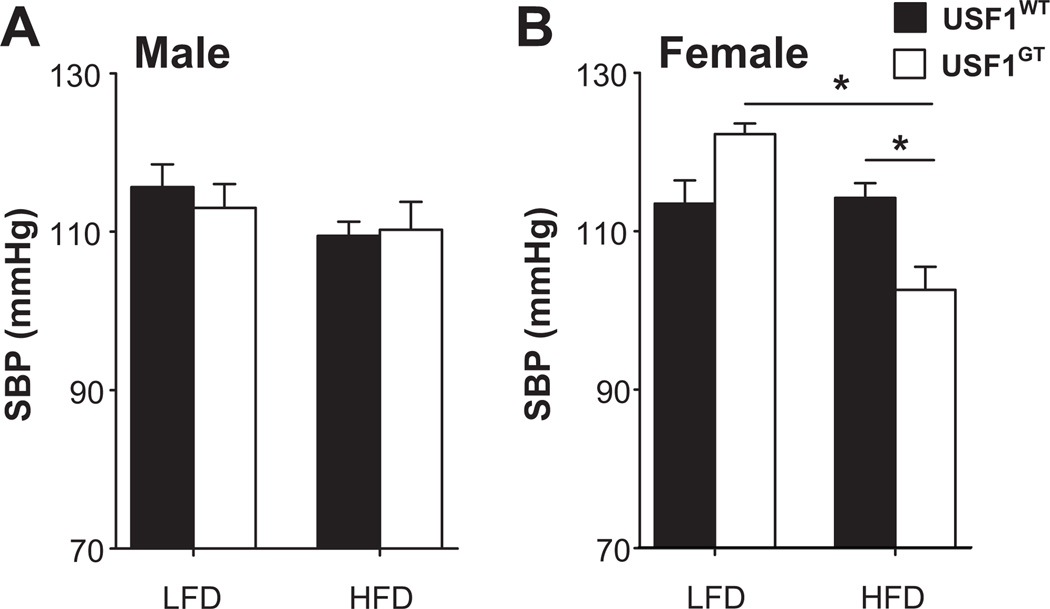
We next determined if the reduction in mAGT expression was sufficient lower BP in female USF1GT mice. We measured SBP in USF1GT mice lacking the hAGT transgene because of the known species-specificity of the renin-AGT enzymatic reaction (and the lack of human renin in the model).21 There was no significant difference in SBP in male or female USF1GT mice fed a low fat diet (Figure 5A). The trend for increased BP in females fed a low fat diet occur concomitantly with a modest but significant increase in plasma aldosterone (see Supplemental Table 1 in the online-only Data Supplement). In males, 6 weeks of high fat feeding increased body weight compared with mice fed a low fat diet (Table S1), but had no effect on SBP (Figure 5A). In females, high fat diet feeding increased body weight in both USF1WT and USF1GT(Table S1), but SBP was blunted in USF1GT mice (Figure 5B). There was no difference in plasma aldosterone in female mice fed a high fat diet (Table S1). Other parameters including food and water intake, plasma fasting glucose, and metabolic rate are presented in Table S1.
Figure 5. Systolic Blood Pressure in USF1GT mice.
SBP was measured by tail cuff in male (n=7–8) and female (n=6–8) USF1WT and USF1GT mice fed a low or high fat diet for 6 weeks starting at 6–8 weeks of age. *, P<0.05 as indicated by ANOVA.
Evidence for USF1 and 2 binding on hAGT promoter
To explore molecular mechanisms underlying the effects of USF1 knockdown in USF1GT liver, we first performed EMSA using nuclear extracts from liver and labeled hAGT promoter oligo probes (containing A or C at position −20, Figure S1A, online-only Data Supplement). One strong major shift product (closed arrowhead) along with one weak shift (open arrowhead) was observed in nuclear extracts from liver derived from USF1WT mice. Both shift products were competed by excess cold wildtype (WT) but not mutant (MUT) probe. Consistent with our previous observation in cultured cells,11 the gel shift signal using the −20C probe was stronger than the signal obtained with the −20A probe. It was initially interesting to note that the same pattern of gel shift products was observed in nuclear extracts from USF1WT and USF1GT liver. Therefore, we performed EMSA using antisera directed against USF1 (α1) and USF2 (α2) to identify the shifted products (Figure S1B). αUSF1 caused a weak supershift and strongly inhibited the formation of the major shift product from wildtype extracts suggesting this complex must contain USF1. This complex also likely contains USF2 because it is retained in USF1GT nuclear extract and supershifts with αUSF2. Consequently, these data suggest that the major product likely contains USF1/USF2 heterodimers, but may also contain USF2/USF2 homodimers, particularly when USF1 is depleted. This is consistent with the very close localization of the USF1/USF2 and USF2/USF2 products in nuclear extracts from a variety of AGT-expressing cells,11 and the observation that the αUSF1 antisera does not reduce or supershift the major product in extracts from USF1GT mice. The minor shifted product does not shift with αUSF1 and is retained in USF1GT suggesting it does not contain USF1. However, this product is effectively supershifted by αUSF2 suggesting it contains USF2. It remains unclear if it is in complex with some other protein. A similar pattern of shifted and super-shifted products was observed when the E-box from the p53 promoter, a known USF target gene, was used (Figure S1C).
Physiological binding between USF1 and the promoters in vivo
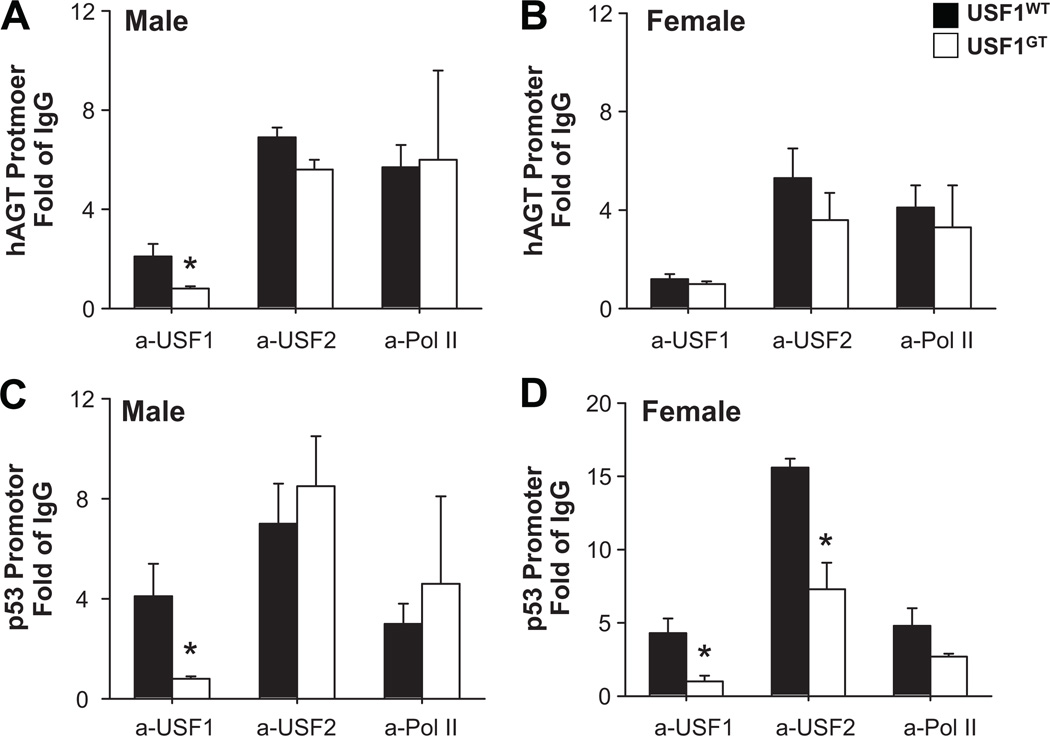
Quantitative ChIP was performed to assess the binding of USF1 and USF2 in chromatin surrounding the hAGT promoter from the liver of male and female mice. In males, a 60% reduction in USF1 binding to the −20 region was observed in USF1GT. USF2, which was more abundant than USF1 at the hAGT promoter was unchanged (Figure 6A). In female liver, the level of USF1 binding was much closer to the IgG background and thus a difference between USF1WT and USF1GT was not clearly evident (Figure 6B). USF2 binding was similarly unaffected (Figure 6B). Because of the low level of USF1 binding in the female mice, where the effect on hAGT was the largest, we repeated the quantitative PCR to query USF1 and USF2 binding to the p53 promoter. USF1 and USF2 have been previously reported to strongly bind to the E-box in the p53 promoter.22 In this experiment, a significant decrease in USF1 binding to the p53 promoter was observed in the liver of male and female mice. Moreover, a decrease in USF2 binding was detected in females (Figure 6D). This correlated with a decrease in p53 expression in the liver of female mice (Figure 4B). There was no change in the level of RNA polymerase II binding consistent with studies showing that the level of poised RNA polymerase II does not directly correlate with the level of transcription.23
Figure 6. Quantitative ChIP of USF1 and USF2 binding in liver chromatin from USF1WT and USF1GT mice.
Relative binding of USF1, USF2 and RNA Polymerase II to the hAGT (A–B) and p53 (C–D) promoters in liver chromatin from male (A, C) and female (B, D) USF1WT and USF1GT mice. N=3–4 in all experiments. *<0.05 vs. USF1WT. All data presented as mean±SEM.
USF2 knockdown by AdshUSF2 in vivo
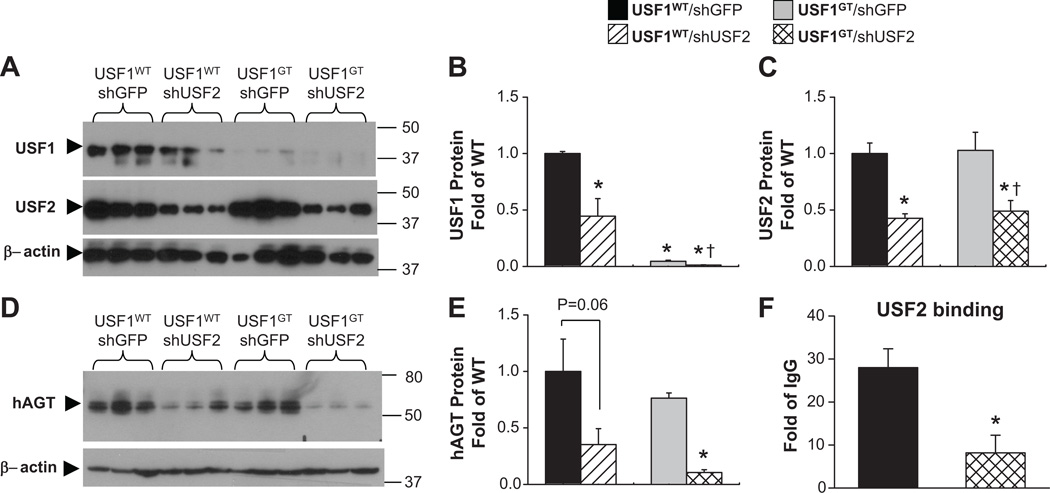
To dissect the mechanisms of USF-dependent regulation on hAGT in vivo, we used adenoviral delivery of shRNA targeting USF2 (AdshUSF2), previously validated to efficiently infect the liver.11 We asked if the preserved expression of hAGT in the liver of male USF1GT was due to retention of USF2 binding as suggested by the EMSA and ChIP studies. USF1WTand USF1GT male mice were treated intravenously with either an AdshUSF2 or a control AdshGFP-expressing virus. USF1 protein levels were reduced by 55% by shUSF2 in USFWT mice but were largely ablated in USF1GT mice (Figure 7A–B). USF2 protein levels in liver were significantly decreased in both USF1WT and USF1GT mice by AdshUSF2 delivery compared to mice infected with AdshGFP (Figures 7A, C). A significant reduction in hAGT protein was observed in both USF1WT and USF1GTby AdshUSF2, but the reduction was greater in mice lacking both USF1 and USF2 (Figures 7D–E). Quantitative ChIP using USF2 antisera confirmed the loss of USF2 binding to hAGT promoter in USF1GT infected with AdshUSF2 virus (Figure 7F).
Figure 7. Effect of intravenous AdshUSF2 on USF1, USF2, and hAGT protein in vivo.
AdshUSF2 or AdshGFP were injected to age matched USF1WTand USFGT male mice (each n≥3). A. Western blots showing USF1, USF2, and β-actin. Quantification of USF1 (B) and USF2 (C) protein expression. D. Western blots showing hAGT and β-actin. E. Quantitation of hAGT protein expression. F. Quantitative ChIP for USF2 binding on hAGT promoter. *<0.05 vs. USF1WT. All data presented as mean±SEM.
Discussion
Studies presented herein examined the relationship between USF1 and USF2 binding to the E-box sequence (CANNTG) overlying the −20 position in the promoter region of hAGT gene using a transgenic mouse expressing hAGT, gene trap mice disrupting USF1 expression, and adenoviruses expressing shRNAs targeting USF2. We conclude from these studies that USF1 and USF2 binding are essential for normal AGT transcriptional regulation, and that USF2 might be able to functionally compensate for loss of USF1, especially in males. Loss of both USF1 and USF2 lead to a significant decline in hepatic hAGT expression.
USF1 or USF2 null mutants have been previously reported.16,17USF1-null mice are viable and fertile and exhibit elevated levels of USF2, which may potentially compensate for the absence of USF1. USF1 and USF2 can homodimerize, but they preferentially heterodimerize in vivo and have similar trans-activating capacities.24 Moreover, different cell types have distinct cell-specific ratios of USF homodimers and heterodimers.16 Consequently, it is possible that USF2 homodimer formation became favored in mice lacking USF1. This is supported by the EMSA studies using antisera directed against USF2 and by the preservation of USF2 binding to the hAGT and p53 promoters in chromatin from the liver in USF1GT mice. Our data are consistent with the hypothesis that USF1-knockdown mice exhibit markedly lower levels of AGT mRNA expression in tissues only if both USF1 and USF2 are decreased simultaneously, such as observed in females or in males after shRNA-mediate ablation of USF2. Thus, USF2 may functionally compensate for decreased USF1 in males. This situation is similar to the regulation of L-type pyruvate kinase and Spot14 by glucose where residual USF2 homodimers can compensate for decreased USF1/2 heterodimers in USF1 knockout mice.25, 26
The effects of USF1-deficiency were much more pronounced in females than in males. Sexual dimorphism in the phenotype of USF1- and USF2-deficient mice has been reported previously. Abnormal brain development and epileptic seizures were observed primarily in female USF1 knockout mice; but on the contrary, the growth defects and shorter life span of USF2-deficient mice were more severe in males than females.16 Overexpression of human USF1 in the mouse effects metabolic traits with decreased body weight, adiposity and total triglyceride levels in males to a greater extent than in females.27 There were no differences in heat production in wildtype or USF1-deficient mice, nor were there differences between males and females in this study. Statistically significant associations of several USF1 polymorphisms with type 2 diabetes mellitus and LDL cholesterol were recently reported in women, but not in men.28 Polymorphisms in USF binding sites upstream of genes in the Familial Combined Hyperlipidemia (FCHL) pathway have been reported,8,29 and a significant correlation between USF1 polymorphism and hyperlipidemia phenotypes in families with hypertension and premature stroke was found in males.9,29 These studies suggest a gender-specific mechanism in the regulation of USF target gene expression in vivo. Similarly, it is well established that AGT gene expression is regulated by sex hormones. For example, estrogen increases AGT mRNA in the liver,30 whereas androgens can induce renal expression of AGT.31 Morgan et al. reported differential modulation of plasma AGT by the −20 polymorphism in healthy pregnancy.32 This modulation may be due to differential binding of the estrogen receptor in competition with other transcription factors to an estrogen receptor response element overlying −20 which affects the level estrogen-induced AGT promoter activity.33
Interestingly, USF1 polymorphisms were reported to be associated with the level of AGT expression in human fat biopsies,34 and we observed that knockdown of USF1 lead to decreased expression of AGT in several adipose tissue depots. Thus, polymorphisms which affect the activity of USF1 or USF2 may modulate the level of AGT in adipose tissues. Interestingly, transgenic studies suggest that adipose AGT can contribute to the circulating pool of AGT and thus can regulate arterial pressure.35 The status of the renin-angiotensin system can also have a profound effect on energy homeostasis and adiposity. Global knockout or pharmacological interference with renin, AGT, ACE, and AT1AR, all result in lower body mass, altered body composition, and/or abnormal adipose development.36–39 Interestingly, although there was no difference in SBP under baseline conditions, female USF1-deficient mice exhibited a decrease in SBP after high fat diet feeding. This is consistent with the observation that females exhibited larger decreases in AGT expression than male. It remains unclear if this is due to decrease AGT expression in the liver (i.e. circulation) or in adipose tissue.
USF proteins bind close to the transcription start site of many genes, and may be recruited to the transcriptional complex to modulate the state of chromatin.14,15 A USF1-dependent increase in transcriptional activity at the β-globin locus involves a complex between USF1, USF2 and chromatin modifying proteins, and is accompanied by localized increases in methylated histone 4 and acetylated histone 3 and histone 4.15 Recent advances in microarray and ChIP-sequencing technologies provide relevant information of transcriptional control, associated with common metabolic disorders. Profiles of USF binding in a liver cell line by ChIP-CHIP demonstrated that there is a positive correlation between binding and target gene expression levels.40 Proximal binding to the transcription start site was critical for the assembly of a transcription complex including transcriptional co-activators such as p300/CBP and enzymes involved in chromatin modification such as histone acetyltransferases PCAF and SRC-1 and arginine methyltransferases such as PRMT1. In our study, loss of USF1 and USF2 binding to the hAGT promoter in chromatin lead to a 90% reduction in hAGT expression suggesting that a similar recruitment of chromatin modifying enzymes may be required to activate hAGT transcription in response to USF binding. However, we did not observe any difference in the binding (using quantitative ChIP) of acetylated (164.8±11.7 vs. 168.12±1.8, fold of IgG) or methylated (1.3±0.1 vs. 1.3±0.1, fold of IgG) histone 3 (lysine 27, H3K27) to the hAGT promoter in AdshUSF2-treated USF1GT compared with liver from normal mice. Therefore, we must consider the possibility that the effects of USF1 and USF2 depletion on AGT expression are indirect. For example, genes potentially regulated by USF (e.g., transcription factors HNF4α and PPARγ) are involved in energy homeostasis which in turn can directly or indirectly regulate hAGT gene expression.
In conclusion, by examining the transcriptional regulation of hAGT gene, we have uncovered novel mechanisms regulating hAGT expression by transcription factors that bind to physiologically relevant promoter sequences (A-20C SNP). This sequence is significant because they overlie polymorphisms which are genetically associated with hypertension and other cardiovascular and metabolic phenotypes. A decrease in USF1, but not USF2 binding was observed in USF1GT mice, suggesting that USF2 might be able to functionally compensate for loss of USF1, especially in males. We provide evidence that the retention of USF2 was critical for regulation of USF target gene regulation.
Perspectives
Essential hypertension is influenced by both genetic and environmental factors. The angiotensinogen (AGT) gene has long been studied as a candidate gene in hypertension, but numerous studies have failed to elucidate the mechanisms by which polymorphisms in AGT cause hypertension. In our study, both USF1 and USF2 play an important role in human AGT regulation, and distinct gender-specific mechanisms are involved in the regulation of their expression in vivo. We hypothesize that the −20C polymorphism results in increased USF recruitment to the AGT promoter, which results in increased AGT expression. This elevation in AGT expression may cause increases in blood pressure and may have metabolic effects.
Supplementary Material
Novelty and Significance.
1) What is New?
Use of a compound transgenic model expressing the −20C allele of human AGT on a USF1-deficient genetic background.
Expression of mouse and human AGT in females in dependent upon USF1.
Expression of mouse and human AGT in males is dependent upon both USF1 and USF2.
2) What is Relevant?
USF1 and USF2 bind to a region of the human AGT promoter which carries a single nucleotide polymorphism which has been genetically associated with hypertension.
The contribution of USF1 and USF2 on human AGT expression and consequently on blood pressure may differ between males and females.
3) Summary
The E-box transcription factors USF1 and USF2 binding are essential for transcriptional regulation of the AGT gene and may mechanistically underlie differential expression of the −20A and −20C alleles.
Acknowledgements
We thank Drs. Justin Grobe, Henry Keen, and Eric Weatherford for generous discussion and review of the manuscript. Transgenic mice were generated at the University of Iowa Transgenic Animal Facility supported in part by grants from the NIH and from the Roy J. and Lucille A. Carver College of Medicine. We thank Norma Sinclair, JoAnne Schwarting, and Patricia Yarolem for genotyping mice.
Sources of Funding: This work was supported through research grants from the NIH to CDS (HL084207, HL048058, and HL061446). The authors also gratefully acknowledge the generous research support of the Roy J. Carver Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None.
References
- 1.Kim HS, Krege JH, Kluckman KD, Hagaman JR, Hodgin JB, Best CF, Jennette JC, Coffman TM, Maeda N, Smithies O. Genetic-Control of Blood-Pressure and the Angiotensinogen Locus. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2735–2739. doi: 10.1073/pnas.92.7.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardes J, Bouhnik J, Clauser E, Corvol P, Menard J. Role of Angiotensinogen in Blood-Pressure Homeostasis. Hypertension. 1982;4:185–189. doi: 10.1161/01.hyp.4.2.185. [DOI] [PubMed] [Google Scholar]
- 3.Menard J, Elamrani AIK, Savoie F, Bouhnik J. Angiotensinogen - An Attractive and Underrated Participant in Hypertension and Inflammation. Hypertension. 1991;18:705–706. doi: 10.1161/01.hyp.18.5.705. [DOI] [PubMed] [Google Scholar]
- 4.Dickson ME, Sigmund CD. Genetic basis of hypertension - Revisiting angiotensinogen. Hypertension. 2006;48:14–20. doi: 10.1161/01.HYP.0000227932.13687.60. [DOI] [PubMed] [Google Scholar]
- 5.Dickson ME, Zimmerman MB, Rahmouni K, Sigmund CD. The-20 and-217 promoter variants dominate differential angiotensinogen haplotype regulation in angiotensinogen-expressing cells. Hypertension. 2007;49:631–639. doi: 10.1161/01.HYP.0000254350.62876.b1. [DOI] [PubMed] [Google Scholar]
- 6.Grobe JL, Dickson ME, Park S, Davis DR, Born EJ, Sigmund CD. Cardiovascular consequences of genetic variation at −6/235 in human angiotensinogen using "humanized" gene-targeted mice. Hypertension. 2010;56:981–987. doi: 10.1161/HYPERTENSIONAHA.110.157354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiago AD, Samani NJ, Candy GP, Brooksbank R, Libhaber EN, Sareli P, Woodiwiss AJ, Norton GR. Angiotensinogen gene promoter region variant modifies body size-ambulatory blood pressure relations in hypertension. Circulation. 2002;106:1483–1487. doi: 10.1161/01.cir.0000029093.93362.fc. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins PN, Heiss G, Ellison RC, Province MA, Pankow JS, Eckfeldt JH, Hunt SC. Coronary artery disease risk in familial combined - Hyperlipidemia and familial hypertriglyceridemia - A case-control comparison from the National Heart, Lung, and Blood Institute Family Heart Study. Circulation. 2003;108:519–523. doi: 10.1161/01.CIR.0000081777.17879.85. [DOI] [PubMed] [Google Scholar]
- 9.Coon H, Xin YP, Hopkins PN, Cawthon RM, Hasstedt SJ, Hunt SC. Upstream stimulatory factor 1 associated with familial combined hyperlipidemia, LDL cholesterol, and triglycerides. Human Genetics. 2005;117:444–451. doi: 10.1007/s00439-005-1340-x. [DOI] [PubMed] [Google Scholar]
- 10.Hilgers KF, Veelken R, Muller DN, Kohler H, Hartner A, Botkin SR, Stumpf C, Schmieder RE, Gomez RA. Renin uptake by the endothelium mediates vascular angiotensin formation. Hypertension. 2001;38:243–248. doi: 10.1161/01.hyp.38.2.243. [DOI] [PubMed] [Google Scholar]
- 11.Dickson ME, Tian X, Liu XB, Davis DR, Sigmund CD. Upstream Stimulatory Factor Is Required for Human Angiotensinogen Expression and Differential Regulation by the A-20C Polymorphism. Circulation Research. 2008;103 doi: 10.1161/CIRCRESAHA.108.180653. 940-U83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarzani R, Bordicchia M, Marcucci P, Minardi D, Muzzonigro G, Dessi-Fulgheri P, Rappelli A. Angiotensinogen promoter variants influence gene expression in human kidney and visceral adipose tissue. Journal of Human Hypertension. 2010;24:213–219. doi: 10.1038/jhh.2009.48. [DOI] [PubMed] [Google Scholar]
- 13.Komulainen K, Alanne M, Auro K, Kilpikari R, Pajukanta P, Saarela J, Ellonen P, Salminen K, Kulathinal S, Kuulasmaa K, Silander K, Salomaa V, Perola M, Peltonen L. Risk alleles of USF1 gene predict cardiovascular disease of women in two prospective studies. Plos Genetics. 2006;2:672–681. doi: 10.1371/journal.pgen.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West AG, Huang SM, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histione modifications by USF proteins at a vertebrate barrier element. Molecular Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Molecular and Cellular Biology. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirito M, Lin Q, Maity T, Sawadogo M. Ubiquitous Expression of the 43-Kda and 44-Kda Forms of Transcription Factor Usf in Mammalian-Cells. Nucleic Acids Research. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casado M, Vallet VS, Kahn A, Vaulont S. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J Biol Chem. 1999;274:2009–2013. doi: 10.1074/jbc.274.4.2009. [DOI] [PubMed] [Google Scholar]
- 18.Yang G, Merrill DC, Thompson MW, Robillard JE, Sigmund CD. Functional expression of the human angiotensinogen gene in transgenic mice. J Biol Chem. 1994;269:32497–32502. [PubMed] [Google Scholar]
- 19.Sinn PL, Davis DR, Sigmund CD. Highly regulated cell type-restricted expression of human renin in mice containing 140- or 160-kilobase pair P1 phage artificial chromosome transgenes. J Biol Chem. 1999;274:35785–35793. doi: 10.1074/jbc.274.50.35785. [DOI] [PubMed] [Google Scholar]
- 20.Lin Q, Luo X, Sawadogo M. Archaic Structure of the Gene Encoding Transcription Factor Usf. J Biol Chem. 1994;269:23894–23903. [PubMed] [Google Scholar]
- 21.Hatae T, Takimoto E, Murakami K, Fukamizu A. Comparative studies on species-specific reactivity between renin and angiotensinogen. Mol Cell Biochem. 1994;131:43–47. doi: 10.1007/BF01075723. [DOI] [PubMed] [Google Scholar]
- 22.Wong RHF, Chang I, Hudak CSS, Hyun S, Kwan HY, Sul HS. A Role of DNA-PK for the Metabolic Gene Regulation in Response to Insulin. Cell. 2009;136:1056–1072. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viollet B, LefrancoisMartinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 25.Vallet VS, Henrion AA, Bucchini D, Casado M, Raymondjean M, Kahn A, Vaulont S. Glucose-dependent liver gene expression in upstream stimulatory factor 2 −/− mice. J Biol Chem. 1997;272:21944–21949. doi: 10.1074/jbc.272.35.21944. [DOI] [PubMed] [Google Scholar]
- 26.Vallet VS, Casado M, Henrion AA, Bucchini D, Raymondjean M, Kahn A, Vaulont S. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. Journal of Biological Chemistry. 1998;273:20175–20179. doi: 10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 27.Wu S, Mar-Heyming R, Dugum EZ, Kolaitis NA, Qi H, Pajukanta P, Castellani LW, Lusis AJ, Drake TA. Upstream transcription factor 1 influences plasma lipid and metabolic traits in mice. Hum Mol Genet. 2010;19:597–608. doi: 10.1093/hmg/ddp526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holzapfel C, Baumert J, Grallert H, Muller AM, Thorand B, Khuseyinova N, Herder C, Meisinger C, Hauner H, Wichmann HE, Koenig W, Illig T, Klopp N. Genetic variants in the USF1 gene are associated with low-density lipoprotein cholesterol levels and incident type 2 diabetes mellitus in women: results from the MONICA/KORA Augsburg case-cohort study, 1984–2002. Eur J Endocrinol. 2008;159:407–416. doi: 10.1530/EJE-08-0356. [DOI] [PubMed] [Google Scholar]
- 29.Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XQJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1) Nature Genetics. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- 30.Stavreus-Evers A, Parini P, Freyschuss B, Elger W, Reddersen G, Sahlin L, Eriksson H. Estrogenic influence on the regulation of hepatic estrogen receptor-alpha and serum level of angiotensinogen in female rats. J Steroid Biochem Mol Biol. 2001;78:83–88. doi: 10.1016/s0960-0760(01)00077-2. [DOI] [PubMed] [Google Scholar]
- 31.Ding Y, Stec DE, Sigmund CD. Genetic evidence that lethality in angiotensinogen-deficient mice is due to loss of systemic but not renal angiotensinogen. J Biol Chem. 2001;276:7431–7436. doi: 10.1074/jbc.M003892200. [DOI] [PubMed] [Google Scholar]
- 32.Morgan L, Crawshaw S, Baker PN, Broughton PF, Kalsheker N. Polymorphism in oestrogen response element associated with variation in plasma angiotensinogen concentrations in healthy pregnant women. J Hypertens. 2000;18:553–557. doi: 10.1097/00004872-200018050-00007. [DOI] [PubMed] [Google Scholar]
- 33.Narayanan CS, Cui Y, Zhao YY, Zhou J, Kumar A. Orphan receptor Arp-1 binds to the nucleotide sequence located between TATA box and transcriptional initiation site of the human angiotensinogen gene and reduces estrogen induced promoter activity. Mol Cell Endocrinol. 1999;148:79–86. doi: 10.1016/s0303-7207(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 34.Naukkarinen J, Gentile M, Soro-Paavonen A, Saarela J, Koistinen HA, Pajukanta P, Taskinen MR, Peltonen L. USF1 and dyslipidemias: converging evidence for a functional intronic variant. Human Molecular Genetics. 2005;14:2595–2605. doi: 10.1093/hmg/ddi294. [DOI] [PubMed] [Google Scholar]
- 35.Batifoulier-Yiannikouris F, Karounos M, Charnigo R, English VL, Rateri DL, Daugherty A, Cassis LA. Adipocyte-specific deficiency of angiotensinogen decreases plasma angiotensinogen concentration and systolic blood pressure in mice. Am J Physiol Regul Integr Comp Physiol. 2012;302:R244–R251. doi: 10.1152/ajpregu.00323.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanai K, Saito T, Kakinuma Y, Kon Y, Hirota K, Taniguchi-Yanai K, Nishijo N, Shigematsu Y, Horiguchi H, Kasuya Y, Sugiyama F, Yagami K, Murakami K, Fukamizu A. Renin-dependent cardiovascular functions and renin-independent blood-brain barrier functions revealed by renin-deficient mice. J Biol Chem. 2000;275:5–8. doi: 10.1074/jbc.275.1.5. [DOI] [PubMed] [Google Scholar]
- 37.Maeda K, Hata R, Bader M, Walther T, Hossmann KA. Larger anastomoses in angiotensinogen-knockout mice attenuate early metabolic disturbances after middle cerebral artery occlusion. J Cereb Blood Flow Metab. 1999;19:1092–1098. doi: 10.1097/00004647-199910000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Xiao HD, Fuchs S, Frenzel K, Cole JM, Bernstein KE. Newer approaches to genetic modeling in mice: tissue-specific protein expression as studied using angiotensin-converting enzyme (ACE) Am J Pathol. 2003;163:807–817. doi: 10.1016/S0002-9440(10)63441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliverio MI, Madsen K, Best CF, Ito M, Maeda N, Smithies O, Coffman TM. Renal growth and development in mice lacking AT1A receptors for angiotensin II. Am J Physiol. 1998;274:F43–F50. doi: 10.1152/ajprenal.1998.274.1.F43. [DOI] [PubMed] [Google Scholar]
- 40.Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.