Abstract
Significance: Epigenetics involves alterations in gene expression without changing the nucleotide sequence. Because some epigenetic changes can be reversed chemically, epigenetics has tremendous implications for disease intervention and treatment. Recent Advances: After epigenetic components in cancer were characterized, genes and pathways are being characterized in other diseases such as diabetes, obesity, and neurological disorders. Observational, experimental, and clinical studies in different diseases have shown that nutrients influence epigenetic regulation. Nutrients such as folic acid that supply methyl groups have been shown to have a protective effect in colon cancer. Critical Issues: Identifying steps during epigenetic regulation and developing intervention and treatment agents are the critical issues in the field. Future Directions: Following completion and validation of key observational studies in nutritional epigenetics, strategies can be developed for cancer control and treatment. Antioxid. Redox Signal. 17, 355–364.
Introduction: Why Nutritional Factors Are Important for Cancer Prevention, and Possible Mechanisms of Action
Cancer is both a genetic and an epigenetic disease (21, 80). Epigenetics involves altered gene expression without any change of gene sequences (2). Several regulatory proteins involved in epigenetics include DNA methyltransferases, methyl-CpG binding proteins, histone modifying enzymes (histone acetyl transferase and histone deacetylase), Polycomb group (PcG) proteins, and chromatin remodeling factors and their multimolecular complexes (20). A number of diseases including cancer are regulated epigenetically (9, 10, 39, 56, 61, 70).
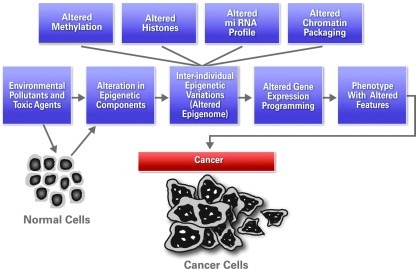
Both genetic and epigenetic events are susceptible to environmental and lifestyle factors such as diet, radiation, exposure to toxins and pollutants, pharmacological intervention agents, and infectious agents (32, 76, 79) (Fig. 1). Unlike behavior or stress, diet is one of the more easily studied and therefore better understood environmental factors in epigenetic change. Alterations in genetic and epigenetic factors can affect the phenotype of cells and organisms.
FIG. 1.
Factors contributing to carcinogenesis. Both genetic and epigenetic regulation of gene expression contributes in cancer development. Factors mentioned here may work independently or in combination. Some factors affect DNA, whereas others affect proteins and nucleic acids simultaneously. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Dietary components play a major role in both cancer prevention and development (12, 66, 67). Well-characterized bioactive food components include tea polyphenol-catechins (green tea), curcumin (turmeric), genistein (soybean), resveratrol (grapes), sulforaphane (SFN, cruciferous vegetables), and other bioactive components such as apigenin (parsley), baicalein (Indian trumpet), cyanidins (grapes), isothiocyanate (cruciferous vegetables), rosmarinic acid (rosemary), and silymarin (milk thistle) (Fig. 2) (55). Intake of certain bioactive food components can modulate cancer risk and tumor development (36). The quantity of food components, frequency of intake, and duration of intake by individuals also play a significant role in cancer development (54).
FIG. 2.
Natural food components with epigenome-altering properties. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Altered diet may have transgenerational effects. Heijmans et al. (28, 29) studied pregnant mothers during the Dutch Hunger Winter of 1944 to 1945 (a severe wartime famine at the end of World War II that affected the western part of the Netherlands). The investigators followed the methylation profiles of the mothers' offspring six decades later and compared them with the profiles of their unexposed, same-sex siblings. The results indicated hypomethylation of insulin-like growth factor 2 (IGF2) and hypermethylation of interleukin-10 (IL-10), LEP, ABCA1, and MEGF. The researchers also observed an association between methylation and gestational time of exposure in 60 individuals at 15 loci (28, 29, 71, 72). Blood samples were used in these studies to analyze methylation profiles. All of these studies indicated the significance of nutritional factors in the development of diseases including cancer.
Several food components can alter tumor cell behavior and cancer risk by influencing key pathways and steps in carcinogenesis, including hormonal regulation, cell signaling, cell cycle control, apoptosis, differentiation, carcinogen metabolism, and/or inflammation (34, 54, 64). Polyphenols (e.g., genistein, which is present in soybeans; and resveratrol, which is present in grapes and peanuts) are known to repress the expression of androgen receptor (AR) (44); reduced levels of AR lower the level of prostate-specific antigen and modulate proliferating cell nuclear antigen, p21, and p27 (30, 59). Indole-3-carbinol (which is abundant in cruciferous vegetables) can inhibit cellular proliferation in human breast cancer cells (73). Xenobiotic compounds, including tobacco-specific carcinogens known to induce lung tumors, can occur following dietary exposure to isothiocyanates from cruciferous vegetables by activating detoxifying phase II enzymes (86). Flaxseed or fish oil (which are rich in n-3 fatty acids) can suppress proinflammatory cytokines, including tumor necrosis factor-α and IL-1-β, which have been linked to increased colon cancer risk (4, 84). These examples demonstrate that some nutritional components show their maximum activity in specific organs, and their targets can be specific as well. Not all tissues respond equally to bioactive food components, as discussed below. Genistein shows specificity for the receptor type; it competes more strongly with estrogen to bind to estrogen receptor-beta (ER-β) than to estrogen receptor-alpha (ER-α) (52, 88). ER-β is expressed extensively in the prostate but to a much lower degree in the colon. Similarly, the effectiveness of genistein against ER-β-mediated processes may be greater in the prostate than in the colon. Zinc deficiency has been reported to be more of a factor in the development of cancer at some organ sites, such as the esophagus, than at others (27, 83). Further mechanistic studies should consider tissue specificity as a parameter in the overall bioefficacy of a specific food component.
Epigenetics and Cancer
During the last decade, advancements were made in understanding the epigenetic regulation of cancer development and identifying modifiable and host factors that contribute to disease development (21, 41, 77, 78, 89). The following section describes major players in epigenetic regulation and key components of the epigenetic machinery.
Components of the epigenetic machinery
The components of the epigenetic machinery are DNA methylation, histone modifications, noncoding RNAs, selected nonhistone proteins, and imprinting (38, 89). The most studied component is methylation. A brief description of each epigenetic component follows.
DNA methylation
DNA methylation involves chemical modifications to the cytosine base in DNA (also called 5-methylcytosine), predominantly in the context of a cytosine-guanine (CpG) dinucleotide (5, 40, 41). Other forms have been identified recently, such as 5′-hydroxymethylcytosine, although the biological significance of this mark has yet to be determined. Approximately 60% of human genes contain a high density of CpG dinucleotides, known as CpG islands. The current dogma describes DNA methylation at the promoter of a gene as a sign of silenced gene expression. It has become apparent over more than a decade that promoter DNA hypermethylation is one of the most common somatic aberrations in cancer development. Animal models that produce characteristics of liver cancer by altering dietary components without using any mutagen or radiation exposure are well established for studying epigenetic influences on liver cancer (49, 62, 63). Dietary selenium also has been shown to affect methylation levels in animal models of cancer (14, 15). DNA was hypomethylated in the liver and colon of animals fed a selenium-deficient diet. These studies demonstrated that the quantity of food components and timing of feeding are both equally important.
Histone modifications
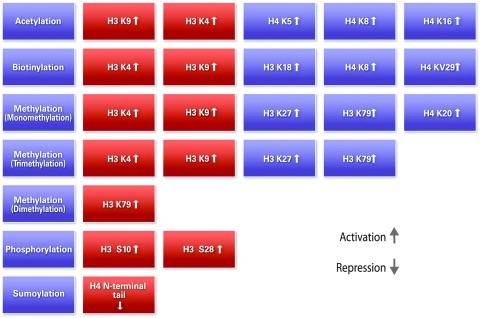
Mammalian DNA is packaged into chromosomes by wrapping the DNA around nucleosomes made up of an octamer of histone proteins. Each of the different histone types has N-terminal protein tails that extend outside of the nucleosome and can be modified by several large families of enzymes (81). Different histone modifications (e.g., acetylation, biotinylation, methylation, phosphorylation, and sumoylation) mark either active (euchromatin) or inactive (heterochromatin) chromatin and define the chromosomal structure and gene expression state of the genes within that chromosomal domain. For example, H3K4me3 has been associated with active transcription, whereas H3K9me3 has been associated with gene repression. The dimeric H3 and H4 form a tetramer, whereas H2A and H2B remain as a dimer. It has been observed that specific histone modifications, namely global loss of acetylation of K16 and trimethylation of K20 of histone H4, are hallmarks of human cancers (22). In addition, the level of histone modifications has been associated with tumor size. More specifically, low levels of H4R3me2, H3K9ac, and H4K16ac were associated with large tumor size and vascular invasion (H4K16ac) (19). Reduced levels of histone modifications such as lysine acetylation (H3K9ac, H3K18ac, and H4K12ac), lysine methylation (H3K4me2 and H4K20me3), and arginine methylation (H4R3me2) were associated with tumors with a poor prognosis, for example, basal-like tumors and HER-2-positive tumors. Different histone modifications and their functions are shown in Figure 3.
FIG. 3.
Histone modifications and their functions. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
miRNA profiling
Epigenetic regulation by RNA-based mechanisms can occur at both the posttranscriptional level and at the level of chromatin. These mechanisms are mediated by small noncoding RNAs, which can induce DNA methylation or histone modifications that result in silenced or enhanced gene expression. Posttranscriptional regulation of gene expression by miRNAs also serves as an important element of the epigenome (8, 25). The involvement of miRNAs in cancer is seen in their essential role of tumor cell differentiation and tumor development. For example, miR-21 was shown to be upregulated in glioblastoma (GBM), impacting the expression of downstream targets such as p53, TGF-β, TIMP3, and PTEN. Another miRNA upregulated in GBM is miR-26a, which is involved in GBM development and proliferation. Downregulated miRs include miR-124, a cell cycle regulator and neuronal differentiator; miR-128, a glioma-proliferation inhibitor; and miR-451, an invasion inhibitor. High-throughput technologies are used to follow miRNA profiling in clinical samples.
Chromatin structure
Chromatin, which is composed of nucleosomes, is the key component of epigenome. Nucleosomes are comprised of histone proteins arranged as octamers associated with 146 bp of DNA via its negatively charged phosphate backbone. The compressed and relaxed state of chromatin affects gene expression.
Imprinting
Paternal and maternal alleles are regulated differently by imprinting. The most studied gene is IGF2 (13, 31). This gene is expressed from the paternal allele and is located within a cluster of imprinted genes on chromosome 11p15. Hypomethylation at a differentially methylated region of IGF2, differentially methylated region 0, is associated with IGF2 loss of imprinting in Wilm's tumor. The maternal allele also is unmethylated in this tumor. As a result, biallelic expression is observed (85).
PcG proteins
The chromatin-associated PcG proteins are needed for accurate axial body patterning during embryonic development (33, 47). PcG proteins silence genes by epigenetic mechanisms (26, 60). The Hox gene has been studied extensively. PcG proteins maintain the silent state of developmentally important genes. The role of miRNAs in targeting PcG proteins to chromatin also has been proposed (69). Few PcG proteins are expressed abnormally in different tumors (47).
Repetitive regions, such as LINE and Alu, are hypermethylated in the normal state and hypomethylated during growth and development. This process prevents chromosomal instability, translocation, and gene disruption caused by the activation of transposons (2, 20).
Models in nutritional epigenetics: folic acid deficiency and cancer
Folate, a cofactor, acts as a carrier of the methyl group and is involved in the generation of S-adenosyl methionine (SAM). In turn, SAM becomes the primary donor of methyl groups for the hypermethylation of genes. Furthermore, dietary factors (e.g., vitamin B6, vitamin B12, and Zn) that feed into the folate cycle have an effect on methyl group availability (66, 67). In animal models, choline or methionine restriction during pregnancy results in hypomethylation of genes involved in brain development (37, 87). Models have been used to follow disease development as a result of over- or undernutrition in utero. For example, vascular disease was developed in rats for two generations when mothers were undernourished (1).
Interaction of bioactive food components with epigenetic components
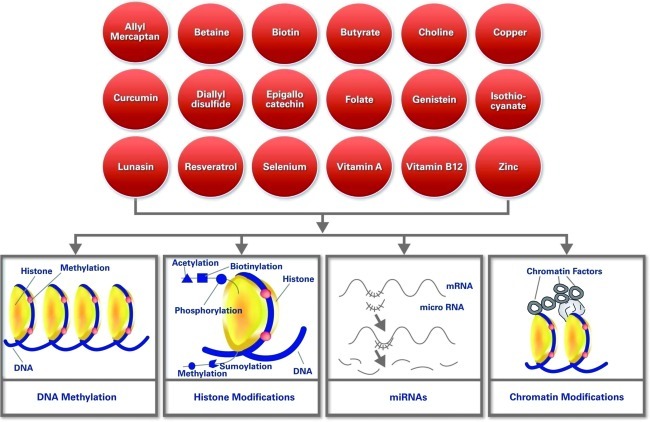
Dietary polyphenols from green tea, turmeric, soybeans, broccoli, and other natural resources have multiple cell-regulatory activities within cancer cells (32). Some dietary polyphenols may exert their chemopreventive effects in part by modulating various components of the epigenetic machinery in humans. The major compound in green tea is catechins, which include (−)-epicatechin (EC), (−)-epicatechin-3-gallate (ECG), (−)-epigallocatechin (EGC), and (−)-epigallocatechin-3-gallate (EGCG). EGCG has been identified as the most effective constituent among all tea bioactive food components. EGCG induces apoptosis and cell cycle arrest in many cancer cells. Another mechanism by which EGCG acts is the inhibition of DNA Methyl Transferase 1 (DNMT1), leading to demethylation and reactivation of methylation-silenced genes. Treatment of human esophageal cells with EGCG has been shown to reduce DNMT1 activity via hypomethylation and re-expression of genes such as p16, retinoic acid receptor β (RARβ), O6-methylguanine methyltransferase (MGMT), and human mutL homologue 1 (hMLH1). In addition, it has been reported that consumption of polyphenols could lead to a decrease in available SAM and an increase in S-adenosyl-L-homocysteine and homocysteine levels, which may affect methylation patterns (14, 55). Few dietary components have been characterized in terms of their involvement via epigenetic regulation and are shown in Figure 4.
FIG. 4.
Dietary components and their interaction with epigenetic regulation. The top part of the figure shows different components that have been reported to interact with epigenetic components. At the bottom part of the figure, major epigenetic components are shown. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Dietary polyphenols and modulation/intervention of epigenetic changes associated with cancer
Tea is the second most consumed beverage worldwide, and tea polyphenols can regulate gene expression by epigenetic mechanisms. The most common compounds in tea are catechins, including (−)-(EC), (−)-ECG, (−)-EGC, and (−)-EGCG. Among these catechins, EGCG accounts for one-half of the polyphenols in green tea (55) that have DNMT1-inhibitory properties. This results in the demethylation of silenced genes and prevention of cancer.
Sirtuins and epigenetic changes
Sirtuins are a group of proteins found in humans that have deacetylase properties (23). In addition, sirtuins are a family of nicotinamide adenine dinucleotide (NAD+)–dependent deacetylases, and they have monoribosyltransferase activity. Sirtuins affect aging, transcription, apoptosis, energy efficiency, and alertness during caloric restriction and stress resistance (43, 65). Seven types of sirtuins have been reported to date (SIRT-1 through SIR-7), and they are localized either in the nucleus or mitochondria. Resveratrol, a potential activator of SIRT-1, is present in plants and has anticancer, antiinflammatory, blood sugar–lowering, and other beneficial cardiovascular effects (3, 75).
One-carbon metabolism
Folate is a water-soluble B vitamin and, in its synthetic form, is called folic acid (74). A number of fortified foods contain folic acid. Its cancer prevention properties have been observed in different cancers, especially colorectal cancer, if a balanced amount of folic acid is consumed (6, 17, 18). Folates, present in high concentrations in green leafy vegetables, maintain DNA stability through their ability to donate one-carbon units for cellular metabolism. Mammals cannot synthesize folate de novo; therefore, they get it either from natural foods (green leafy vegetables), supplemented foods, or from microbial breakdown during digestion (35). The methionine cycle starts from 5-methyltetrahydrofolate, which remethylates homocysteine to methionine. In the next step, methionine is metabolized to SAM, which controls transcription and protein expression due to its ability to methylate cytosine in DNA. Folate deficiency may contribute to carcinogenesis by altering the processes just described. Mitochondrial DNA stability and mitochondrial functions also are affected by dietary folate status (7, 11, 82).
Early exposure and cancer risk (maternal nutrition and placental development)
Genetic susceptibility, environmental factors, age, and microenvironment all play a significant role in the development of cancer. Fewer studies have been conducted on the effects of these factors on the risk of cancer for pregnant women and fetuses. When mother is exposed to adverse conditions, the fetal nutrition may cause alterations in structure, physiology, and metabolism that predispose individuals to metabolic, endocrine, and cardiovascular diseases as adults. Because of the ethical issues involved, most of these studies have been observational studies only.
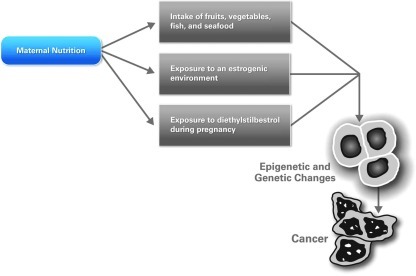
Selected dietary components consumed during early pregnancy may influence postnatal risk of cancer development, although all dietary components are not harmful (32, 35). In those cases where adverse effects on fetal development were observed, a proposed mechanism includes methylation of genes due to dietary food components in the mother's diet (14) (Fig. 5). Both hypermethylation and hypomethylation of selected genes were observed. Genes that were overexpressed included Klf6, Klf9, Nid2, Ntn4, Per1, and Txnip, and genes that were repressed included Bcar3, Cldn12, Csf1, Jag1, Lgals3, Lypd3, Nme1, Ptges2, Ptgs1, and Smarcb1. In animal models, deficiencies of macronutrients during placental growth have been shown to affect fetal growth (58, 68). Most of the genes that contribute to reduced fetal growth are regulated by imprinting, and the maternal allele is affected in these cases. Functionally, the nutrient transport from mother to fetus via the placenta is affected dramatically by the hypomethylation of genes in the embryonic trophectoderm. Note that undernutrition due to a limited food supply, severe nausea and vomiting, early or closely spaced pregnancies, multiple pregnancies, and placental dysfunction is quite common in some populations (48).
FIG. 5.
Cancer risk due to maternal diet. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Examples of nutritional epigenetics and cancer (nutritional intervention and epigenetic mechanisms)
Most of the clinical samples used for assessing the risk of and diagnosing cancer are from either blood or tissues, making it convenient to identify peripheral blood-based biomarkers for disease risk and prognosis. Researchers should make use of the many peripheral blood resource collections from around the world that have sample sizes that are appropriate for these types of investigation (46). Several studies have provided evidence for epigenetic traits in blood as potential cancer risk markers (45). As a first step to understanding the normal pattern of DNA methylation, a comprehensive map was constructed on a single Asian individual, at single base pair resolution using the latest high-throughput sequencing-based approaches (47). Because this was done for the first time on a human genome sequence, it can serve as a first reference. However, the mapping of many more individuals will be needed to provide a comprehensive map of normal epigenetic variation with which to compare cancer-specific epigenetic traits.
Epigenetic inhibitors from natural products
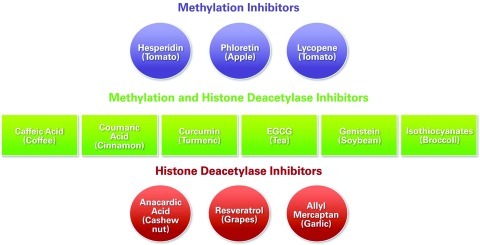
Several natural nutrient products have interesting biological properties and structural diversity; such products often are leading drug candidates for the treatment of cancer and other diseases. Epigenetic inhibitors (and activators of different steps) and their natural sources are described here. Parsley's apigenin is a DNMT inhibitor; ginger's allyl mercaptan is an Histone De-Acetylase (HDAC) inhibitor; turmeric's curcumin is a DNMT, HAT, and HDAC inhibitor; green tea's EGCG is a Histone Acetyl Transferase (HAT) and DNMT inhibitor; the soybean's genistein is a DNMT and HDAC inhibitor and an HDAC activator; lycopene, from tomatoes, is a demethylase; resveratrol, from grapes, is a DNMT inhibitor and an SIRT-1 activator; milk thistle's silymarin is an SIRI1 activator; and sulforaphane from cruciferous vegetables is an HDAC and DNMT1 inhibitor (14). Zeste homolog 2 gene, EZH2, is overexpressed in different cancers including breast cancer, and PcG protein levels can influence EZH2 activity. Recently, Dimri et al. (16) demonstrated that dietary omega-3 polyunsaturated fatty acids can regulate EZH2 expression in breast cancer by reducing the levels of PcG proteins by posttranslational modifications. Note that histone modifications triggered by Polycomb repressor complex signaling play an important role during embryonic stem cell differentiation; alterations in normal patterns may contribute to different diseases.
Policies for determining nutrition supplement levels and implementing potential cancer prevention and therapy techniques
In nutritional epigenetics, the duration of exposure to bioactive food components should demonstrate a physiologically relevant change in a molecular target involved with cancer prevention. Investigators routinely use high-throughput screening technologies such as microarray analyses to examine the global effects of bioactive food components on gene expression patterns. These studies include the impact of super-nutritional exposures and their effects on growth or apoptosis. It is important to note, however, that blood levels of bioactive food components do not accurately reflect tissue concentrations, and research designs should employ concentrations that coincide with levels that reach the target tissue through dietary means without producing negative consequences. In other words, it is critical that proposed studies employ dietary rather than pharmacological concentrations of bioactive components.
A number of articles have been written about DNMT and HDAC inhibitors and their implications for cancer prevention and therapy (50, 53, 57). A few natural edible plant products have been shown to be capable of inhibiting DNMTs or HDACs (24, 51). When these nutrients are supplemented in the human diet, they may activate oncogenes. Due to the lack of specificity of these inhibitors, care should be taken in planning such trials (42). Regulatory agencies such as the U.S. Food and Drug Administration evaluate such data and provide policies and guidelines for the use of dietary products.
Challenges and potential solutions in nutrient measurement technologies
Current dietary assessment methods and systems depend largely on time- and resource-intensive self-reporting and recall methods. When diet is assessed via self-reporting, the cognitive challenges involved in recalling and reporting quantities, types, and preparation of foods eaten often compromise assessment accuracy. Dietary assessment methods that do not rely solely on self-reporting and recall could enhance the accuracy and efficiency of dietary intake data collection and contribute to an improved understanding of the diet–disease relationship. Technologic and analytic advances during the past decade have led to the development of more objective methods for assessing dietary intake. Leading examples include sophisticated dietary image or short video capture devices that maybe housed on a mobile phone platform and paired with speech recognition, text interface, and/or geospatial location. With the advent of electronic medical records and focus on the epidemic of obesity and related comorbidities, clinicians, researchers, and practitioners increasingly are interested in using objective measures to monitor patient/participant behavior as a tool for chronic disease prevention and management and health research.
The development of an easily deployable architecture for image-based dietary data transfer, storage, analysis, and reporting will support the potential to increase our understanding of the relationship between diet and cancer risk. Software systems capable of managing and analyzing the rich media collected by mobile sensors currently are limited, however. The complexity of data management and analysis needed to provide image-based measures of dietary intake presents a significant barrier to the integration of these measures into clinical practice and trials, epidemiological research, and behavioral monitoring applications. To overcome current barriers and facilitate the integration of image-based dietary measures into applications including electronic medical records and other health information systems, research should be conducted with a focus on developing an easily deployable architecture for data collection, transfer, storage, analysis, and reporting of dietary intake. Specific topics for further research may include development of (i) a mobile application to facilitate and control the collection and transfer of dietary images or video and any associated information such as annotations, probes, or geospatial location; (ii) a standardized dietary-rich media database architecture and procedures to import and store data transferred from the mobile application; (iii) transparent and modifiable analytic tools that can incorporate existing and evolving methods to generate individual- and group-level dietary intake measures from dietary images and associated data; and (iv) reporting systems to communicate outputs to patients/subjects, electronic medical records, health surveillance systems, and/or researchers.
Data processing applications and analytic tools maybe derived from established methods for dietary imaging analysis and common practices. Automated food item identification, quantity estimation, and consumed volume reconstruction can be achieved based on pre- and postmeal videos, digital images, and photos. Links should be created to established nutrient databases, such as the U.S. Department of Agriculture's (USDA) Food and Nutrient Database for Dietary Studies, USDA's MyPyramid Equivalents Database, Global Positioning System, and the Gladson Nutrition Database. Furthermore, data linkages should be established and validated within the Dietary Intake Summary Database. This will facilitate the identification of individual-level data characteristics on a per meal, daily, and/or weekly basis.
The Current Landscape and Perspectives, and Where Do We Go from Here?
Cancer has been associated with inherited genetic sequences but also results from epigenetic changes. Topics for future research in this area include validation of current putative diet-related surrogate endpoints, development of novel endpoints, and incorporation and integration of information from epigenomics and genomics.
Epigenetic alterations in tumor suppressor genes and in genes that are involved in controlling cell proliferation, DNA repair and metastasis, and hormone-receptor expression have been shown to play a role in tumor causation and progression. Dietary components also contribute to these processes. This information has implications for cancer epidemiology. The prevalence of epigenetic alterations may provide a basis for understanding the unequal cancer burden in early onset disease, disease aggressiveness, and the poor outcomes observed in various racial and ethnic populations. Identifying these differences in epigenetic processes in various diverse populations may enable the development of epigenetic biomarkers of cancer risk and the design of more effective therapeutic interventions. Epigenetic research has the potential to enhance our understanding of the determinants of the cancer burden among diverse populations and, ultimately, for reducing cancer health disparities.
Research in nutritional epigenetics may help to answer questions such as how bioactive food components regulate epigenetic events in different diseases; how bioactive food components alter epigenetic patterns and restore gene function; how these components circumvent and compensate for pathways that are altered during disease development; how gene-specific epigenetic inhibitors can be developed; how temporality in the epigenetic profile caused by bioactive food components can be measured; and how epigenomic and genomic data can be integrated to develop personalized medicine approaches.
Other challenges include the large number of input variables, relatively few intermediate markers and measurements, limited outcome measurements, lack of in silico models, and the dynamic nature of nutrients. Single-pathway approaches should be expanded to a genome-wide approach to measure epigenetic changes (45). Another aspect that attracts attention is variation in food processing and preparation techniques, which also contributes to differences in the bioactivities of food components.
Dietary recommendations at the population level will continue to be made, but customized dietary recommendations at the individual level are the expectation for the future. Combined efforts including changes in lifestyle (exercise), controlled and selective nutrition, and epigenetic drugs could bring about the reversal of diseases or at least slow down disease processes and enhance survival. Our current knowledge of the human epigenome and genome with respect to dietary components may make this possible.
Abbreviations Used
- AR
androgen receptor
- DNMT1
DNA methyl transferase 1
- EC
epicatechin
- ECG
epicatechin-3-gallate
- EGC
epigallocatechin
- EGCG
epigallocatechin-3-gallate
- ER
estrogen receptor
- GBM
glioblastoma
- HAT
histone acetyl transferase
- HDAC
histone de-acetylase
- hMLH1
human mutL homologue 1
- IGF2
insulin-like growth factor 2
- IL
interleukin
- MGMT
O6-methylguanine methyltransferase
- NAD
nicotinamide adenine dinucleotide
- PcG
polycomb group
- RARβ
retinoic acid receptor β
- SAM
S-adenosyl methionine
- USDA
U.S. Department of Agriculture
Acknowledgment
I am thankful to Joanne Brodsky for reading the manuscript and providing her suggestions.
References
- 1.Anderson CM. Lopez F. Zimmer A. Benoit JN. Placental insufficiency leads to developmental hypertension and mesenteric artery dysfunction in two generations of Sprague-Dawley rat offspring. Biol Reprod. 2006;74:538–544. doi: 10.1095/biolreprod.105.045807. [DOI] [PubMed] [Google Scholar]
- 2.Ballestar E. Esteller M. Epigenetic gene regulation in cancer. Adv Genet. 2008;61:247–267. doi: 10.1016/S0065-2660(07)00009-0. [DOI] [PubMed] [Google Scholar]
- 3.Beumer JH. Tawbi H. Role of histone deacetylases and their inhibitors in cancer biology and treatment. Curr Clin Pharmacol. 2010;5:196–208. doi: 10.2174/157488410791498770. [DOI] [PubMed] [Google Scholar]
- 4.Bommareddy A. Arasada BL. Mathees DP. Dwivedi C. Chemopreventive effects of dietary flaxseed on colon tumor development. Nutr Cancer. 2006;54:216–222. doi: 10.1207/s15327914nc5402_8. [DOI] [PubMed] [Google Scholar]
- 5.Bryzgunova OE. Morozkin ES. Yarmoschuk SV. Vlassov VV. Laktionov PP. Methylation-specific sequencing of GSTP1 gene promoter in circulating/extracellular DNA from blood and urine of healthy donors and prostate cancer patients. Ann N Y Acad Sci. 2008;1137:222–225. doi: 10.1196/annals.1448.039. [DOI] [PubMed] [Google Scholar]
- 6.Carr DF. Whiteley G. Alfirevic A. Pirmohamed M. FolATED study team. Investigation of inter-individual variability of the one-carbon folate pathway: a bioinformatic and genetic review. Pharmacogenomics J. 2009;9:291–305. doi: 10.1038/tpj.2009.29. [DOI] [PubMed] [Google Scholar]
- 7.Chang CM. Yu CC. Lu HT. Chou YF. Huang RF. Folate deprivation promotes mitochondrial oxidative decay: DNA large deletions, cytochrome c oxidase dysfunction, membrane depolarization and superoxide overproduction in rat liver. Br J Nutr. 2007;97:855–863. doi: 10.1017/S0007114507666410. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y. Gelfond JA. McManus LM. Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chobanian AV. Improved hypertension control: cause for some celebration. JAMA. 2010;303:2082–2083. doi: 10.1001/jama.2010.692. [DOI] [PubMed] [Google Scholar]
- 10.Chobanian AV. Bakris GL. Black HR. Cushman WC. Green LA. Izzo JL. Jones DW. Materson BJ. Oparil S. Wright JT., Jr. Roccella EJ. Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure, National Heart, Lung, and Blood Institute; and National High Blood Pressure Education Program Coordinating Committee. Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 11.Chou YF. Huang RF. Mitochondrial DNA deletions of blood lymphocytes as genetic markers of low folate-related mitochondrial genotoxicity in peripheral tissues. Eur J Nutr. 2009;48:429–436. doi: 10.1007/s00394-009-0031-0. [DOI] [PubMed] [Google Scholar]
- 12.Coppedè F. The complex relationship between folate/homocysteine metabolism and risk of Down syndrome. Mutat Res. 2009;682:54–70. doi: 10.1016/j.mrrev.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Dammann RH. Kirsch S. Schagdarsurengin U. Dansranjavin T. Gradhand E. Schmitt WD. Hauptmann S. Frequent aberrant methylation of the imprinted IGF2/H19 locus and LINE1 hypomethylation in ovarian carcinoma. Int J Oncol. 2010;36:171–179. [PubMed] [Google Scholar]
- 14.Davis CD. Ross SA. Dietary components impact histone modifications and cancer risk. Nutr Rev. 2007;65:88–94. doi: 10.1111/j.1753-4887.2007.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 15.Davis CD. Uthus EO. Finley JW. Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr. 2000;130:2903–2909. doi: 10.1093/jn/130.12.2903. [DOI] [PubMed] [Google Scholar]
- 16.Dimri M. Bommi PV. Sahasrabuddhe AA. Khandekar JD. Dimri GP. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis. 2010;31:489–495. doi: 10.1093/carcin/bgp305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donkena KV. Yuan H. Young CY. Vitamin Bs, one carbon metabolism and prostate cancer. Mini Rev Med Chem. 2010;10:1385–1392. doi: 10.2174/138955710793564106. [DOI] [PubMed] [Google Scholar]
- 18.Duthie SJ. Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J Inherit Metab Dis. 2011;34:101–109. doi: 10.1007/s10545-010-9128-0. [DOI] [PubMed] [Google Scholar]
- 19.Elsheikh SE. Green AR. Rakha EA. Powe D G. Ahmed RA. Collins HM. Soria D. Garibaldi JM. Paish CE. Ammar AA. Grainge MJ. Ball GR. Abdelghany MK. Martinez-Pomares L. Heery DM. Ellis IO. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraga MF. Ballestar E. Villar-Garea A. Boix-Chornet M. Espada J. Schotta G. Bonaldi T. Haydon C. Ropero S. Petrie K. Iyer NG. Pérez-Rosado A. Calvo E. Lopez JA. Cano A. Calasanz MJ. Colomer D. Piris MA. Ahn N. Imhof A. Caldas C. Jenuwein T. Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 23.Fraga MF. Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Ganesan A. Nolan L. Crabb SJ. Packham G. Epigenetic therapy: histone acetylation, DNA methylation and anti-cancer drug discovery. Curr Cancer Drug Targets. 2009;9:963–981. doi: 10.2174/156800909790192428. [DOI] [PubMed] [Google Scholar]
- 25.Garzon R. Calin GA. Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 26.Gieni RS. Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem Cell Biol. 2009;87:711–746. doi: 10.1139/O09-057. [DOI] [PubMed] [Google Scholar]
- 27.Han CT. Schoene NW. Lei KY. Influence of zinc deficiency on Akt-Mdm2-p53 and Akt-p21 signaling axes in normal and malignant human prostate cells. Am J Physiol Cell Physiol. 2009;297:C1188–C1199. doi: 10.1152/ajpcell.00042.2009. [DOI] [PubMed] [Google Scholar]
- 28.Heijmans BT. Tobi EW. Lumey LH. Slagboom PE. The epigenome: archive of the prenatal environment. Epigenetics. 2009;4:526–531. doi: 10.4161/epi.4.8.10265. [DOI] [PubMed] [Google Scholar]
- 29.Heijmans BT. Tobi EW. Stein AD. Putter H. Blauw GJ. Susser ES. Slagboom PE. Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong SK. Kim JH. Lin MF. Park JI. The Raf/MEK/extracellular signal-regulated kinase 1/2 pathway can mediate growth inhibitory and differentiation signaling via androgen receptor downregulation in prostate cancer cells. Exp Cell Res. 2011;317:2671–2682. doi: 10.1016/j.yexcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jelinic P. Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211:261–268. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- 32.Johnson IT. Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food Chem Toxicol. 2008;46:1346–1359. doi: 10.1016/j.fct.2007.09.101. [DOI] [PubMed] [Google Scholar]
- 33.Jones A. Wang H. Polycomb repressive complex 2 in embryonic stem cells: an overview. Protein Cell. 2010;1:1056–1062. doi: 10.1007/s13238-010-0142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keijer J. Bekkenkamp-Grovenstein M. Venema D. Dommels YE. Bioactive food components, cancer cell growth limitation and reversal of glycolytic metabolism. Biochim Biophys Acta. 2011;1807:697–706. doi: 10.1016/j.bbabio.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Kim KC. Friso S. Choi SW. DNA methylation, an epigenetic mechanism connecting folate to healthy embryonic development and aging. J Nutr Biochem. 2009;20:917–926. doi: 10.1016/j.jnutbio.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YS. Milner JA. Bioactive food components and cancer specific metabolomic profiles. J Biomed Biotech. 2011:21113295. doi: 10.1155/2011/721213. PMID. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovacheva VP. Davison JM. Mellott TJ. Rogers AE. Yang S. O'Brien MJ. Blusztajn JK. Raising gestational choline intake alters gene expression in DMBA-evoked mammary tumors and prolongs survival. FASEB J. 2009;23:1054–1063. doi: 10.1096/fj.08-122168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ku G. McManus MT. Behind the scenes of a small RNA gene-silencing pathway. Hum Gene Ther. 2008;19:17–26. doi: 10.1089/hum.2007.1226. [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni A. Chavan-Gautam P. Mehendale S. Yadav H. Joshi S. Global DNA methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell Biol. 2011;30:79–84. doi: 10.1089/dna.2010.1084. [DOI] [PubMed] [Google Scholar]
- 40.Kumar D. Verma M. Methods in cancer epigenetics and epidemiology. Methods Mol Biol. 2009;471:273–288. doi: 10.1007/978-1-59745-416-2_14. [DOI] [PubMed] [Google Scholar]
- 41.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 42.Lane AA. Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 43.Lara E. Mai A. Calvanese V. Altucci L. Lopez-Nieva P. Martinez-Chantar ML. Varela-Rey M. Rotili D. Nebbioso A. Ropero S. Montoya G. Oyarzabal J. Velasco S. Serrano M. Witt M. Villar-Garea A. Imhof A. Mato JM. Esteller M. Fraga MF. Salermide, a sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene. 2009;28:781–791. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 44.Li Y. Wang Z. Kong D. Li R. Sarkar SH. Sarkar FH. Regulation of Akt/FOXO3a/GSK-3beta/AR signaling network by isoflavone in prostate cancer cells. J Biol Chem. 2008;283:27707–27716. doi: 10.1074/jbc.M802759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N. Ye M. Li Y. Yan Z. Butcher LM. Sun J. Han X. Chen Q. Zhang X. Wang J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–212. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Li Y. Zhu J. Tian G. Li N. Li Q. Ye M. Zheng H. Yu J. Wu H. Sun J. Zhang H. Chen Q. Luo R. Chen M. He Y. Jin X. Zhang Q. Yu C. Zhou G. Sun J. Huang Y. Zheng H. Cao H. Zhou X. Guo S. Hu X. Li X. Kristiansen K. Bolund L. Xu J. Wang W. Yang H. Wang J. Li R. Beck S. Wang J. Zhang X. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin YW. Chen HM. Fang JY. Gene silencing by the Polycomb group proteins and associations with cancer. Cancer Invest. 2011;29:187–195. doi: 10.3109/07357907.2010.512605. [DOI] [PubMed] [Google Scholar]
- 48.Link A. Balaguer F. Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopatina NG. Vanyushin BF. Cronin GM. Poirier LA. Elevated expression and altered pattern of activity of DNA methyltransferase in liver tumors of rats fed methyl-deficient diets. Carcinogenesis. 1998;19:1777–1781. doi: 10.1093/carcin/19.10.1777. [DOI] [PubMed] [Google Scholar]
- 50.Lustberg MB. Ramaswamy B. Epigenetic targeting in breast cancer: therapeutic impact and future direction. Drug News Perspect. 2009;22:369–381. doi: 10.1358/dnp.2009.22.7.1405072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma X. Ezzeldin HH. Diasio RB. Histone deacetylase inhibitors: current status and overview of recent clinical trials. Drugs. 2009;69:1911–1934. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 52.Marik R. Allu M. Anchoori R. Stearns V. Umbricht CB. Khan S. Potent genistein derivatives as inhibitors of estrogen receptor alpha-positive breast cancer. Cancer Biol Ther. 2011;11:883–892. doi: 10.4161/cbt.11.10.15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marson CM. Histone deacetylase inhibitors: design, structure-activity relationships and therapeutic implications for cancer. Anticancer Agents Med Chem. 2009;9:661–692. doi: 10.2174/187152009788679976. [DOI] [PubMed] [Google Scholar]
- 54.Matsuoka T. Adair JE. Lih FB. Hsi LC. Rubino M. Eling TE. Tomer KB. Yashiro M. Hirakawa K. Olden K. Roberts JD. Elevated dietary linoleic acid increases gastric carcinoma cell invasion and metastasis in mice. Br J Cancer. 2010;103:1182–1191. doi: 10.1038/sj.bjc.6605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meeran SM. Ahmed A. Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenetics. 2010;1:101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Millis RM. Epigenetics and hypertension. Curr Hypertens Rep. 2011;13:21–28. doi: 10.1007/s11906-010-0173-8. [DOI] [PubMed] [Google Scholar]
- 57.Milner JA. Nutrition and cancer: essential elements for a roadmap. Cancer Lett. 2008;269:189–198. doi: 10.1016/j.canlet.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 58.Nafee TM. Farrell WE. Carroll WD. Fryer AA. Ismail KM. Epigenetic control of fetal gene expression. BJOG. 2008;115:158–168. doi: 10.1111/j.1471-0528.2007.01528.x. [DOI] [PubMed] [Google Scholar]
- 59.Pitchakarn P. Suzuki S. Ogawa K. Pompimon W. Takahashi S. Asamoto M. Limtrakul P. Shirai T. Induction of G1 arrest and apoptosis in androgen-dependent human prostate cancer by Kuguacin J, a triterpenoid from Momordica charantia leaf. Cancer Lett. 2011;306:142–150. doi: 10.1016/j.canlet.2011.02.041. [DOI] [PubMed] [Google Scholar]
- 60.Piunti A. Pasini D. Epigenetic factors in cancer development: polycomb group proteins. Future Oncol. 2011;7:57–75. doi: 10.2217/fon.10.157. [DOI] [PubMed] [Google Scholar]
- 61.Plagemann A. Harder T. Brunn M. Harder A. Roepke K. Wittrock-Staar M. Ziska T. Schellong K. Rodekamp E. Melchior K. Dudenhausen JW. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol. 2009;587:4963–4976. doi: 10.1113/jphysiol.2009.176156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pogribny IP. Ross SA. Wise C. Pogribna M. Jones EA. Tryndyak VP. James SJ. Dragan YP. Poirier LA. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 63.Poirier LA. Methyl group deficiency in hepatocarcinogenesis. Drug Metab Rev. 1994;26:185–199. doi: 10.3109/03602539409029790. [DOI] [PubMed] [Google Scholar]
- 64.Powolny AA. Bommareddy A. Hahm ER. Normolle DP. Beumer JH. Nelson JB. Singh SV. Chemopreventative potential of the cruciferous vegetable constituent phenethyl isothiocyanate in a mouse model of prostate cancer. J Natl Cancer Inst. 2011;103:571–584. doi: 10.1093/jnci/djr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez RM. Fraga MF. Aging and cancer: are sirtuins the link? Future Oncol. 2010;6:905–915. doi: 10.2217/fon.10.57. [DOI] [PubMed] [Google Scholar]
- 66.Ross SA. Evidence for the relationship between diet and cancer. Exp Oncol. 2010;32:137–142. [PubMed] [Google Scholar]
- 67.Ross SA. Dwyer J. Umar A. Kagan J. Verma M. Van Bemmel DM. Dunn BK. Introduction: diet, epigenetic events and cancer prevention. Nutr Rev. 2008;66(Suppl 1):S1–S6. doi: 10.1111/j.1753-4887.2008.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strakovsky RS. Zhou D. Pan YX. A low-protein diet during gestation in rats activates the placental mammalian amino acid response pathway and programs the growth capacity of offspring. J Nutr. 2010;140:2116–2120. doi: 10.3945/jn.110.127803. [DOI] [PubMed] [Google Scholar]
- 69.Szulwach KE. Li X. Smrt RD. Li Y. Luo Y. Lin L. Santistevan NJ. Li W. Zhao X. Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Timp W. Levchenko A. Feinberg AP. A new link between epigenetic progenitor lesions in cancer and the dynamics of signal transduction. Cell Cycle. 2009;8:383–390. doi: 10.4161/cc.8.3.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tobi EW. Heijmans BT. Kremer D. Putter H. Delemarre-van de Waal HA. Finken MJ. Wit JM. Slagboom PE. DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics. 2011;6:171–176. doi: 10.4161/epi.6.2.13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tobi EW. Lumey LH. Talens RP. Kremer D. Putter H. Stein AD. Slagboom PE. Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Traka MH. Spinks CA. Doleman JF. Melchini A. Ball RY. Mills RD. Mithen RF. The dietary isothiocyanate sulforaphane modulates gene expression and alternative gene splicing in a PTEN null preclinical murine model of prostate cancer. Mol Cancer. 2010;9:189–194. doi: 10.1186/1476-4598-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ulrich CM. Folate and cancer prevention—where to next? Counterpoint. Cancer Epidemiol Biomarkers Prev. 2008;17:2226–2230. doi: 10.1158/1055-9965.EPI-07-2952. [DOI] [PubMed] [Google Scholar]
- 75.Vaquero A. The conserved role of sirtuins in chromatin regulation. Int J Dev Biol. 2009;53:303–322. doi: 10.1387/ijdb.082675av. [DOI] [PubMed] [Google Scholar]
- 76.Verma M. Viral genes and methylation. Ann N Y Acad Sci. 2003;983:170–180. doi: 10.1111/j.1749-6632.2003.tb05972.x. [DOI] [PubMed] [Google Scholar]
- 77.Verma M. The human epigenome and cancer. In: Khoury M, editor; Bedrosian S, editor; Gwinn M, editor; Higgins J, editor; Ioannidis J, editor; Little J, editor. Human Genome Epidemiology. 2nd. New York: Oxford University Press; 2010. pp. 551–578. [Google Scholar]
- 78.Verma M. Kumar D. Diagnosing cancer using histone analysis. In: Tollefsbol T., editor. Cancer Epigenetics. New York: CRC Press; 2009. pp. 347–357. [Google Scholar]
- 79.Verma M. Maruvada P. Srivastava S. Epigenetics and cancer. Crit Rev Clin Lab Sci. 2004;41:585–607. doi: 10.1080/10408360490516922. [DOI] [PubMed] [Google Scholar]
- 80.Verma M. Seminara D. Arena FJ. John C. Iwamoto K. Hartmuller V. Genetic and epigenetic biomarkers in cancer: improving diagnosis, risk assessment, and disease stratification. Mol Diagn Ther. 2006;10:1–15. doi: 10.1007/BF03256438. [DOI] [PubMed] [Google Scholar]
- 81.Villagra A. Sotomayor EM. Seto E. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 2010;29:157–173. doi: 10.1038/onc.2009.334. [DOI] [PubMed] [Google Scholar]
- 82.Vu TH. Nguyen AH. Hoffman AR. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum Mol Genet. 2010;19:901–919. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wan SG. Taccioli C. Jiang Y. Chen H. Smalley KJ. Huang K. Liu XP. Farber JL. Croce CM. Fong LY. Zinc deficiency activates S100A8 inflammation in the absence of COX-2 and promotes murine oral-esophageal tumor progression. Int J Cancer. 2011;129:331–345. doi: 10.1002/ijc.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams D. Verghese M. Walker LT. Boateng J. Shackelford L. Chawan CB. Flax seed oil and flax seed meal reduce the formation of aberrant crypt foci (ACF) in azoxymethane-induced colon cancer in Fisher 344 male rats. Food Chem Toxicol. 2007;45:153–159. doi: 10.1016/j.fct.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 85.Wu MY. Kuo CS. Lin CY. Lu CL. Syu Huang RF. Lymphocytic mitochondrial DNA deletions, biochemical folate status and hepatocellular carcinoma susceptibility in a case- control study. Br J Nutr. 2009;102:715–721. doi: 10.1017/S0007114509243054. [DOI] [PubMed] [Google Scholar]
- 86.Yan H. Zhu Y. Liu B. Wu H. Li Y. Wu X. Zhou Q. Xu K. Mitogen-activated protein kinase mediates the apoptosis of highly metastatic human non-small cell lung cancer cells induced by isothiocyanates. Br J Nutr. 2011;23:1–13. doi: 10.1017/S0007114511002315. [DOI] [PubMed] [Google Scholar]
- 87.Zeisel SH. Nutritional genomics: defining the dietary requirement and effects of choline. J Nutr. 2011;141:531–534. doi: 10.3945/jn.110.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang M. Liu X. Holman CD. Effect of dietary intake of isoflavones on the estrogen and progesterone receptor status of breast cancer. Nutr Cancer. 2010;62:765–773. doi: 10.1080/01635581003605979. [DOI] [PubMed] [Google Scholar]
- 89.Zhao R. DeCoteau JF. Geyer CR. Gao M. Cui H. Casson AG. Loss of imprinting of the insulin-like growth factor II (IGF2) gene in esophageal normal and adenocarcinoma tissues. Carcinogenesis. 2009;30:2117–2122. doi: 10.1093/carcin/bgp254. [DOI] [PubMed] [Google Scholar]