Summary
Cellular mRNAs exist in messenger ribonucleoprotein (mRNP) complexes, which undergo transitions during the lifetime of the mRNAs and direct posttranscriptional gene regulation. A final posttranscriptional step in gene expression is the turnover of the mRNP, which involves degradation of the mRNA and recycling of associated proteins. How tightly associated protein components are released from degrading mRNPs is unknown. Here we demonstrate that the ATPase activity of the RNA helicase Upf1 allows disassembly of mRNPs undergoing nonsense-mediated mRNA decay (NMD). In the absence of Upf1 ATPase activity, partially degraded NMD mRNA intermediates accumulate in complex with NMD factors and concentrate in processing bodies. Thus, disassembly and completion of turnover of mRNPs undergoing NMD requires ATP hydrolysis by Upf1. This uncovers a previously unappreciated and potentially regulated step in mRNA decay, and raises the question of how other mRNA decay pathways release protein components of substrate mRNPs.
Keywords: mRNP, RNA helicase, mRNA turnove, nonsense-mediated decay (NMD), Upf1, processing bodies
Introduction
mRNA decay is a critical step in the regulation of gene expression. The stability of mRNAs can vary by orders of magnitude and is dictated by the composition of the messenger ribonucleoprotein (mRNP) (Balagopal and Parker, 2009; Moore, 2005). How decay-promoting mRNP components activate mRNA turnover is poorly understood. Several studies have shown evidence that the recruitment of mRNA decay enzymes is a critical step in mRNA turnover (Cho et al., 2009; Gherzi et al., 2004; Lykke-Andersen and Wagner, 2005; Moraes et al., 2006). Yet, it is unknown how recruited mRNA decay enzymes access the mRNA through stably associated protein components of the mRNP. In an analogous manner, early models for transcription activation focused on the recruitment of RNA polymerase, while later studies demonstrated the importance of chromatin modification and remodeling (Campos and Reinberg, 2009). Does the mRNP constitute an obstacle to mRNA turnover as chromatin does to transcription?
Evidence primarily from the yeast Saccharomyces cerevisiae suggests that mRNA degradation generally initiates with removal of the mRNA poly(A)-tail by deadenylases, which stimulates either mRNA decapping and subsequent 5’-to-3’ exonucleolytic decay by Xrn1 (Doma and Parker, 2007; Garneau et al., 2007) or degradation in the 3’-to-5’ direction by the exosome (Schmid and Jensen, 2008). In addition, some mRNA decay pathways trigger endonucleolytic cleavage followed by 3’-to-5’ and 5’-to-3’ exonucleolytic decay of the mRNA fragments by the exosome and Xrn1, respectively (Wilusz, 2009). However, while much has been learned about the enzymes that degrade mRNAs, it remains unknown how the mRNA decay enzymes negotiate the mRNP.
Nonsense-mediated mRNA decay (NMD) is an mRNA turnover pathway that targets mRNAs with premature translation termination codons (PTCs) for rapid degradation, thereby suppressing protein expression from aberrant mRNAs, as well as a subset of normal NMD-regulated mRNAs (Amrani et al., 2006; Behm-Ansmant et al., 2007; Chang et al., 2007; Isken and Maquat, 2007; Muhlemann et al., 2008; Rebbapragada and Lykke-Andersen, 2009). How a termination codon is recognized as premature remains under investigation, but appears to occur when a ribosome terminates translation sufficiently upstream of a normal 3’ UTR to prevent 3’ UTR-associated proteins, including cytoplasmic poly(A) binding protein (PABPC), from stimulating a proper terminati7on event (Amrani et al., 2006; Muhlemann et al., 2008; Rebbapragada and Lykke-Andersen, 2009). This initiates the assembly of an NMD mRNP with the recruitment of the NMD factor Upf1 and its co-factors Upf2 and Upf3 to the terminating ribosome. In vertebrates, NMD is strongly stimulated when an exon junction complex (EJC), which interacts with the Upf complex, is positioned downstream of the termination event (Behm-Ansmant et al., 2007; Isken and Maquat, 2007; Moore and Proudfoot, 2009; Muhlemann, 2008; Rebbapragada and Lykke-Andersen, 2009). In metazoans, the NMD mRNP is further modulated by a phosphorylation-dephosphorylation cycle of Upf1 mediated by the kinase Smg1 and by protein phosphatase 2A in association with the NMD factors Smg5 and Smg7 (Fukuhara et al., 2005; Glavan et al., 2006; Ohnishi et al., 2003; Page et al., 1999; Yamashita et al., 2001). The assembled NMD mRNP subsequently recruits mRNA decay enzymes to initiate mRNA degradation. Dependent on the specific organism, decay can initiate by decapping, deadenylation and/or endonucleolytic cleavage (Mühlemann and Lykke-Andersen, 2010). Recent evidence suggests that in human and Drosophila cells, decay of the NMD substrate is primarily initiated by endonucleolytic cleavage by the NMD factor Smg6 followed by 3’-to-5’ and 5’-to-3’ exonucleolytic decay of the mRNA fragments by the exosome and Xrn1, respectively (Gatfield and Izaurralde, 2004; Glavan et al., 2006; Huntzinger et al., 2008; Eberle et al., 2009). However, it remains an unresolved question how the NMD factors are recycled from the degrading NMD mRNP. Are they released by the activity of the mRNA decay enzymes or do they require active removal prior to, or during mRNA decay?
A central component of the NMD pathway, Upf1, belongs to helicase superfamily 1 and shows RNA-dependent ATPase and 5’-to-3’ RNA helicase activities in vitro (Bhattacharya et al., 2000; Chamieh et al., 2008; Cheng et al., 2007; Czaplinski et al., 1995). The ATPase activity of Upf1 is critical to the NMD pathway (Kashima et al., 2006; Weng et al., 1996a); however its specific role remains unresolved. Although helicases were first described as ATPases that unwind polynucleotide duplexes, several helicases of superfamily 2 have more recently been shown to function as RNPases that promote ATP-dependent mRNP remodeling in the absence of double stranded RNA (Fairman et al., 2004; Jankowsky et al., 2001). Early studies implicated the Upf1 ATPase at the translation termination step of yeast NMD (Weng et al., 1998), but more recent observations in yeast show that ATPase-deficient mutant Upf1 accumulates with NMD substrates in cytoplasmic mRNP granules called processing bodies (PBs) (Cheng et al., 2007; Sheth and Parker, 2006). This suggests that the failure of Upf1 to hydrolyze ATP causes the accumulation of an NMD mRNP in association with mRNA decay factors. Here, we demonstrate that the Upf1 ATPase stimulates the removal and recycling of NMD factors from mRNPs targeted for NMD. This is required for the completion of exonucleolytic decay of the NMD substrate. In the absence of Upf1 ATPase activity, NMD factors become trapped with partially degraded 3' NMD mRNP intermediates. This demonstrates the importance of mRNP disassembly in mRNA turnover, and raises the questions of whether this is a regulated step in NMD and to what extent mRNP disassembly is a critical step in other mRNA decay pathways.
Results
The 3’ fragment generated upon endonucleolytic cleavage of NMD mRNA substrates accumulates in the presence of ATPase-deficient Upf1
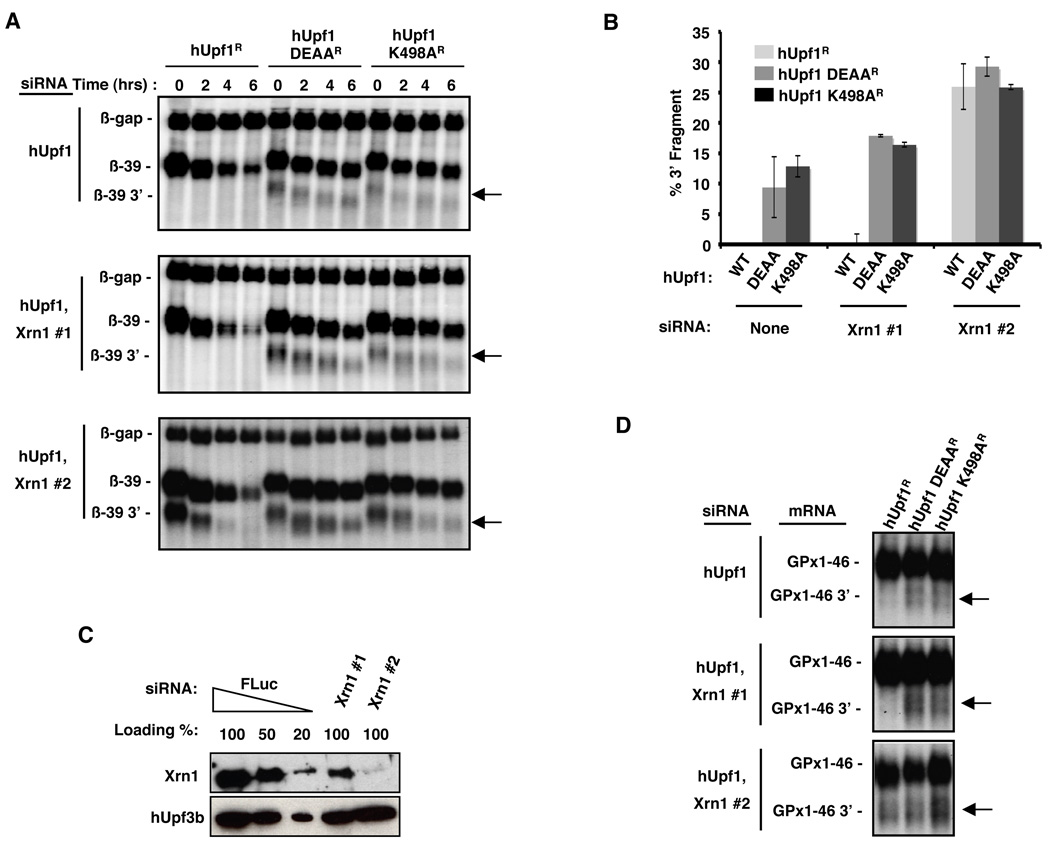
To investigate the function of Upf1 ATPase activity in NMD, we tested the effect of impairing human (h)Upf1 ATP binding and hydrolysis on the degradation of NMD substrate mRNAs. A β-globin mRNA with a PTC at position 39 (β-39) was subjected to pulse-chase mRNA decay assays in human HeLa Tet-off cells, in which endogenous hUpf1 was depleted with an siRNA and replaced with exogenous siRNA-resistant wild-type hUpf1 (hUpf1R), or mutants thereof that fail to hydrolyze (hUpf1 DEAAR) or fail to bind (hUpf1 K498AR) ATP (Bhattacharya et al., 2000; Cheng et al., 2007). As expected, the β-39 NMD substrate is significantly more stable in the presence of hUpf1 ATPase mutants than with wild-type hUpf1 (Figure 1A, top panel; see band labeled β-39). Surprisingly however, a fast migrating mRNA species (indicated by an arrow in Figure 1A) accumulates when hUpf1 ATPase mutant proteins are expressed, but is not observed in the presence of wild-type hUpf1 (Figure 1A, top panel; quantified in Figure 1B). This product corresponds to the 3’ fragment of the NMD substrate following endonucleolytic cleavage by Smg6, because it is not observed with a probe specific to the 5’ end of β-globin mRNA and is strongly reduced under Smg6 knockdown conditions (Supplemental Figures S1A–C). In contrast to the 3' fragment, no 5’ fragment was detectable upon hUpf1 ATPase mutant expression (Figures 1A and S1D). Thus, ATPase-deficient hUpf1 allows endonucleolytic cleavage of the NMD substrate followed by exonucleolytic decay of the resulting 5’ product, but impairs the degradation of the 3’ product.
Figure 1. The 3’ fragment generated upon endonucleolytic cleavage of NMD substrates accumulates when hUpf1 fails to hydrolyze ATP.
(A) Northern blots showing the decay of β-globin mRNA with a PTC at position 39 (β-39) in HeLa Tet-off cells depleted of endogenous hUpf1 using an siRNA and expressing exogenous siRNA-resistant wild-type hUpf1 (hUpf1R), or hUpf1 ATPase (hUpf1 DEAAR) or ATP-binding (hUpf1 K498AR) mutants. siRNAs targeting Xrn1 were included in the experiments in the bottom two panels. Time points above each lane represent the elapsed time following transcriptional shut-off of β-39 mRNA by tetracycline addition. The 3’ endonucleolytic cleavage fragment of β-39 (β-39 3’) is indicated by arrows.
(B) Quantification showing the percent β-39 3’ mRNA fragment of total β-39 mRNA immediately after the transcriptional pulse (t=0) for each condition indicated. Percent and standard deviation are calculated from three experiments.
(C) Western blots showing Xrn1 levels in HeLa Tet-off cells treated with a control siRNA (FLuc) (100%, 50% or 20% total protein was loaded) or with the two Xrn1 siRNAs used in (A) (Xrn1 #1 or Xrn1 #2). hUpf3b served as a loading control.
(D) Northern blots showing GPx1 mRNA with a PTC at position 46 (GPx1-46) after a 6 hour transcriptional pulse in HeLa Tet-off cells treated as described in (A).
See also Figure S1.
How can the failure of hUpf1 to bind or to hydrolyze ATP specifically affect the NMD substrate 3’ decay intermediate? One possibility is that the 3’ intermediate requires Upf1 ATPase activity to be accessible to Xrn1, the 5'-to-3' exonuclease that normally degrades this fragment (Gatfield and Izaurralde, 2004; Huntzinger et al., 2008; Eberle et al., 2009). If so, the same fragment should accumulate upon depletion of Xrn1 both in the presence of wild-type and ATPase-deficient hUpf1. To test this, Xrn1 was depleted with siRNAs that modestly (Xrn1 #1) or strongly (Xrn1 #2) reduce Xrn1 levels (Figure 1C) and the effect on the decay of the β-39 mRNA was monitored. As seen in Figure 1A (middle panel), when Xrn1 is modestly depleted, the β-39 mRNA 3’ fragment accumulates strongly in the presence of ATPase-deficient hUpf1, but not with wild-type hUpf1. Only when Xrn1 is strongly depleted does the 3’ β-39 mRNA fragment accumulate in cells expressing wild-type hUpf1 (Figure 1A, bottom panel; quantified in Figure 1B). However, even under these conditions the resulting 3’ mRNA fragment is rapidly degraded with an apparent half-life 2- to 4-fold shorter than that observed when hUpf1 ATPase mutants are expressed. A similar pattern of NMD substrate 3’ fragment accumulation was observed when a different NMD substrate, GPx1-46, was tested (Figure 1D). These observations are not a result of globally impaired Xrn1 activity, because Xrn1-mediated degradation of the 3’ fragment of a β-globin reporter mRNA subjected to endonucleolytic cleavage by endogenous let-7 microRNA is not impaired in the presence of ATPase-deficient hUpf1 (Figure S1E). Thus, while it is well established that the Upf1 protein plays a key role in the recognition step of NMD (Amrani et al., 2006; Kashima et al., 2006; Muhlemann et al., 2008; Ohnishi et al., 2003; Rebbapragada and Lykke-Andersen, 2009), the observations shown here suggest that the ATPase activity of Upf1 is required at a later step in NMD (Figure 1). Consistent with this, when mRNA decay is initiated by tethered hUpf1, thereby bypassing the Upf1 recruitment step of NMD (Lykke-Andersen et al., 2000), ATP binding-deficient hUpf1 causes accumulation of a 3’ fragment that is not observed with tethered wild-type Upf1 unless Xrn1 is efficiently knocked down (Figure S1F).
ATPase-deficient hUpf1 accumulates on the 3’ NMD intermediate
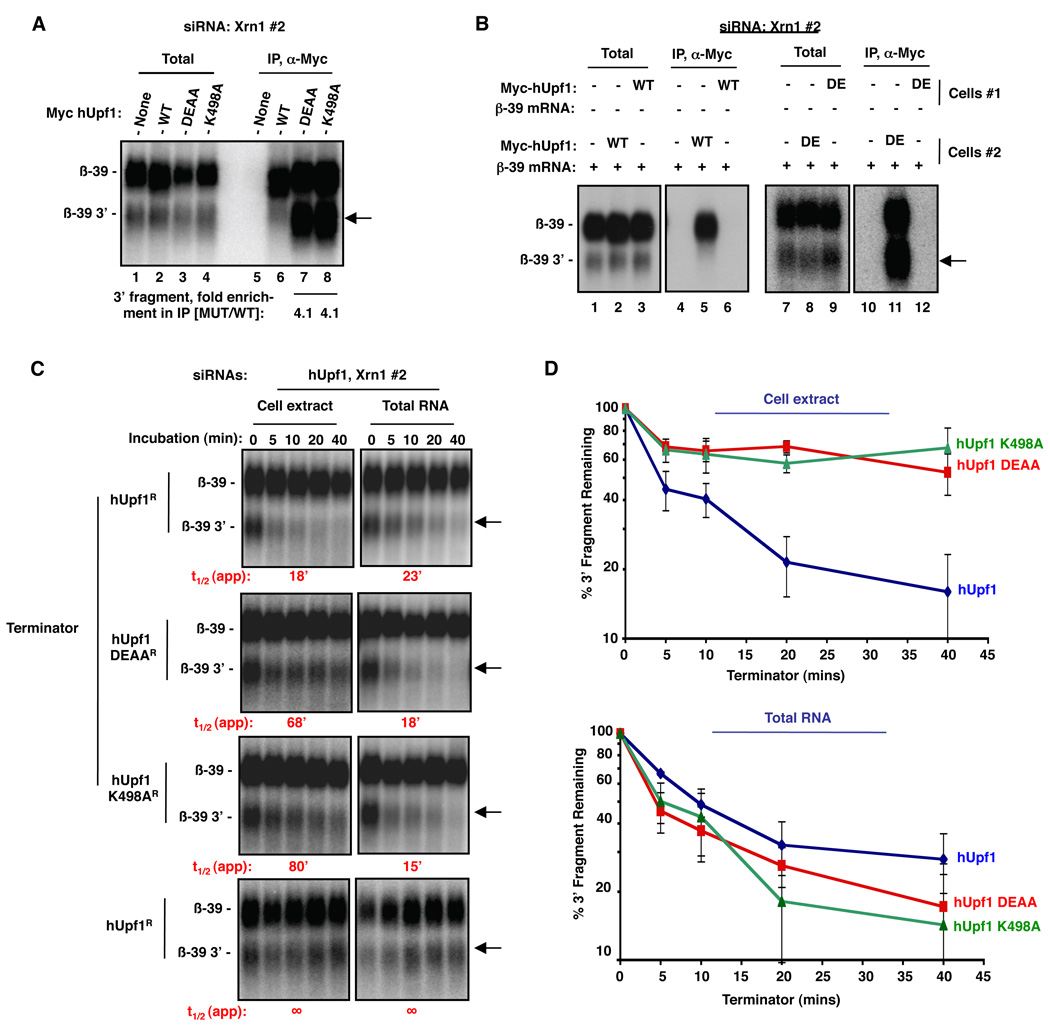
How does the Upf1 ATPase stimulate degradation of the 3’ NMD fragment by Xrn1? One possibility is that the Upf1 ATPase triggers removal of protein from the 3’ NMD mRNP intermediate, thereby allowing access for Xrn1. If so, it is predicted that wild-type hUpf1 should cycle off the 3’ NMD intermediate, whereas ATPase-deficient hUpf1 should fail to do so. This was tested using hUpf1 immunoprecipitation (IP) followed by Northern blotting for associated NMD substrate mRNA under strong Xrn1 knock-down conditions. As seen in Figure 2A, both wild-type and mutant hUpf1 proteins associate with full-length β-39 NMD substrate produced by a short transcriptional pulse (lanes 6–8). However, the association of the accumulating 3’ β-39 fragment with ATPase-deficient hUpf1 is strongly enhanced (4.1-fold relative to full-length β-39) as compared with wild-type hUpf1 (Figure 2A; compare lanes 7 and 8 to lane 6; band marked by arrow). These interactions occur in the cell and do not form after cell lysis, as β-39 mRNA does not co-purify with wild-type or mutant hUpf1 when expressed in separate cells and combined during cell lysis (Figure 2B; compare lanes 6 to 5 and 12 to 11), and the mRNA substrate does not co-purify with the antibody in the absence of exogenous hUpf1 (Figure 2A, lane 5; Figure 2B, lanes 4, 10). These observations are consistent with the idea that ATPase activity is not critical for recruitment of hUpf1 to the NMD substrate, but is required for the release of hUpf1 from the 3’ fragment that forms after initiation of mRNA decay.
Figure 2. The 3’ NMD endonucleolytic cleavage fragment is stuck with ATPase-deficient Upf1 and is resistant to 5'-to-3' exonucleolytic decay in vitro.
(A) Northern blot for β-39 mRNA from pellet (lanes 5–8) or 5% of total extract (lanes 1–4) fractions from anti-myc IP assays from cells transiently expressing myc-tagged hUpf1 proteins indicated on the top or no exogenous protein (none). Cells were treated with Xrn1 #2 siRNA to promote the accumulation of the β-39 mRNA 3’ fragment.
(B) Same as (A), but β-39 mRNA was expressed either in the same cells as wild-type (WT) or DEAA mutant (DE) hUpf1 (lanes 2, 5, 8, 11), or in different cells and mixed prior to extract preparation (lanes 3, 6, 9, 12). Lanes 1–3, 7–9: 5% of total extracts; Lanes 4–6, 10–12: IP pellets. Lanes 1, 4, 7 and 10 are from cells not expressing Myc-hUpf1. All cells were treated with Xrn1 #2 siRNA to promote the accumulation of the β-39 mRNA 3’ fragment.
(C) Northern blots showing in vitro Terminator-mediated decay of β-39 3' mRNA fragment from extracts (left panels) or total RNA (right panels) from HeLa Tet-off cells depleted of endogenous hUpf1 using an siRNA and expressing exogenous siRNA-resistant wild-type hUpf1 (hUpf1R) or hUpf1 ATPase mutants. An siRNA targeting Xrn1 (Xrn1 #2) was included in all experiments. Time points above each lane represent the time of Terminator incubation. Bottom panels: incubation in the absence of Terminator.
(D) Quantification for each of the experiments in (C). Percent and standard deviation are calculated from three experiments.
See also Figure S2.
The 3’ NMD mRNP fragment generated in the presence of ATPase-deficient Upf1 is resistant to 5’-to-3’ exonucleolytic decay in vitro
If the 3' NMD intermediate that forms in the presence of hUpf1 ATPase mutants is resistant to Xrn1 due to a failure in mRNP disassembly, it should become sensitive to 5'-to-3' exonucleolytic decay if protein is removed from the mRNP. To test this in vitro, β-39 mRNA was expressed along with exogenous wild-type or ATPase-deficient hUpf1 proteins in HeLa Tet-off cells depleted for endogenous Xrn1 and hUpf1. Cells were subsequently permeabilized and the resulting cell extracts were incubated with the Terminator enzyme, a commercially available 5’-to-3’ exonuclease. As seen in Figure 2C (left panels), while the 3’ NMD mRNA fragment that accumulates due to Xrn1 knock-down in the presence of wild-type hUpf1 is degraded efficiently (t1/2≈18 mins), a large fraction (60–70%) of the same RNA produced in cells expressing ATPase-deficient Upf1 proteins is highly resistant to 5'-to-3' exonucleolytic decay (quantified in Figure 2D, top panel). By contrast, when mRNPs were disrupted and protein removed from the cell extracts by phenol extraction prior to incubation with the nuclease, the 3’ NMD fragment is degraded efficiently regardless of the ability of Upf1 to hydrolyze ATP (Figure 2C, right panels; quantified in Figure 2D, bottom panel). As expected, full-length β-39 mRNA is resistant to the Terminator enzyme, which is specific for 5’ monophosphate-containing RNA and thus does not target capped RNA (Figure 2C; upper bands), and the 3’ fragment does not degrade in the absence of Terminator (bottom panels). When the endonuclease RNase A was used in place of Terminator, all RNAs rapidly degrade (Figure S2). Thus, when Upf1 fails to hydrolyze ATP, the 3' NMD fragment generated by Smg6-mediated endonucleolytic cleavage becomes trapped in an mRNP that includes hUpf1 and is resistant to exonucleolytic decay from the 5’ end.
The 3’ NMD intermediate accumulates in PBs in the presence of ATPase-deficient Upf1
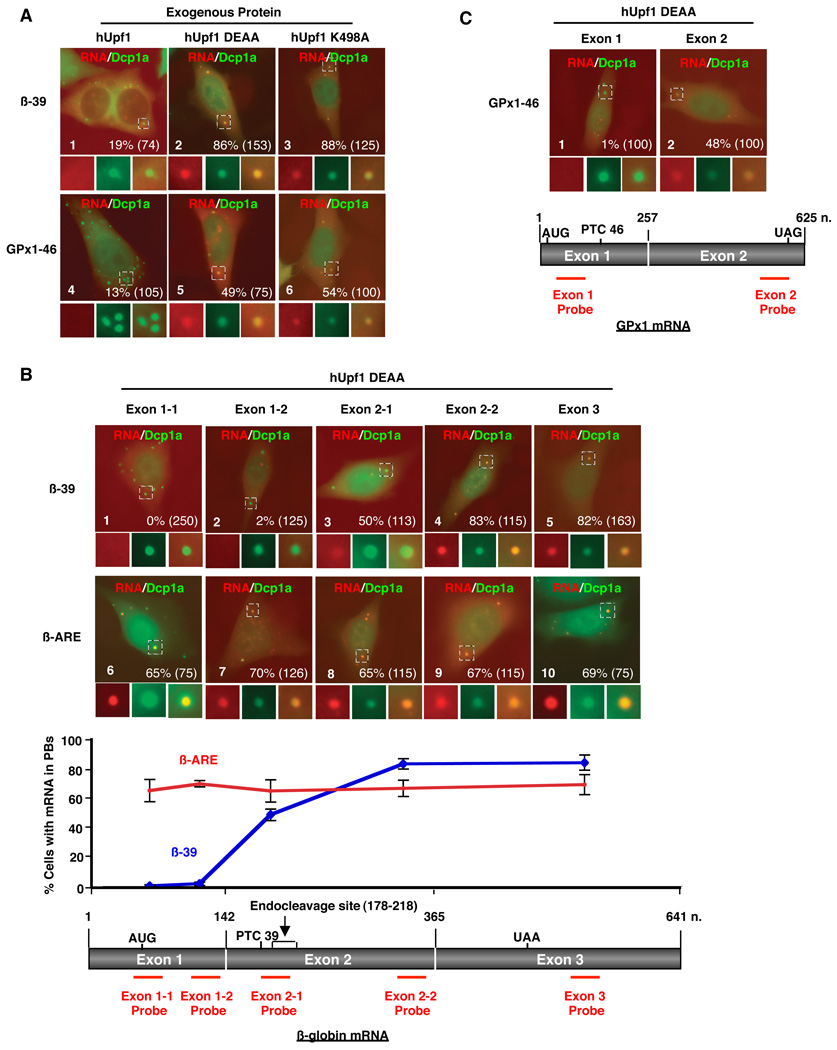
A number of studies have demonstrated that cytoplasmic mRNPs that accumulate in association with 5’-to-3’ mRNA decay complexes concentrate in PBs (Eulalio et al., 2007; Franks and Lykke-Andersen, 2008; Parker and Sheth, 2007). Thus, if hUpf1 ATPase activity is critical for NMD mRNP disassembly during mRNA decay, it is predicted that the NMD intermediate should accumulate in PBs when hUpf1 fails to hydrolyze ATP. Indeed, as seen in the fluorescence in situ hybridization (FISH) assays in Figure 3A, both β-39 (panels 2 and 3) and GPx1-46 (panels 5 and 6) mRNAs accumulate strongly in PBs in the presence of ATPase-deficient hUpf1, but are rarely detected when wild-type hUpf1 is expressed (panels 1 and 4). This is consistent with previous observations in yeast (Sheth and Parker, 2006). By contrast, wild-type β-globin mRNA accumulated only at very low levels in PBs upon mutant hUpf1 expression (Figure S3).
Figure 3. The 3’ NMD intermediate accumulates in PBs in the presence of ATPase-deficient hUpf1.
(A), (B) and (C) FISH assays showing localization of β-39, β-ARE and GPx1-46 mRNAs in HeLa cells in which endogenous hUpf1 was replaced with exogenous hUpf1, hUpf1 DEAA or hUpf1 K498A as indicated above the panels. Individual Texas-Red-labeled 50-nt probes targeting various regions of β-globin and GPx-1 mRNAs were used in (B) and (C) as indicated below images, whereas equimolar amounts of all probes were used in the experiments in (A). GFP-hDcp1a was used as a PB marker. Merged images are displayed (RNA:red, GFP-hDcp1a:green) while selected enlarged regions are shown unmerged below. Percentage of cells displaying mRNA signal in PBs is shown in the bottom right corner of images (with the number of cells counted from at least three experiments in parentheses), and graphed for individual probes against β-39 or β-ARE mRNA below cell images in (B). Note: plasmids that express β-39, β-ARE and GPx1-46 mRNAs also express GFP; thus some nuclear staining can be observed in the green channel.
See also Figure S3.
We next used individual probes hybridizing to different regions along the β-globin mRNA to ask which part of the NMD substrate accumulates in PBs. Remarkably, while the region 3’ of the PTC of β-39 mRNA was readily detectable in PBs in the presence of ATPase-deficient hUpf1, the 5’ end remained completely undetectable in PBs (Figure 3B, compare panels 4 and 5 with panels 1 and 2; quantifications below), despite the fact that the full-length mRNA under these conditions is 6- to 10-fold more abundant than the 3’ fragment (Figures 1A and 1B). A probe that hybridizes across the mapped Smg6 endonucleolytic cleavage sites (Eberle et al., 2009) modestly detects the mRNA in PBs (panel 3). The observed differences in PB detection are not due to different efficiencies of the FISH probes, since in contrast to the β-39 mRNA, each FISH probe equally detected in PBs a β-globin mRNA targeted for the ARE-mRNA decay pathway (β-ARE) (compare panels 6–10 with panels 1–5; quantifications below). The observed localization pattern is not unique to the β-39 mRNA, as in the presence of ATPase-deficient hUpf1 the 3’ end of an unrelated NMD substrate, GPx1-46, could also be observed in PBs in contrast to its 5’ end (Figure 3C). Thus the 3' NMD mRNA decay intermediate that accumulates when Upf1 fails to hydrolyze ATP forms an mRNP that concentrates in PBs.
Multiple NMD factors accumulate in PBs in the absence of Upf1 ATPase activity
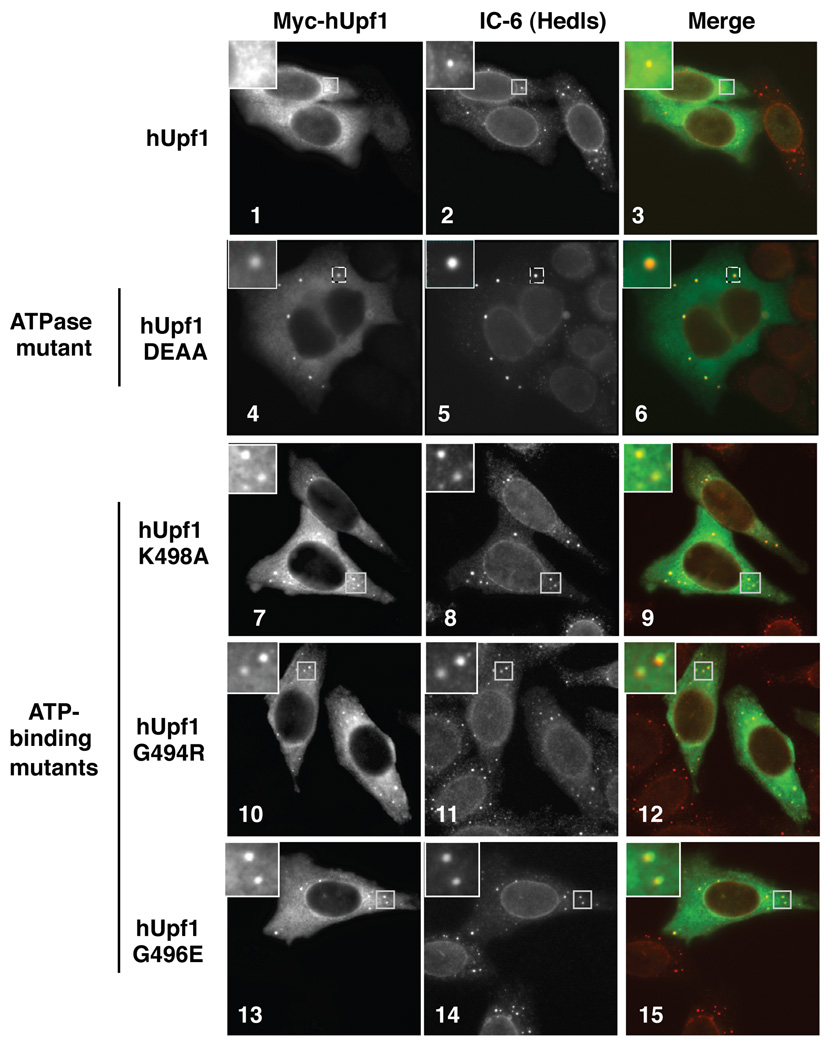
What are the protein components of the accumulating 3’ NMD mRNP intermediate? Based on the observations above, such proteins are predicted, 1) to accumulate in PBs in the presence of ATPase-deficient Upf1, 2) to co-purify more strongly with ATPase-deficient Upf1 than with wild-type Upf1 in co-immunoprecipitation (co-IP) assays, and 3) to co-purify the NMD mRNA 3’ fragment when immunoprecipitated. We tested these predictions for multiple NMD factors. Consistent with hUpf1 being part of the 3’ NMD mRNP and with previous observations in yeast and human cells (Sheth and Parker, 2006; Cheng et al., 2007; Cho et al., 2009; Stalder and Muhlemann, 2009), indirect immunofluorescence assays revealed that ATP binding- and ATPase-deficient mutant hUpf1 proteins, but not wild-type hUpf1, accumulate strongly in PBs (Figure 4, compare panels 4, 7, 10, 13 to panel 1). This is consistent with the observation that ATPase activity is required for the release of hUpf1 from the degrading NMD mRNP (Figure 2A).
Figure 4. Mutant hUpf1 proteins deficient in ATP binding or ATP hydrolysis accumulate in PBs.
Indirect immunofluorescence assays showing localization of myc-tagged wild-type hUpf1, ATPase mutant (DEAA), or ATP-binding mutants (K498A, G494R, G496E) hUpf1 proteins transiently expressed in HeLa cells (left panels). Human IC-6 serum, which detects the decapping factor Hedls and the nuclear envelope component Lamin, was used as a PB marker (middle panels). Merged images (hUpf1: green; IC-6: red) are shown in the right panels. Enlarged images of the indicated boxed areas are shown in the upper left corner for each image.
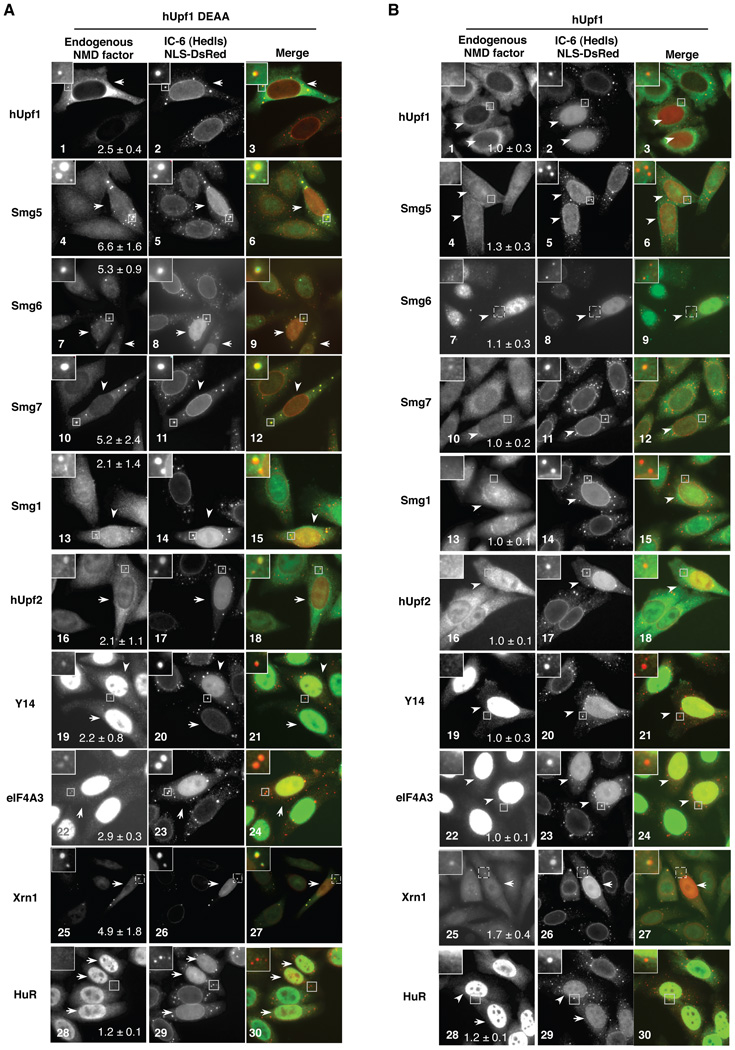
What about other NMD factors? Remarkably, exogenously expressed ATPase-deficient hUpf1 (Figure 5A), but not wild-type hUpf1 (Figure 5B), induces strong accumulation of endogenous Smg5, Smg6 and Smg7 in PBs (panels 4, 7 and 10; transfected cells identified by the co-expression of NLS-DsRed are marked by arrowheads) but has no observable effect on the localization of an unrelated RNA binding protein, HuR (panel 28). Smg1, hUpf2 and the EJC components Y14 and eIF4A3 more modestly accumulate in PBs (panels 13, 16, 19, and 22), whereas hUpf3a and hUpf3b were only rarely observed in PBs (our unpublished observations). None of the NMD factors localized strongly in PBs in untransfected cells (Figures 5A and B; cells not indicated by arrowheads), which in all cases looked similar to those expressing exogenous wild-type hUpf1 (Figure 5B). Similarly to NMD factors, Xrn1 consistently showed enhanced accumulation in PBs in cells expressing ATPase-deficient hUpf1 (Figure 5A, panel 25; cell marked by arrowhead) as compared with cells expressing exogenous wild-type hUpf1 (Figure 5B, panel 25) or no exogenous hUpf1 (Figures 5A and B, panel 25; unmarked cells). Thus, multiple NMD factors and Xrn1 co-accumulate with NMD intermediates in PBs in the presence of ATPase-deficient hUpf1 (Figure 5).
Figure 5. Multiple NMD factors accumulate in PBs in the presence of ATPase-deficient hUpf1.
(A) and (B) Indirect immunofluorescence assays showing localization in HeLa cells of endogenous NMD factors as indicated on the left, or a protein not involved in NMD, HuR, in the presence of exogenously expressed (A) hUpf1 DEAA, or (B) wild-type hUpf1. Middle panels show human IC-6 serum as a PB marker and DsRed with a nuclear localization signal to mark transfected cells (indicated by white arrowheads). Merged images (NMD factor: green; IC-6/NLS-DsRed: red) are shown in right panels. An enlarged cell section representing the boxed area of each image is shown in the upper left corner. The average enrichment of the protein factor in PBs over the general cytoplasm was quantified in transfected cells and given with standard deviation in each of the panels on the left.
ATPase-deficient hUpf1 shows enhanced co-purification with multiple NMD factors
We next tested the prediction that proteins that require Upf1 ATPase activity for release from the NMD mRNP should co-purify more strongly with ATPase-deficient hUpf1 mutant proteins than with wild-type hUpf1 in co-IP assays. In striking correlation with the immunofluorescence assays above, the co-IP assays in Figure 6A (lanes 1–8), in which cell extracts where not treated with RNase, show strong enrichment of endogenous Smg5 and Smg7, and exogenous Smg6, in hUpf1 ATPase mutant protein complexes as compared with wild-type hUpf1 complexes (compare lanes 3 and 4 with lane 2; see also Figure S4). Other NMD factors, hUpf2, hUpf3b and eIF4A3 show modestly enhanced accumulation with hUpf1 ATPase mutant complexes (Figure 6A), which correlates well with their moderate accumulation in PBs under the same conditions (Figure 5A; our anti-hSmg6 and anti-hSmg1 antibodies failed to detect the endogenous proteins on Western blots). These observations are consistent with previous observations of enhanced association of ATP binding-deficient hUpf1 with Smg7, hUpf3a and hUpf2 (Kashima et al., 2006). β-actin served as a negative control and did not co-purify with wild-type or mutant hUpf1 proteins (Figure 6A, bottom panel), and none of the endogenous NMD factors non-specifically co-purified with the IP resin (lane 1). When the same assays were repeated in the presence of RNase, Smg5 and Smg7 were the only NMD factors that remained enriched in the mutant hUpf1 complexes, suggesting the accumulation of an RNA-independent interaction between these factors when hUpf1 fails to hydrolyze ATP (Figure 6A, lanes 9–16). In addition to NMD factors, both Xrn1 and PABPC1 showed enhanced association with ATPase-deficient hUpf1 proteins over wild-type hUpf1, although this was more evident in the presence than in the absence of RNase-treatment (compare lanes 11,12 to 10 and 3,4 to 2). Unlike Xrn1 and NMD factors, PABPC1 was not observed to concentrate in PBs upon ATPase-deficient hUpf1 expression (our unpublished observations), perhaps due to the high cytoplasmic abundance of PABPC1 overwhelming detection in PBs. Taken together, these observations are consistent with the idea that the hUpf1 ATPase stimulates disassembly of the NMD mRNP. However, some NMD factors show stronger accumulation than others in the trapped mRNP complexes (Figures 5 and 6).
Figure 6. Multiple NMD factors accumulate in complex with ATPase-deficient hUpf1 and the NMD substrate 3’ fragment.
(A) Western blots for the proteins indicated on the left from pellet (left panels) or 2% of total extract (right panels) fractions from anti-myc IP assays from HEK 293T cells transiently expressing proteins shown on the top, or no exogenous protein (none). Cell extracts in lanes 9–16 were treated with RNase A prior to IP.
(B) Northern blots for β-39 mRNA isolated from pellets (IP; top panels) or 5% total extract (Total; bottom panels) fractions from immunoprecipitation reactions for tagged exogenous, or in the case of hUpf2, endogenous, NMD, EJC or PABPC1 factors, as shown on the top, in the presence of co-expressed wild-type (WT), or ATPase-deficient (DEAA) hUpf1. Endogenous hUpf1 and Xrn1 were knocked down using siRNAs. (−) indicates a reaction using anti-HA beads in the absence of HA-tagged protein. Anti-FLAG and anti-Myc beads looked similar (not shown). Below each panel is shown the calculated enrichment of the 3’ fragment relative to full-length β-39 mRNA in IP pellets in the presence of mutant hUpf1 (DEAA) over that in the presence of wild-type hUpf1. Representative of three independent experiments.
See also Figure S4.
NMD factors are associated more strongly with the 3’ NMD fragment in the presence of ATPase-deficient Upf1
Finally, to test whether NMD factors can be directly observed in complex with the NMD 3’ intermediate, individual NMD factors were immunoprecipitated from cells depleted of Xrn1 and expressing the β-39 NMD substrate as well as exogenous wild-type or ATPase-deficient hUpf1 in place of endogenous hUpf1. As seen in the Northern blots in Figure 6B, all tested NMD factors, EJC components and PABPC1 are found in complex with the 3’ NMD intermediate (upper panels, band marked by arrow). For the tested NMD factors, the association with the 3’ intermediate relative to that of full-length β-39 mRNA was enhanced 2.1- to 6.2-fold in the presence of ATPase-deficient over wild-type hUpf1 (quantifications shown below blots). By contrast, EJC components and PABPC1 showed little or no difference in their association with the 3’ fragment whether or not hUpf1 can hydrolyze ATP (right panels). These observations suggest that NMD factors are released from the 3’ fragment by the action of the Upf1 ATPase, whereas release of EJC components and PABPC1 appear to require Xrn1 activity.
Discussion
The Upf1 ATPase allows NMD mRNP disassembly
Here we have provided several lines of evidence that ATP hydrolysis by Upf1 is critical for the disassembly and completed degradation of mRNPs undergoing NMD (Figure 7). First, mutant Upf1 proteins unable to bind or to hydrolyze ATP cause impaired degradation of NMD substrates and accumulation of a 3’ intermediate (Figure 1). Second, the 3’ intermediate (Figure 2A) and multiple NMD factors (Figure 6) accumulate in complex with Upf1 when it fails to bind or hydrolyze ATP. Third, the NMD mRNA intermediate (Figure 3) and multiple NMD factors (Figures 4 and 5) accumulate in PBs in the presence of ATP binding- or ATPase-deficient mutant Upf1. The accumulation of the 3’ intermediate in the presence of ATPase-deficient Upf1 is likely a result of the inability of Xrn1 to degrade the RNA in the absence of mRNP disassembly (Figure 7). Consistent with this, Xrn1 appears to be trapped with the NMD mRNP that accumulates upon expression of ATPase-deficient Upf1, as evidenced by the enhanced association of Xrn1 with Upf1 complexes and with PBs under those conditions (Figures 5A and 6) (Cho et al., 2009; Isken et al., 2008). Moreover, the 3’ NMD mRNP generated in the presence of ATPase-deficient Upf1 is resistant to 5’-to-3’ exonucleolytic decay in vitro unless protein is first removed by phenol extraction (Figure 2C). It is unclear where on the accumulating 3’ NMD mRNP that Upf1 and the NMD complex are positioned. Site-specific RNase H cleavage followed by IP-Northern assays indicated that ATPase-deficient Upf1 is associated with both 5’ and 3’ fragments of the β-39 NMD 3’ mRNP (our unpublished observations), perhaps reflecting interactions of Upf1 with the EJC and PABPC1 (Figure 6A) as well as directly with the RNA. Based on our observations, a simple hypothesis for why NMD shuts down in the presence of ATPase-deficient Upf1 (Figure 1A) (Kashima et al., 2006; Weng et al., 1996a, b) is that the entrapment of NMD factors on partially degraded NMD mRNPs renders the pathway non-catalytic due to the failure of NMD factor recycling. Alternatively, the Upf1 ATPase could be rate-limiting for a more upstream mRNP remodeling step, in which case the accumulation of full-length NMD substrate and 3’ intermediates in the presence of ATPase-deficient Upf1 reflects a stronger defect in 5’-to-3’ decay than in endonucleolytic cleavage. The effect of the Upf1 ATPase on other mRNA decay activities triggered by NMD, decapping and deadenylation, remains to be tested. In either case, our studies illustrate the importance of mRNP disassembly in mRNA turnover.
Figure 7. mRNP disassembly during NMD.
How mRNP disassembly dependent on Upf1 ATPase activity is required for completion of NMD and recycling of NMD factors. See Discussion for details.
While most NMD factors accumulate in PBs (Figure 5) and in association with Upf1 (Figure 6) when Upf1 fails to hydrolyze ATP, Smg5, Smg6 and Smg7 show stronger accumulation than do Upf2, Upf3 and EJC proteins. These weaker associated NMD proteins may either be more loosely associated with the NMD mRNP intermediate, are found at lower stoichiometry in the complex, or are found only on a subset of substrates that require Upf1 ATPase activity for mRNP disassembly. Consistent with the latter idea, Upf1 has been implicated independently of Upf2 and Upf3 in the degradation of mRNAs other than NMD substrates, including histone mRNAs (Kaygun and Marzluff, 2005) and mRNAs associated with Staufen (Kim et al., 2005). Moreover, evidence has been presented for Upf2-, Upf3- and EJC-independent NMD pathways in human cells (Bühler et al., 2006; Chan et al., 2007; Gehring et al., 2005). The relatively weak accumulation of EJC components could also be a result of EJC disassembly by the recently discovered EJC disassembly activity of the protein PYM (Gehring et al., 2009).
The mechanism by which the Upf1 ATPase leads to NMD mRNP disassembly remains to be determined. Upf1 could act as a processive RNPase that uses ATPase activity to traverse the mRNA while displacing NMD factors and other RNA-binding proteins from the NMD substrate (Fairman et al., 2004; Jankowsky and Bowers, 2006). Alternatively, Upf1 could remain stationary and hydrolyze ATP to release itself and other associated factors from the mRNA (Ballut et al., 2005). Yet another possibility is that ATP hydrolysis by Upf1 acts upstream of a chain of mRNP remodeling events that in the end lead to NMD mRNP disassembly. The observations that Upf1 has highest affinity for RNA in the absence of ATP and shows ATP-dependent 5’ to 3’ RNA translocation activity in vitro (Cheng et al., 2007; Weng et al., 1998), favor the former possibility. However, the observation that the level of NMD intermediate associated with PABPC1 and EJC components, in contrast to NMD factors, is independent of Upf1 ATPase activity (Figure 6B), suggests that these factors are not released directly by the Upf1 ATPase, but rather at a downstream step, perhaps by the activity of Xrn1 (Figure 7). Either way, our observations demonstrate a previously unappreciated step in mRNA decay by which mRNP disassembly allows the completion of exonucleolytic decay and the recycling of mRNP components. The specific mRNP components responsible for blockage of exonucleolytic decay of the NMD substrate in the presence of ATPase-deficient Upf1 remain to be determined. Possible candidates could be the NMD factors themselves or, perhaps, unreleased ribosomes or ribosomal subunits.
Is mRNP disassembly a regulated step in NMD?
Taken together, our observations uncover a previously unappreciated ATP-dependent mRNP disassembly step in mRNP turnover. A key question is what controls the timing of mRNP disassembly in NMD, as slow disassembly would cause accumulation of decay intermediates whereas rapid disassembly could potentially release the NMD mRNP even before it initiates decay. The ATPase activity of human Upf1 is stimulated by the Upf2-Upf3 complex (Chamieh et al., 2008), and the yeast Upf1 ATPase is repressed by translation release factors eRF3 and eRF1 (Czaplinski et al., 1998). Thus, a transition in the NMD mRNP in which Upf1 is released from eRFs and associates with Upf2-Upf3 may precede activation of the Upf1 ATPase and subsequent mRNP disassembly. Consistent with this, ATP binding-deficient Upf1 has been observed to co-purify less efficiently than wild-type Upf1 with eRF1 and eRF3 (Czaplinski et al., 1998; Kashima et al., 2006; Isken et al., 2008), suggesting that it becomes trapped in a complex lacking eRFs. Moreover, analyses of NMD complexes stalled by NMD factor mutation or depletion have indicated a transition in the human NMD mRNP from a complex between Upf1, Smg1 and eRFs (called SURF) to a complex of NMD factors lacking eRFs (called DECID) (Kashima et al., 2006). In addition, the phosphorylation and dephosphorylation of metazoan Upf1 seems to be coordinated with the Upf1 ATPase, as ATPase-deficient Upf1 accumulates in a hyperphosphorylated form (Isken et al., 2008; Kashima et al., 2006; Page et al., 1999), which has been reported to prevent translation re-initiation on the NMD mRNP (Isken et al., 2008).
Why would mRNP disassembly be under such tight control during NMD? This could possibly ensure that NMD factors are released only after mRNA decay factors have already been recruited and/or mRNA decay initiated. This also raises the possibility that ATPase-mediated mRNP disassembly could serve as a previously proposed proof-reading step in the NMD pathway (Sheth and Parker, 2006), in which rapid hydrolysis of ATP by Upf1 would allow the release of the NMD machinery from the mRNA even before initiation of mRNA decay, thus allowing mRNAs wrongly targeted for NMD to be released prior to decay (Figure 7). Several lines of evidence suggest that the composition of the mRNP downstream of the translation termination event controls NMD (Amrani et al., 2006; Muhlemann, 2008; Rebbapragada and Lykke-Andersen, 2009). A key question for future studies is whether the downstream mRNP controls NMD not just by NMD factor recruitment as has been generally assumed, but also in part by regulating the Upf1 ATPase.
Is mRNP disassembly critical for mRNA turnover pathways other than NMD?
Another important question for future studies is whether mRNP disassembly is a critical step in mRNA decay pathways other than NMD. There have been several observations of mRNA and mRNP structures impairing exonucleolytic decay. For example, in S. cerevisiae, both 5’-to-3’ and 3’-to-5’ exonucleolytic decay is impaired by strong RNA secondary structures (Vreken and Raue, 1992; Decker and Parker, 1993; Muhlrad et al., 1995), and 5’-to-3’ exonucleolytic decay is inhibited by ribosomes stalled by cycloheximide or by rare codons (Beelman and Parker, 1994; Cereghino et al., 1995; Hu et al., 2009). In C. elegans, 5’-to-3’ decay intermediates of lin-41 mRNA targeted by let-7 microRNA have been observed with the 5’ end mapping immediately upstream of the let-7 binding sites (Bagga et al., 2005). Even a heterologous RNA binding protein, the MS2 coat protein, appears capable of stalling 5’-to-‘3 exonucleolytic decay in C. elegans (Liu et al., 2003). In addition to exonucleolytic decay, PABPC and the cap-binding protein, eIF4E, can inhibit initiation of mRNA decay by deadenylation and decapping, respectively (Schwartz and Parker, 2000; Tucker et al., 2002). Thus, disassembly of the mRNP is likely to be a critical step in both the initiation and the completion of mRNA turnover. Future studies should reveal the extent to which helicases are involves in these processes. Several helicase proteins have been identified in association with mRNA decay enzymes, including Rck/p54 of the decapping complex and Ski2 of the exosome (Anderson and Parker, 1998; Coller et al., 2001; Fischer and Weis, 2002; Fenger-Grøn et al., 2005), as well as in association with bacterial and mitochondrial exonucleases (Carpousis, 2009). Future studies should reveal whether such helicases are important for mRNP disassembly to allow for processivity of their associated mRNA decay enzymes, and whether pathway-specific mRNP disassembly factors are common in mRNA turnover pathways in addition to NMD.
Experimental Procedures
mRNA Decay and RNA Immunoprecipitation Assays
Expression of NMD reporter β-39 or GPx1-46 mRNAs was induced for 6 hours by incubation in tetracycline-free medium of HeLa Tet-off cells, depleted of endogenous hUpf1 and/or Xrn1 using siRNAs, and transiently transfected with plasmids expressing tetracycline-regulated β-39 or GPx1-46 mRNAs, and constitutively expressed control β-GAP mRNAs (Figure 1 only), as well as plasmids expressing siRNA-resistant wild-type or mutant (DEAA or K498A) hUpf1 protein, and in Figure 6B, other tagged NMD factors as indicated (see Supplemental Experimental Procedures for details). In endogenous mRNA decay assays (Figure 1), total RNA was prepared from cells using Trizol reagent (Invitrogen), 0, 2, 4, or 6 hours after addition of 1 µg/ml tetracycline to repress NMD reporter mRNA transcription. In in vitro decay assays mediated by Terminator (Figure 2C), cell extracts prepared in hypotonic gentle lysis buffer, or total RNA prepared from extracts using Trizol, were incubated with Terminator 5’-to-3’ exonuclease (Epicentre) for 0, 5, 10, 20 or 40 minutes followed by RNA preparation using Trizol. In RNA-immunoprecipitation assays (Figures 2A,B and 6B), cell extracts prepared in isotonic lysis buffer were subjected to immunoprecipitation against the indicated NMD factors, and RNA from immunoprecipitated samples was isolated using Trizol. NMD substrate levels were analyzed by Northern blotting.
Indirect Immunofluorescence and Fluorescence In Situ Hybridization (FISH) Assays
Human HeLa cells transiently expressing wild-type or mutant myc-tagged hUpf1 proteins were fixed with formaldehyde and permeabilized with Triton-X100 (Figures 4 and 5) or ethanol (Figure 3). For indirect immunofluorescence assays, cells were incubated with antibodies against Myc-tag (Figure 4) or against endogenous NMD factors, Xrn1 or HuR (Figure 5) as well as with human IC-6 serum, which recognizes endogenous Hedls (P-body marker) and Lamin, followed by fluorescently labeled secondary antibodies (anti-mouse or -rabbit: Alexa 488; anti-human: Texas Red). Cells in Figure 5 express nuclear DsRed to mark transfected cells. For FISH assays, cells were hybridized with a mixture of (Figures 3A, C), or individual (Figure 3B), TexasRed-5’-labeled 50-nucleotide NMD substrate mRNA antisense DNA probes. Cells for FISH assays express GFP-tagged hDcp1a to mark P-bodies (see Supplemental Experimental Procedures for details).
Co-Immunoprecipitation Assays
Lysates from HEK 293T cells transiently expressing Myc-tagged wild-type or mutant (DEAA or K498A) hUpf1 were subjected, in the presence or absence of RNase A, to anti-Myc immunoprecipitation followed by Western blotting for endogenous NMD factors, Xrn1, PABPC1 or β-actin, or in the case of Smg6, co-expressed HA-tagged Smg6.
Supplementary Material
Acknowledgments
We thank Drs. Tom Blumenthal (University of Colorado), Melissa Moore (University of Massachusetts Medical Center) and Sebastien Durand (UCSD) for comments on the manuscript. Alex Choe and Claire Egan are thanked for technical support and Joachim Weischenfeldt for production of the antigen for anti-Smg1 antibodies. Drs. Marv Fritzler, Ed Chan and Donald Bloch are thanked for human IC-6 serum. Dr. Oliver Mühlemann is thanked for the HA-Smg6 construct. Work on P-bodies in our laboratory is supported by funding from grant R01 GM077243 from the National Institutes of Health to J.L.-A. T.M.F. has been supported by National Institutes of Health NRSA Institutional Training Grant no. GM-07135 from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AP. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3’UTR length. Nat Struc Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: Annotating Chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2009;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Cereghino GP, Atencio DP, Saghbini M, Beiner J, Scheffler IE. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisia: Sequence elements in the 5’ untranslated region of the Ip mRNA play a dominant role. Mol Biol Cell. 1995;6:1125–1143. doi: 10.1091/mbc.6.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of posttranscriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D "RNA helicases" without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kuersten S, Huang T, Larsen A, MacMorris M, Blumenthal T. An uncapped RNA suggests a model for Caenorhabditis elegans polycistronic pre-mRNA processing. RNA. 2003;9:677–687. doi: 10.1261/rna.2128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O. Recognition of nonsense mRNA: towards a unified model. Biochem Soc Trans. 2008;36:497–501. doi: 10.1042/BST0360497. [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Lykke-Andersen J. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 2010;7:28–32. doi: 10.4161/rna.7.1.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schwarz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J. Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol Cell. 2007;27:780–792. doi: 10.1016/j.molcel.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA. 2009;15:1265–1273. doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P, Raué HA. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3’-terminal part of the coding region. Mol Cell Biol. 1992;12:2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996a;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996b;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. RNA stability: is it the endo' the world as we know it? Nat Struct Mol Biol. 2009;16:9–10. doi: 10.1038/nsmb0109-9. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.