Abstract
The first synthesis of 1-deaza-pyridoxal 5’-phosphate (2-formyl-3-hydroxy-4-methylbenzyl phosphate) is described. The chemoenzymatic approach described here is a reliable route to this important isosteric pyridoxal phosphate analogue. This work enables elucidation of the role of the pyridine nitrogen in pyridoxal 5’-phosphate dependent enzymes.
Pyridoxal 5’-phosphate (PLP) dependent enzymes comprise more than 140 enzyme commission numbers.1 They catalyze a wide variety of reactions including decarboxylation, racemization, transamination, β elimination and retro aldol cleavages.1,2,3 Yet despite this versatility, mechanistic similarities are found. All reactions diverge from the common “external aldimine” intermediate1 (Figure 1) formed by the displacement of a lysine residue (which forms the “internal aldimine”) by the amino group of the amino acid substrate. Most reactions are thought to proceed through formation of the “quinonoid” intermediate,1,2,3 a Cα carbanion stabilized by delocalization with the conjugated π system of the cofactor (Figure 1). Indeed, the protonated pyridine nitrogen of the cofactor acting as an electron sink in carbanion stabilization is considered to be the hallmark of PLP catalyzed reactions.2
Figure 1.
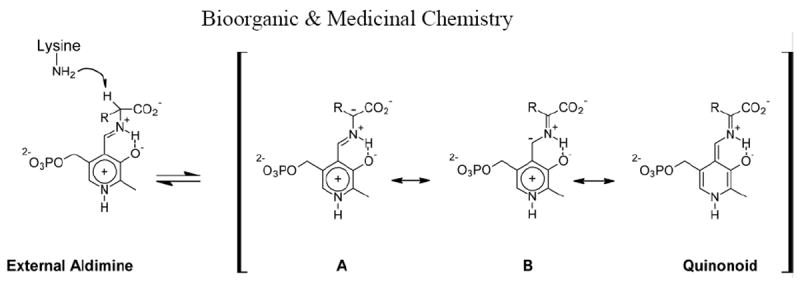
Carbanion formation and stabilization by PLP.
Recently, however, both experimental and computational studies call into question the crucial importance of the protonated pyridine nitrogen.1,4a,b,c The structures of several enzyme active sites preclude protonation of the pyridine nitrogen.4d,e Alternatively, the protonated Schiff base, through electrostatic stabilization of the carbanion, may play a central role (Figure 1, structures A and B).1
Studies of catalysis by PLP analogues are a direct experimental approach to this fundamental debate. At one extreme the pyridine nitrogen of PLP and pyridine-based PLP analogues may be quaternized, (1, Figure 2), enforcing a positive charge. At the other extreme are carbocyclic salicylaldehyde analogues (3, Figure 2), which have neither the potential for protonation nor the greater electronegativity of nitrogen (vs. carbon) in the aromatic ring. The charge of the pyridine nitrogen on PLP itself (2, Figure 2) depends on pH (non-enzymatic) and the acidity of active site residue(s) interacting with it (enzymatic).
Figure 2.
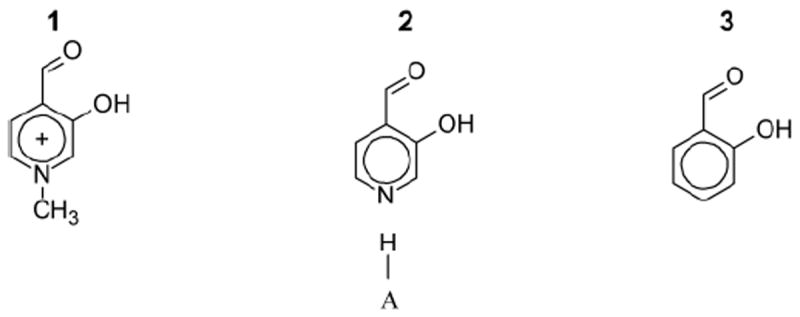
Simplest pyridine and salicylaldehyde based analogues of PLP. H—A in structure 2 denotes that the charge of the pyridine nitrogen may vary by pH (non-enzymatic conditions) and the acidity of active site residue(s) interacting with it (enzymatic conditions).
Non-enzymatic transamination5 and racemization6 catalyzed by the above mentioned analogues have been studied. However, the constraints of active sites optimized for PLP preclude the use of analogues structurally dissimilar from PLP in enzyme catalyzed reactions. While N-methyl PLP, which contains a quaternized pyridine nitrogen, has a long history of enzymatic study, 1,7 the lack of an isosteric salicylaldehyde analogue of PLP (“deazaPLP”; 2-formyl-3-hydroxy-4-methylbenzyl phosphate; 8, Scheme 1) has hampered investigation of the role of the pyridine nitrogen in enzymatic reactions. Surprisingly, the literature contains few attempts at obtaining deazaPLP8 and, to the best knowledge of the authors, until now there has been no successful synthesis reported.
Scheme 1.
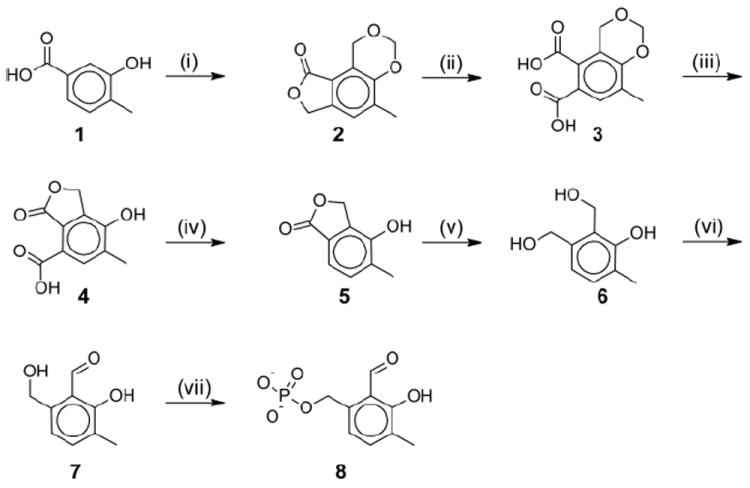
Reagents and conditions: (i) HCL, formaldehyde, 80°C; (ii) KOH, KMnO4, 5-10°C; (iii) HCL, reflux; (iv) copper chromite, quinoline, 180°C; (v) borane dimethyl sulfide, THF, reflux; (vi) MnO2, ethyl acetate, room temp.; (vii) ATP, MgCl2, pyridoxal kinase.16
The general strategy employed here in the synthesis of deazaPLP centers on oxidation and phosphorylation of triol 6 (Scheme 1) to the final product. Therefore a reliable route to 6 was sought out. Reduction of lactone 5 would provide the desired compound. A search of the literature revealed a previous published synthesis9 which proved to be a reliable route to lactone 5. Steps (i)-(iv) were carried out as described in the original article except as noted.16 All yields proved to be close to those published with the exception of 2, which provided only half the expected product.
Reduction of 5 with borane dimethyl sulfide provided triol 6 with yields of ~50%. Attempts to obtain 6 by treatment of 5 with LiBH4 proved unsuccessful. Phosphorylation of triol 6 by pyridoxal kinase10,11 proved to be a facile and reliable route to the phosphorylated triol with yields typically at 60-70%. Surprisingly, oxidation of the phosphorylated triol by manganese dioxide to the final product was not successful.
The presence of the negatively charged phosphate group precluded the use of organic solvents for oxidation of the phosphorylated triol. On the other hand, water is a suboptimal solvent choice for manganese dioxide oxidations due to competition from solvent for adsorption to manganese dioxide12 and the propensity for over-oxidation to the acid. The obvious alternative was to oxidize triol 6 to 7, followed by phosphorylation to 8. Oxidation to the desired product occurred readily, but only with freshly prepared manganese dioxide.13 Commercially available manganese dioxide was ineffective and decreases in yield were noted with the use of manganese dioxide over one week old. Yields for 7 were 10-20%.
Phosphorylation of 7 was carried out with pyridoxal kinase, with yields similar to those with 6, although removal of contaminating manganese with cation exchange resin was necessary. The overall yield for 8 was 0.2 %.
While the presence of two benzylic alcohols presented the complication of a potential mixture of oxidation products13 from step (vi), comparison of the UV-vis absorption spectra of salicylaldehyde and 3-hydroxybenzaldehyde with deazaPLP indicated that the desired isomer was obtained. Successful enzymatic phosphorylation of 7 further supports this conclusion. Final confirmation of the structure of 8 was made by 1D nOe NMR experiments14 (see supplemental material for NMR and mass spectra).
To test the enzymatic binding properties of deazaPLP, apo aspartate aminotransferase was prepared according to literature methods15 and reconstituted with deazaPLP in a split-cell cuvette to give difference absorption spectra due to deazaPLP binding. The resulting spectra vs. time show a decrease in absorbance at ~350nm and an increase in absorbance at ~ 420 nm due to the bathochromic shift on internal aldimine formation (Figure 3). Fluorescence quenching (280nm/340nm) titrations of apo aspartate aminotransferase with deazaPLP indicate the dissociation constant to be less than 1 μM (data not shown).
Figure 3.
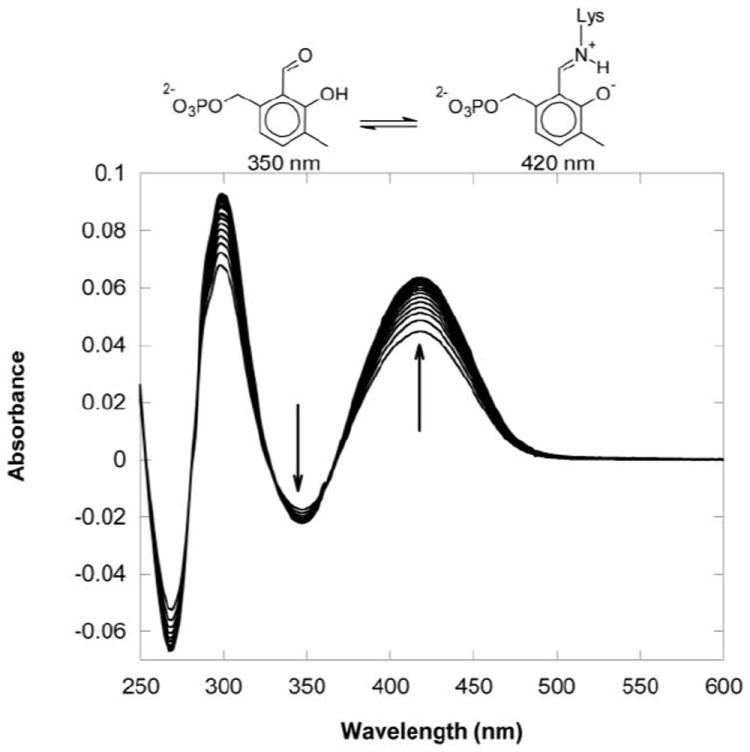
Difference UV-vis spectra vs. time (over ~30 min) showing formation of the internal aldimine of aspartate aminotransferase with deazaPLP.
In summary, a reliable synthesis of the isosteric, carbocyclic analogue of PLP (8) has been described. It binds tightly to aspartate aminotransferase and forms the internal aldimine with the active site lysine. The availability of this analogue will contribute to the ongoing debate concerning the source of the catalytic prowess of PLP enzymes.
Supplementary Material
Acknowledgments
This work was supported by Grant GM54779 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Toney MD. Arch Biochem Biophys. 2005;433:279. doi: 10.1016/j.abb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 2.Eliot AC, Kirsch JF. Annu Rev Biochem. 2004;73:383. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi H. J Biochem. 1995;118:463. doi: 10.1093/oxfordjournals.jbchem.a124931. [DOI] [PubMed] [Google Scholar]
- 4.(a) Bach RD, Canepa C, Glukhovtsev MN. J Am Chem Soc. 1999;121:6542. [Google Scholar]; (b) Richards JP, Amyes TL, Crugeiras J, Rios A. Curr Opin Chem Biol. 2009;13:475. doi: 10.1016/j.cbpa.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Major DT, Nam K, Gao J. J Am Chem Soc. 2006;128:8114. doi: 10.1021/ja062272t. [DOI] [PubMed] [Google Scholar]; (d) Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. J Biol Chem. 1988;263:17857. [PubMed] [Google Scholar]; (e) Shaw JP, Petsko GA, Ringe D. Biochemistry. 1997;36:1329. doi: 10.1021/bi961856c. [DOI] [PubMed] [Google Scholar]
- 5.(a) Auld DS, Bruice TC. J Am Chem Soc. 1967;89:2098. [Google Scholar]; (b) Dixon DE, Bruice TC. Biochemistry. 1973;12:4762. doi: 10.1021/bi00747a031. [DOI] [PubMed] [Google Scholar]; (c) Ikawa M, Snell EE. J Am Chem Soc. 1954;76:653. [Google Scholar]; (d) Maley JR, Bruice TC. Arch Biochem Biophys. 1970;136:187. doi: 10.1016/0003-9861(70)90340-1. [DOI] [PubMed] [Google Scholar]
- 6.(a) Ando M, Emoto S. Bull Chem Soc Jpn. 1969;42:2628. [Google Scholar]; (b) Dixon DE, Bruice TC. Biochemistry. 1973;12:4762. doi: 10.1021/bi00747a031. [DOI] [PubMed] [Google Scholar]; (c) Olivard J, Metzler DE, Snell EE. J Biol Chem. 1952;199:669. [PubMed] [Google Scholar]; (d) Weinstein GN, O’Connor MJ, Holm RH. Inorg Chem. 1970;9:2104. [Google Scholar]
- 7.(a) Gong J, Hunter GA, Ferreira GC. Biochemistry. 1998;37:3509. doi: 10.1021/bi9719298. [DOI] [PubMed] [Google Scholar]; (b) Onuffer JJ, Kirsch JF. Protein Eng. 1994;7:413. doi: 10.1093/protein/7.3.413. [DOI] [PubMed] [Google Scholar]; (c) Yano T, Hilnoue Y, Chen VJ, Metzler DM, Miyahara I, Hirotsu K, Kagamiyama H. J Mol Biol. 1993;234:1218. doi: 10.1006/jmbi.1993.1672. [DOI] [PubMed] [Google Scholar]
- 8.Oseledchik VS, Karpeiskii MYa, Florent’ev VL. Bull Acad Sci USSR. 1973;22:1271. [Google Scholar]
- 9.Charlesworth EH, Anderson HJ, Thompson NS. Can J Chem. 1953;31:65. [Google Scholar]
- 10.Pyridoxal kinase was a gift from Professors Verne Schirch and Martin Safo at Virginia Tech University.
- 11.diSalvo ML, Schirch V. Protein Expression Purif. 2004;36:300. doi: 10.1016/j.pep.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Smith MB. Organic Synthesis. McGraw-Hill; New York: 2002. [Google Scholar]
- 13.Constantinides I, Macomber RS. J Org Chem. 1992;57:6063. [Google Scholar]
- 14.Crews P, Rodriguez J, Jaspers M. Organic Structure Analysis. Oxford University Press; New York: 1998. [Google Scholar]
- 15.Toney MD, Kirsch JF. Biochemistry. 1991;30:7461. doi: 10.1021/bi00244a014. [DOI] [PubMed] [Google Scholar]
- 16.Notes on synthesis. 1: (25 grams) purchased from Alfa Aesar. 2: judged sufficiently pure by 1H NMR to proceed without any purification. 3: purified by anion exchange; acidification of fractions with conc. HCL precipitated out the desired compound. 4: judged sufficiently pure by 1H NMR to proceed without any purification. Step (iv): after extraction with ether, volume reduced by one half and extracted 3 times with 5 mL portions of 1 M KOH. Combined extracts acidified with conc. HCL to precipitate crude product. 5: purified by silica gel: 100% ethyl acetate, Rf = 0.8. Step (v): 2.2 equivalents of borane dimethyl sulfide added to solution of 5 (400 mM) in THF. Addition of borane dimethyl sulfide to THF solution already at 60 °C doubled yields of 6. 6: purified by silica gel (1:1 hexane:ethyl acetate to 100% ethyl acetate), Rf = 0.6 (in 100% ethyl acetate). Step (vi): typically 50-100 mg MnO2 added to 1.5 mL 50-100 mM 6 in ethyl acetate. Vigorously shaken for 20-25 minutes at room temperature. Extracted 3 times with 2-3 mL 100 mM KOH. Treated with cation exchange resin to remove manganese. Concentration of 6 estimated by using extinction coefficient for phenol14 (1500 M-1, cm-1). Step (vii): concentration of 7 adjusted to 1 mM or less (estimated from 344 nm absorbance using extinction coefficient of salicylaldehyde, 3300 M-1 cm-1). Added 4-fold excess of ATP and MgCl2, adjusted solution to pH 8-9 and added pyridoxal kinase to 20 μM. Reaction wrapped in foil to exclude light and stirred gently overnight at room temperature. Passed through a 10 kD cut off filter to remove protein prior to purification. 8: purified by anion exchange: 0-10 minutes, 100% water; 10-100 minutes, 0-20% 1 M NH4HCO3, pH 8.0; flow rate of 3.5 mL/min (column vol ~25 mL). Final product repeatedly lyophilized to remove NH4HCO3. Potassium salt prepared by cation exchange followed by lyophilization. Final product stored at −80 °C.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


