Abstract
A nuclease-resistant RNA enzyme, constructed entirely from L-ribonucleotides, was shown to undergo ligand-dependent, self-sustained replication with exponential growth. The catalytic motif is based on a previously described RNA ligase that can undergo either self- or cross-replication, but had been limited in its application to ligand sensing due to its susceptibility to degradation by ribonucleases. The self-replicating RNA enzyme and its RNA substrates were prepared synthetically from either D- or L-nucleoside phosphoramidites. The D and L reaction systems undergo isothermal, ligand- dependent exponential amplification in the same manner, but only the L system is impervious to ribonucleases and can operate, for example, in the presence of human serum. This system has potential for the quantitative detection of various ligands that are present within biological or environmental samples. In addition, this work provides the first demonstration of the self-sustained exponential amplification of non-biological molecules.
Recently RNA enzymes were devised that undergo self-sustained exponential amplification at a constant temperature and in the absence of any proteins or other biological materials.1 These enzymes subsequently were engineered to operate as “aptazymes” that undergo amplification dependent on the presence of a target ligand.2 The exponential growth rate depends on the concentration of the ligand, enabling one to determine the ligand concentration in an unknown sample. In an alternative reaction format, a non-replicating, ligand-dependent RNA enzyme generates a seed concentration of replicating enzymes that subsequently undergo ligand-independent amplification, thus extending the dynamic range of the system to much lower concentrations of the ligand.3 This system is analogous to quantitative PCR (qPCR) for the detection of target nucleic acids,4,5 but can be generalized to any ligand that can be recognized by an aptamer domain that is linked to the catalytic domain of the replicating enzymes.
Practical application of the ligand-dependent amplification system has been limited by the modest amplification rate of the RNA enzymes and by their instability in biological samples due to rapid degradation by ribonucleases. The ribonucleases must first be removed from the sample, for example by phenol extraction or protease digestion, but such procedures are time consuming and not applicable if the target ligand itself is a protein. The usual approach when employing RNA molecules in a diagnostic or therapeutic context is to replace the standard ribonucleotides with nuclease-resistant nucleotide analogs, typically with a modification at the 2´-hydroxyl position. Care must be taken not to disrupt the functional properties of the RNA, especially when making many substitutions. In the case of the replicating RNA enzymes, which contain ~70 nucleotides and have been optimized with regard to their functional properties, it would be challenging to achieve wholesale substitution without diminishing function.
A synthetic approach provides a different solution to this problem, which is to construct the entire RNA replication system as its enantiomeric twin. The enantiomeric system would have the same catalytic properties, but being composed of non-biological L-RNA would be completely resistant to ribonucleases. The principle of utilizing L-RNA to achieve nuclease resistance has already been applied to L-aptamers, termed “Spiegelmers”, which have been selected to bind various biological proteins and thereby block the protein’s function.6–8 L-aptamers have been developed for therapeutic applications, including three compounds that are currently in clinical trials.9–11 The increasing use of L-aptamers, as well as L-ribose derivatives that are employed as antiviral agents,12 have made readily available the L-nucleoside phosphoramidites needed to prepare synthetic L-RNA oligomers.
The RNA self-replication cycle (Figure 1a) involves an RNA enzyme (E) that catalyzes the ligation of two RNA substrates (A and B), one bearing a 3´-hydroxyl and the other a 5´-triphosphate, to form a ligated product that is identical to the starting enzyme.13 The E•E complex dissociates spontaneously, providing two enzyme molecules to begin the next replication cycle. This doubling process continues at a constant temperature until the supply of substrates is exhausted. If additional substrates are provided, for example by a serial transfer procedure, then exponential growth can be continued indefinitely. The same behavior is expected for either the D or L reaction system because no other chiral components are involved in the replication process. The self-replicating RNA enzyme employed in this study (Figure 1b) differs slightly compared to that described previously,13 containing two mutations that previously were shown to enhance the rate of cross-replication1 and similarly proved beneficial in the self-replication format.
Figure 1.
Self-sustained replication of an RNA enzyme. (a) The self-replication cycle, involving an RNA enzyme (E) that binds two RNA substrates (A and B) and catalyzes their ligation to form a new copy of E. The E•E complex dissociates to provide two copies of E to begin the next replication cycle. (b) Sequence and secondary structure of the E•A•B complex. Curved arrow indicates the site of ligation. Boxed region indicates the central stem-loop that can be replaced by an aptamer domain. The theophylline aptamer domain and the chemical structure of theophylline are shown at the bottom left.
D-RNA polymers can be prepared either enzymatically by transcription of a DNA template or synthetically by solid-phase synthesis, whereas L-RNA polymers can only be prepared synthetically. The D form of the E, A, and B molecules were prepared both enzymatically using T7 RNA polymerase and synthetically using 2´-t-butyldimethylsilyl (TBDMS) RNA phosphoramidites (see Supporting Information). The 2´-triisopropylsilyloxymethyl (TOM) protecting group is preferred for the solid-phase synthesis of long RNA oligomers, but is not commercially available in the L series. Thus to maintain parity with the L-RNA molecules, which were synthesized using TBDMS RNA phosphoramidites, the D-RNA molecules were synthesized in the same manner. There is a requirement for a 5´-triphosphate on the B substrates, which is installed automatically during in vitro transcription, but must be added chemically following solid-phase synthesis. This was done for both the D- and L-molecules using established methods for the 5´-triphosphorylation of mononucleosides,14,15 which were optimized for application to RNA oligomers16,17 (Horning & Joyce, unpublished data). The D and L form of the A substrates were labeled at their 5´-end with hexachlorofluorescein and fluorescein, respectively, to facilitate quantitation of the reaction products.
The D reaction system employing in vitro transcribed components exhibited robust exponential growth. A reaction mixture containing 2.5 µM A, 20 µM B, and 0.25 µM E, which was incubated in the presence of 25 mM MgCl2 at pH 8.5 and 42 °C, showed sigmoidal growth indicative of exponential amplification subject to the finite supply of substrates (Figure S1). The exponential growth rate was 1.1 h−1, corresponding to a doubling time of 38 min, and the reaction reached a maximum extent of 76% after 6 h. Exponential amplification could be initiated even in the absence of starting concentrations of E, albeit with a time lag. The tetramolecular complex of two A and two B molecules is catalytically active and can spontaneously generate E molecules that catalyze subsequent ligations reactions.13
The in vitro transcribed and synthetic D-RNA substrates were interchangeable, although activity was consistently higher with the transcribed form of the A substrate (Figure S2). This may reflect the lower quality of materials resulting from solid-phase synthesis of the 52-nucleotide RNA. In contrast, the synthetic form of the B substrate exhibited slightly higher activity compared to the in vitro transcribed form. The B substrate contains only 14 nucleotides, which is easily accessible by solidphase synthesis using TBDMS phosphoramidites. The in vitro transcribed materials may contain sequence heterogeneity at the reactive 5´-terminus,18,19 which is not the case for synthetic materials.
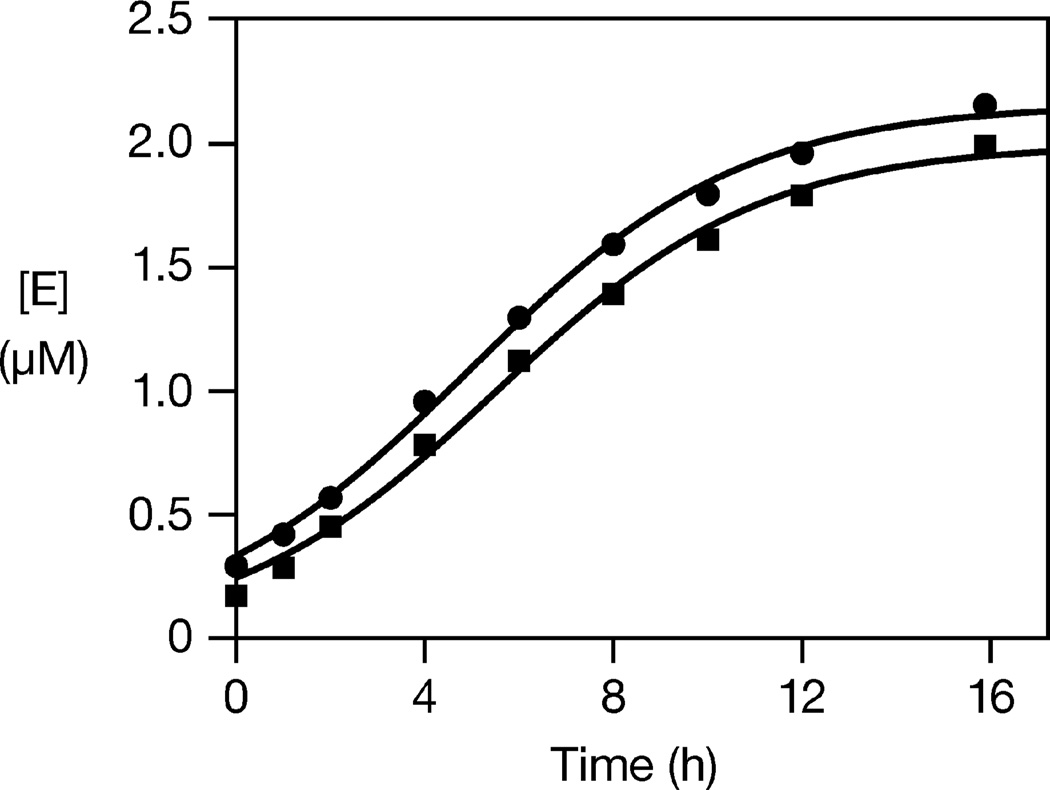
The fully synthetic D and L self-replicating systems were compared in exponential amplification reactions employing 2.5 µM A and 20 µM B, under the same conditions as above, with or without a starting amount of E. The starting concentration of E was adjusted to compensate for slight differences in the specific activity of the D-versus L-enzymes, which likely result from differences in the quality of the corresponding materials (Figure S3). Employing a starting concentration of either 0.36 µM DE or 0.25 µM L-E, the reactions exhibited robust amplification, with exponential growth rates of 0.36 and 0.34 h−1, respectively (Figure 2).
Figure 2.
Exponential amplification of d- (squares) and l-(circles) self-replicating RNA enzymes. The yield of E was determined at various times and the data were fit to the logistic growth equation: [E] = a / (1 + be−c t), where a is the final extent, b is the degree of sigmoidicity, and c is the exponential growth rate. Reaction conditions: 2.5 µM A, 20 µM B, either 0.36 µM d-E or 0.25 µM l-E, 25 mM MgCl2, pH 8.5, 42 °C.
The self-replicating D- and L-RNA enzymes next were configured as aptazymes by replacing their central stem-loop by an aptamer domain that specifically recognizes theophylline (Figure 1b).20 This ligand was chosen because it is achiral and therefore binds to the D- and L-aptamers in the same manner. In the absence of theophylline the aptamer domain is unstructured and unable to support the active conformation of the enzyme, while in the presence of theophylline the central stem-loop closes around the ligand to render the enzyme catalytically active. It was only necessary to synthesize the aptamer-modified form of the A substrates (Atheo). The B substrates are universal and the aptamer-modified form of the enzyme, Etheo, can be generated from its corresponding substrates (Atheo and B).
The D- and L-aptazymes indeed operate in a theophylline-dependent manner (Figure S4). The reaction mixtures contained 10 µM Atheo, 20 µM B, and either 0 or 5 mM theophylline, under the same reaction conditions as above, without any starting amount of E or Etheo. There was no production of Etheo in the absence of theophylline, whereas both the D- and L-aptazymes exhibited exponential amplification in the presence of theophylline.
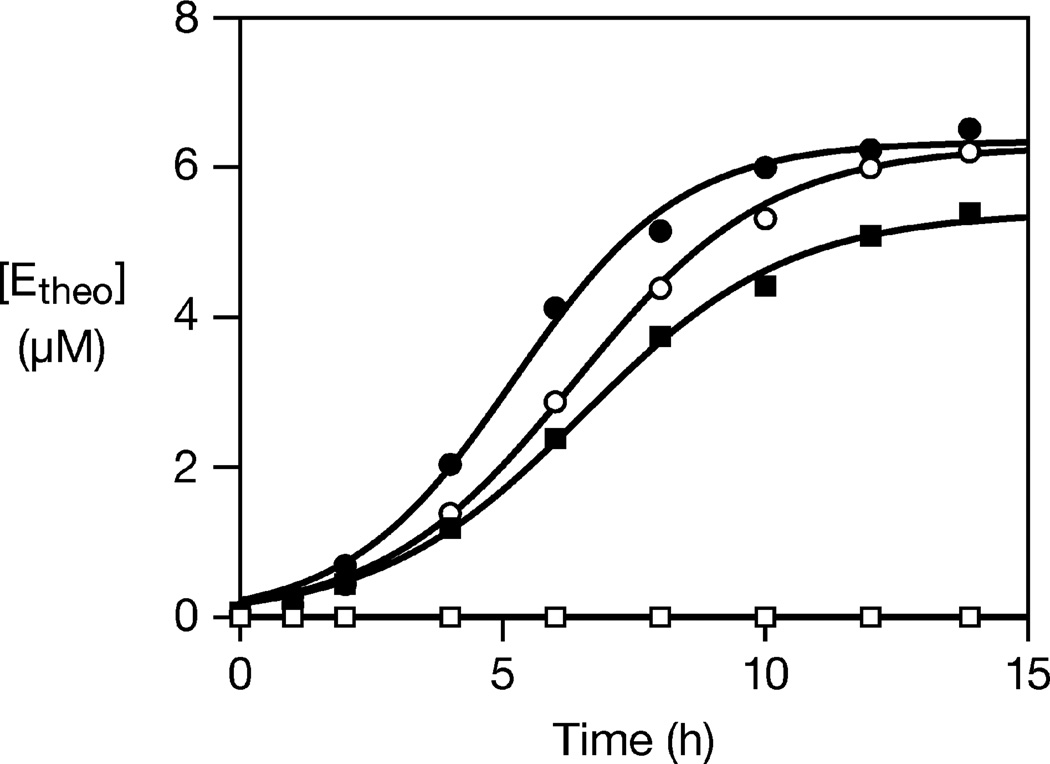
The same theophylline-dependent amplification reactions were carried out in the presence of 10% human serum, which contains abundant ribonucleases (Figure 3). Even in the presence of theophylline, there was no product detected for the D reaction system, and the substrates were completely degraded within 30 s. The L reaction system, in contrast, exhibited exponential growth at a rate of 0.54 h−1, corresponding to a doubling time of 77 min. This rate was only slightly diminished compared to the rate of exponential growth in the absence of human serum, which was 0.63 h−1. L-RNA does not readily interact with biological macromolecules, and thus it is not surprising that the L reaction system is unperturbed by the components of human serum.
Figure 3.
Ligand-dependent exponential amplification of d- (squares) and l- (circles) aptazymes in either the presence (open symbols) or absence (filled symbols) of 10% human serum. The data were fit to the logistic growth equation, as described in the legend to Figure 2. Reaction conditions: either 17.5 µM d-Atheo and 27.5 µM d-B, or 10 µM l-Atheo and 20 µM l-B, 5 mM theophylline, 25 mM MgCl2, pH 8.5, 42 °C.
The prior system for ligand-dependent exponential amplification could not operate in the presence of crude biological samples and could not be used to detect proteins in those samples. By stepping into the mirror of biology and constructing a self-replicating system based on L-RNA molecules, it should now be possible to detect various biological materials without concern for degradation by ribonucleases. The aptamer domain must also be nuclease resistant, and this can be achieved in three ways. The first is to retrofit an existing D-aptamer with nuclease-resistant nucleotide analogs, as has been done for most RNAs that have been developed for therapeutic applications.21–23 The second approach is to generate a D-aptamer starting from a pool of random-sequence, nuclease-resistant RNA analogs, as is becoming the norm for diagnostic applications.24 The third option is to develop Spiegelmers, which are obtained by selecting a D-RNA aptamer that binds to the enantiomer of the intended target, then constructing the corresponding L-RNA aptamer to bind the actual target.6–8 All of these approaches have become routine and are compatible with the modular nature of the ligand-dependent self-replicating RNA enzyme, although the third option likely will be the most straightforward to implement in the L system. Even so, some optimization will be required when linking the aptamer domain to the catalytic domain of the enzyme to achieve maximal ligand sensitivity.2
The ligand-dependent exponential amplification system is not yet suitable for broad practical application, primarily because the rate of amplification is too slow. Efforts are underway to use in vitro evolution methods to optimize the catalytic properties of the self-replicating RNA enzyme, something that had not been pursued aggressively in the past because of the concern for achieving both improved function and nuclease resistance. Now with the ability to operate as either a D or L reaction system, any functional improvements that are achieved using D-RNA can immediately be realized in a nuclease-resistant format. A fluorescence detection method for monitoring the course of exponential amplification was reported previously,2 and is now being supplanted by other fluorescence methods that directly measure the reaction products.25 It is unlikely that the ligand-dependent amplification system will ever be as broadly applicable as the enzyme-linked immunosorbent assay (ELISA),26 but may be preferred for ligands that must be recognized with high specificity or that require multiplexed analysis.2
In addition to addressing potential applications for ligand detection, this study provides the first demonstration of self-sustained exponential amplification of non-biological molecules. The central process of biology is the exponential amplification of genetic information. Synthetic biologists have sought to capture this and other biological processes for their own purposes, but have not strayed far from natural biological systems. The previously described D-RNA enzymes that undergo self-sustained exponential amplification operate outside of biology, but are composed of the same materials found in biology. This work takes a further step away from natural biology by operating entirely with L-RNA. Just as the D reaction system has been extended to populations of replicators that transmit genetic information to their progeny and can undergo Darwinian evolution, it now will be possible to construct self-sustained evolving systems based on L-RNA.
Supplementary Material
ACKNOWLEDGMENT
We thank Michael Robertson for advice regarding the design of the self-replicating form of the RNA enzyme. This work was supported by Grant No. GM065130 from the National Institutes of Health. C.O. was supported by Ruth L. Kirschstein National Research Service Award No. F32CA165430 from the National Institutes of Health. D.P.H. was supported by a National Defense Science and Engineering Graduate Fellowship from the Department of Defense and by a Graduate Fellowship from the Fannie and John Hertz Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Materials and methods, and four figures showing the time course of reaction of various self-replicating and simple ligase RNA enzymes. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lincoln TA, Joyce GF. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam BJ, Joyce GF. Nat. Biotechnol. 2009;27:288–292. doi: 10.1038/nbt.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam BJ, Joyce GF. J. Am. Chem. Soc. 2011;133:3191–3197. doi: 10.1021/ja111136d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang AM, Doyle MV, Mark DF. Proc. Natl. Acad. Sci. USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riedy MC, Timm EA, Jr, Stewart CC. BioTechniques. 1995;18:70–74. [PubMed] [Google Scholar]
- 6.Klussmann S, Nolte A, Bald R, Erdmann VA, Furste JP. Nat. Biotechnol. 1996;14:1112–1115. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- 7.Nolte A, Klussmann S, Bald R, Erdmann VA, Furste JP. Nat. Biotechnol. 1996;14:1116–1119. doi: 10.1038/nbt0996-1116. [DOI] [PubMed] [Google Scholar]
- 8.Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Erfurth J, Burgstaller P, Klussmann S. Chem. Biol. 2002;9:351–359. doi: 10.1016/s1074-5521(02)00111-4. [DOI] [PubMed] [Google Scholar]
- 9.Wlotzka B, Leva S, Eschgfaller B, Burmeister J, Kleinjung F, Kaduk C, Muhn P, Hess-Stumpp H, Klussmann S. Proc. Natl. Acad. Sci. USA. 2002;99:8898–8902. doi: 10.1073/pnas.132067399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helmling S, Maasch C, Eulberg D, Buchner K, Schröder W, Lange C, Vonhoff S, Wlotzka B, Tschöp MH, Rosewicz S, Klussmann S. Proc. Natl. Acad. Sci. USA. 2004;101:13174–13179. doi: 10.1073/pnas.0404175101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darisipudi MN, Kulkarni OP, Sayyed SG, Ryu M, Migliorini A, Sagrinati C, Parente E, Vater A, Eulberg D, Klussmann S, Romagnani P, Anders HJ. Am. J. Pathol. 2011;179:116–124. doi: 10.1016/j.ajpath.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okano K. Tetrahedron. 2009;65:1937–1949. [Google Scholar]
- 13.Paul N, Joyce GF. Proc. Natl. Acad. Sci. USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig J. In: Biophosphates and their Analogues — Synthesis, Structure, Metabolism and Activity. Bruzik KS, Stec WJ, editors. Amsterdam: Elsevier Science Publishers; 1987. pp. 201–204. [Google Scholar]
- 15.Gaur RK, Krupp G. Methods Mol. Biol. 1997;74:99–110. doi: 10.1385/0-89603-389-9:99. [DOI] [PubMed] [Google Scholar]
- 16.Lebedev AV, Koukhareva II, Beck T, Vaghefi MM. Nucleosides Nucleotides Nucleic Acids. 2001;20:1403–1409. doi: 10.1081/NCN-100002565. [DOI] [PubMed] [Google Scholar]
- 17.Paul N, Springsteen G, Joyce GF. Chem. Biol. 2006;13:329–338. doi: 10.1016/j.chembiol.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Pleiss JA, Derrick ML, Uhlenbeck OC. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helm M, Brulé H, Giegé R, Florentz C. RNA. 1999;5:618–621. doi: 10.1017/s1355838299982328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 21.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 22.Beigelman L, et al. J. Biol. Chem. 1995;270:25702–25708. doi: 10.1074/jbc.270.43.25702. [DOI] [PubMed] [Google Scholar]
- 23.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjić N. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 24.Gold L. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. (68 additional authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 26.Engvall E, Perlman P. Immunochem. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.