Abstract
Background. Although human immunodeficiency virus (HIV) infection and antiretroviral therapy (ART) affect mitochondrial DNA (mtDNA) content and function, comprehensive evaluations of their effects on mitochondria in muscle, adipose tissue, and blood cells are limited.
Methods. Mitochondrial DNA quantification, mitochondrial genome sequencing, and gene expression analysis were performed on muscle, adipose tissue, and peripheral blood mononuclear cell (PBMC) samples from untreated HIV-positive patients, HIV-positive patients receiving nucleoside reverse transcriptase inhibitor (NRTI)–based ART, and HIV-negative controls.
Results. The adipose tissue mtDNA/nuclear DNA (nDNA) ratio was increased in untreated HIV-infected patients (ratio, 353) and decreased in those receiving ART (ratio, 162) compared with controls (ratio, 255; P < .05 for both comparisons); the difference between the 2 HIV-infected groups was also significant (P = .002). In HIV-infected participants, mtDNA/nDNA in adipose tissue correlated with the level of activation (CD38+/HLA-DR+) for CD4+ and CD8+ lymphocytes. No significant differences in mtDNA content were noted in muscle or PMBCs among groups. Exploratory DNA microarray analysis identified differential gene expression between patient groups, including a subset of adipose tissue genes.
Conclusions. HIV infection and ART have opposing effects on mtDNA content in adipose tissue; immune activation may mediate the effects of HIV, whereas NRTIs likely mediate the effects of ART.
Combination antiretroviral therapy (ART) has led to improvements in the morbidity and mortality associated with human immunodeficiency virus (HIV) infection, but chronic ART use can result in a variety of drug-related toxicities. Nucleoside reverse transcriptase inhibitors (NRTIs), which inhibit the RNA-dependent DNA polymerase activity of HIV reverse transcriptase, are nearly universally included in current ART regimens [1]. Unfortunately, NRTIs can inhibit mitochondrial DNA (mtDNA) polymerase γ, which is required for mtDNA replication, resulting in a reduction in mtDNA content per cell [2, 3] and potentially impairing mitochondrial function [4, 5].
First observed in muscle tissues of patients receiving zidovudine therapy [6], mtDNA depletion and consequent mitochondrial dysfunction have subsequently been postulated as mechanisms for other NRTI toxicity syndromes [4, 5, 7]. A low mtDNA/nuclear DNA (nDNA) ratio in PBMCs has been described with NRTI-associated symptomatic hyperlactemia [8, 9]. Mitochondrial DNA depletion has been demonstrated in subcutaneous adipose tissue of patients receiving NRTI-containing therapy, suggesting that NRTI-mediated mtDNA depletion plays a role in the pathogenesis of lipodystrophy [10–12]. Clinical studies have confirmed the association of specific NRTI agents with lipoatrophy [13, 14] and in vitro studies have shown that NRTIs such as zidovudine, stavudine, and didanosine result in mtDNA depletion [3].
Mitochondrial dysfunction is not consistently associated with mtDNA depletion [15, 16], and significant reductions in mtDNA have been described in asymptomatic subjects [17, 18]. Therefore, other mechanisms leading to mitochondrial dysfunction have been proposed as contributing to NRTI toxicity syndromes, including alterations in mitochondrial mass or gene expression, that do not rely on inhibition of DNA polymerase γ. NRTIs may decrease mtRNA transcription without an associated depletion of mtDNA [19]. HIV infection itself may cause mitochondrial toxicity [20, 21], although the mechanism has not been established.
To better understand the effects of HIV and ART on mitochondrial number and function, we undertook a cross-sectional study to examine histopathology, mtDNA content, and the expression of mitochondrial-related genes in muscle, adipose tissue, and peripheral blood mononuclear cells (PBMCs) of HIV-infected adults and HIV-negative control subjects.
METHODS
Study Participants
Three groups of volunteers aged 18–55 years were enrolled: HIV-infected adults receiving ART, HIV-infected adults not receiving ART, and healthy, HIV-negative adults. ART recipients were required to have received a stable regimen containing 3 drugs, including at least 2 nucleoside or nucleotide analogues, for ≥12 consecutive months. Patients receiving protease inhibitor therapy were excluded to minimize potential confounding effects. HIV-infected participants not receiving ART could not have received any ART in the 12 months preceding enrollment. Patients with a history of muscle disorder or diabetes were excluded. The protocol was approved by the National Insitute of Allergy and Infectious Diseases (NIAID) Institutional Review Board, and all participants provided written informed consent.
Participants underwent baseline laboratory testing, including creatine kinase, thyroid-stimulating hormone, fasting insulin, lactate, lymphocyte subset analysis, and HIV antibody status. HIV-infected participants had HIV viral load quantification (Quantiplex HIV-1 RNA 3.0. Assay, Bayer Diagnostics). PBMCs for mtDNA quantitation were isolated from whole blood collected in Cell Preparation Tubes (Becton Dickinson) or were obtained by Ficoll-Hypaque density gradient centrifugation of cells obtained by lymphapheresis and were washed 3 times to minimize platelet contamination. Participants completed a series of questionnaires assessing fatigue and quality of life (Piper Fatigue Scale, Beck Depression Inventory II, HIV-related Fatigue Scale, and Medical Outcomes Study: Short Form 36) [22–28].
Muscle and Adipose Tissue Biopsy
Muscle biopsies of the nondominant quadriceps or biceps muscle were performed according to standard procedures; subcutaneous adipose tissue was obtained simultaneously. Muscle tissues were sectioned with one section fixed in formalin, and another processed immediately for histopathology (below); additional sections were placed in 2.5% glutaraldehyde for electron microscopy [29] or RNAlater (Ambion) for RNA extraction, or were frozen for DNA extraction.
For histopathology, muscle samples were dissected free of adipose and connective tissue, mounted in Cryomatrix (Shandon), frozen directly in isopenthane cooled with liquid nitrogen, and stored at −80°C. Serial transverse sections (5 μm) were cut using a cryostat at −20°C, and routine hematoxylin-eosin staining, enzyme histochemistry, and immunohistochemistry were performed on cut sections.
DNA and RNA Isolation
Tissue samples were disrupted in a TissueLyser (Qiagen) prior to DNA or RNA extraction. Total DNA was extracted from PBMCs using the QIAamp DNA kit and from adipose tissue and muscle using the QIAamp DNA tissue kit. Total RNA was isolated from PBMCs using RNeasy Mini QIAshredder Kit, from adipose tissue using RNeasy Lipid Tissue Mini Kit, and from muscle using the RNeasy Fibrous Tissue Mini Kit (Qiagen).
mtDNA Quantification
Separate quantitative real-time polymerase chain reactions were performed for a mitochondrial gene, cytochrome c oxidase subunit I (COX1), using previously published primer and probe sequences [8], and a nuclear gene, β-globin, using the following primers and probes: BetaGLOBIN-FOR (5′–GTCCAAGCTAGGCCC-3′); BetaGLOBIN-REV (5′-CCAGATGCTCAAGGCCC-3′); Beta-Up1 (5′-ATCATGTTCATACCTCTTATCTTCCTCCCAC-Fluo-3′); and Beta-Dwn2 (5′-Red640-GCTCCTGGGCAACGTGCT-3′). Fluorescence resonance energy transfer probes were used to detect the amplicon. Results are expressed as the ratio between mtDNA and nDNA copy number.
Microarray
A custom microarray chip (Affymetrix) was used to study gene expression according to the manufacturer’s recommended protocols [30]. The huMITOchip includes 51 probe sets interrogating the 37 genes unique to the mitochondrial genome, as well as 4774 probe sets targeting nuclear genes involved in mitochondrial function, muscle function, lipid metabolism, and inflammatory pathways [30].
Partek Genomics Software 2.3 (Partek) was used for statistical analysis. The raw data were normalized using the quantile normalization method [31]. To neutralize variation by tissue, the normalized gene expression values in each tissue type were further standardized with a median-shift strategy (ie, subtracting each gene expression value from the median expression value of the corresponding cohort). A 2-way analysis of variance model (ie, tissue, disease group) was designed to identify differentially expressed genes with absolute fold change ≥0.5 (log 2 scale) and P values ≤.05. Selected genes were clustered by hierarchical clustering. The DAVID Bioinformatics Resources (version 2.1; available at: http://david.abcc.ncifcrf.gov/) were used to annotate the enriched biology associated with each gene cluster [32].
Mitochondrial DNA Sequencing and Haplogroup Determination
DNA sequencing of the mitochondrial genome was performed using the Affymetrix GeneChip Mitochondrial Resequencing Array 2.0, according to the manufacturer’s recommended protocols. Affymetrix GCOS 1.4 and GSEQ 4.0 software were used for image and initial sequence analysis with default settings. Then, comparing to reference mitochondrial sequences of the revised Cambridge Reference Sequence (rCRS) (accession no. NC_12920), single-nucleotide polymorphisms (SNPs) were screened using a previously reported method [33]. The selected SNPs were further characterized using MitoMap and mitowheel databases (http://www.mitomap.org and http://www.mitowheel.org). Mitochondrial haplogroups were categorized by Haplogroup Prediction tool (http://nnhgtool.nationalgeographic.com) with neighbor-joining distance calculated by MEGA4, based on Phylotree database (http://www.phylotree.org).
Statistical Analysis
Median values and interquartile ranges (IQRs) are presented for continuous data. Nonparametric tests including the Mann-Whiney U test, Spearman rank correlation, and Wilcoxon signed-rank test were used throughout. Fisher exact test was used to compare the rate of histopathologic abnormalities between groups. All tests were 2-tailed. Analyses were not corrected for multiple comparisons in this exploratory study. P values of <.05 were considered statistically significant. Prism version 5.0b (GraphPad Software) was used for statistical analyses. Microarray analyses were performed as above.
RESULTS
Subject Characteristics
Between May 2005 and April 2007, 61 subjects underwent muscle and adipose tissue biopsy: 29 HIV-infected adults receiving ART, 17 HIV-infected adults not receiving ART, and 15 HIV-negative controls. Table 1 summarizes the demographic and baseline characteristics of study participants. The HIV-infected group was slightly older and included more men; median time since HIV diagnosis was 11.9 years. Median CD4 T-cell count and viral load were 532 cells/μL and <50 copies/mL, respectively, in the ART group and 419 cells/μL and 5315 copies/mL, respectively, in the no ART group. For the ART group, median duration of ART at biopsy was 8.2 years; the specific components of the antiretroviral regimens are summarized in Table 2. Ten of the 29 participants (34%) receiving ART had a clinical diagnosis of lipoatrophy (patient report confirmed by physician assessment) and 8 (28%) had a history of ART-associated peripheral neuropathy. Ten of 17 subjects in the no ART group were ART naive; 7 had prior treatment experience, with a median of 3.0 years since last ART (range, 1.1–7.3 years). Prior ART included thymidine analogues for 6 of these 7 subjects; however, mtDNA copies/nDNA did not differ between the subset of previously treated participants and those who had never received ART.
Table 1.
Selected Demographic, Clinical, and Laboratory Parameters for the Cohort
| Characteristic | HIV Negative | HIV Positive | ||
|---|---|---|---|---|
| All | No ART | On ART | ||
| n = 15 | n = 46 | n = 17 | n = 29 | |
| Age, year (range) | 43.2 (22.6–51.7) | 44.8 (22.1–55.6) | 44.5 (33.7–55.6) | 45.6 (22.1–55.6) |
| Female sex, no. (%) | 7 (46.7) | 13 (28.2) | 6 (35) | 7 (24) |
| Race/ethnicity, no. (%) | ||||
| White | 9 (60) | 19 (41.3) | 5 (29.4) | 14 (48.3) |
| Black | 3 (20) | 20 (43.5) | 12 (70.6) | 8 (27.6) |
| Hispanic | 2 (13.3) | 7 (15.2) | 0 (0) | 7 (24.1) |
| Asian/Pacific Islander | 1 (6.6) | 0 (0) | 0 (0) | 0 (0) |
| Duration of HIV infection, year | NA | 11.9 (1.0–23.1) | 11.3 (1.0–23.1) | 12.5 (2.7–20.6) |
| CD4, cells/mL (range) | 932 (297–1710) | 480 (73–1192) | 419 (73–929) | 532 (293–1192) |
| HIV viral load, copies/mLa(range) | NA | <50 (<50–165 352) | 5315 (61–165 352) | <50 (<50–507) |
| Chronic hepatitis C, clinical diagnosis (%) | 0 (0) | 5 (11) | 2 (12) | 3 (10) |
| Lipoatrophy, clinical diagnosisb (%) | 0 (0) | 10 (22) | 0 (0) | 10 (34) |
| Peripheral neuropathy, current, clinical diagnosisc (%) | 0 (0) | 8 (17) | 0 (0) | 8 (28) |
Values represent median (range) unless otherwise indicated.
Abbreviations: ART, antiretroviral therapy; HIV, human immunodeficiency virus.
HIV viral load measured by branched DNA, version 3 (Chiron); lower limit of detection of 50 copies/mL.
The presence and severity of lipoatrophy was determined using the HIV Outpatient Study (HOPS) severity scale [34]. The degree of lipoatrophy in the face, arms, legs, and buttocks was rated as absent, mild (noticeable on close inspection), moderate (noticeable by patient and physician), or severe (noticeable to a casual observer). Subjects with at least 1 moderate or severe lipoatrophic area were considered to have lipoatrophy.
The presence of peripheral neuropathy was determined by patient report of symptoms consistent with HIV-associated sensory neuropathy (tingling, pins-and-needles sensation, stabbing pain in the distal extremities) confirmed by objective evidence on neurologic examination (change in pinprick, vibration or temperature sensibility, deep tendon reflexes).
Table 2.
Characteristics of Antiretroviral Regimens at Biopsy for Patients on Antiretroviral Therapy (n = 29)
| Parameter | Value |
|---|---|
| Median duration of current regimen, y (range) | 2.3 (0.3–8.2) |
| Median total antiretroviral exposure, y (range) | 8.2 (0.3–15.2) |
| Proportion for whom current regimen is their first (%) | 2/29 (7%) |
| Antiretroviral regimen at biopsy | |
| 2-class regimen | 26/29 (90%) |
| 3-drug regimen | 23 |
| 4-drug regimen | 3 |
| Nucleoside/nucleotide reverse transcriptase inhibitors only | 3/29 (10%) |
| AZT/3TC/ABC | 2 |
| DDI/D4T/3TC | 1 |
| Nucleoside/nucleotide reverse transcriptase inhibitors | |
| AZT | 10 |
| D4T | 3 |
| DDI | 4 |
| 3TC or FTC | 25 |
| ABC | 8 |
| TDF | 15 |
| Nonnucleoside reverse transcriptase inhibitors | |
| EFV | 18 |
| NVP | 8 |
Values represent the no. (%) of patients unless otherwise indicated.
Abbreviations: ABC, abacavir; AZT, zidovudine; 3TC, lamivudine; DDI, didanosine; D4T, stavudine; EFV, efavirenz; NVP, nevirapine; TDF, tenofovir.
mtDNA Assessment in Adipose Tissue, Muscle, and PMBCs
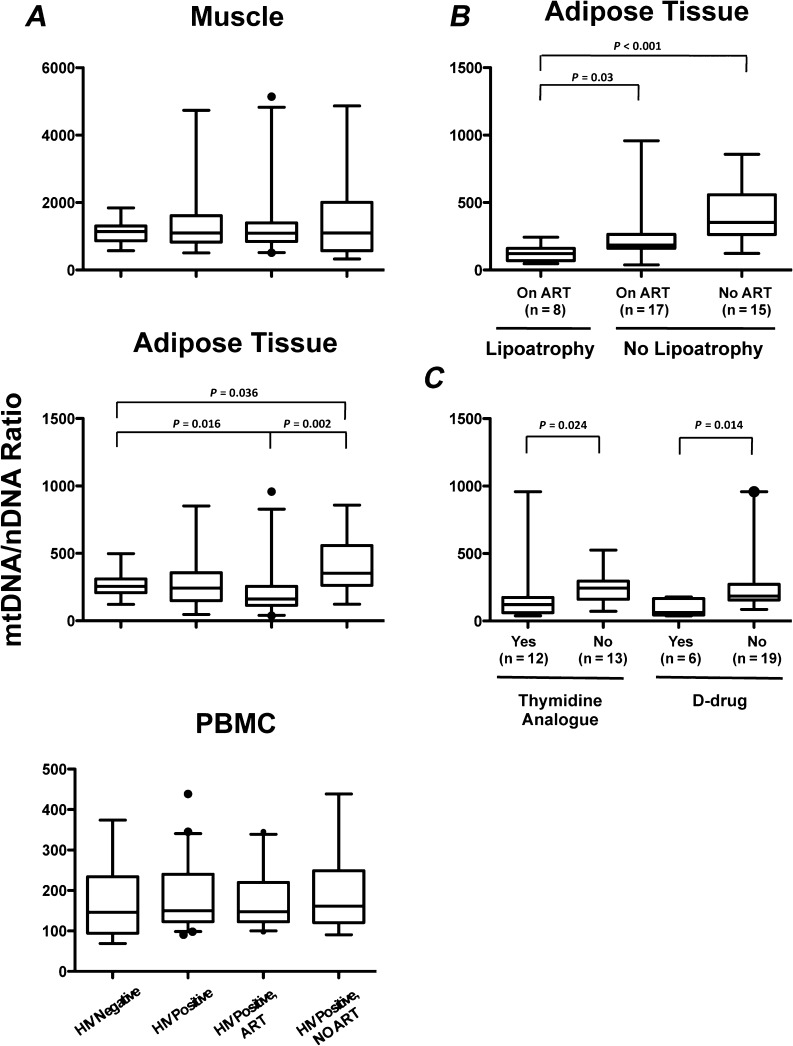
A primary aim of the study was to compare mtDNA levels in the different tissue compartments among the 3 patient groups. The mtDNA/nDNA ratio in adipose tissue (Figure 1) was significantly increased in HIV-infected adults not receiving ART (ratio, 353) compared with HIV-negative controls (ratio, 255, P = .036). In contrast, mtDNA content in adipose tissue was significantly decreased in HIV-infected adults receiving ART (ratio, 161) compared with either HIV-uninfected controls (P = .016) or HIV-positive adults not receiving ART (P = .002). Additionally, mtDNA content in adipose tissue was significantly decreased in HIV-infected adults with lipoatrophy (all receiving ART, n = 8; ratio = 141) compared with HIV-infected adults without lipoatrophy, on or off ART (n = 32; ratio = 260, P = .024) (Figure 1). In patients receiving ART, mtDNA content in adipose tissue was significantly decreased in those currently receiving thymidine analogues (zidovudine or stavudine, ratio = 122) or D-drugs (didanosine or stavudine; ratio, 63) compared with HIV patients not receiving these medications (ratio, 243, P = .024 and .014, respectively; Figure 1). No significant differences were seen in the mtDNA content in muscle or PBMCs (Figure 1) when comparing HIV-negative and HIV-positive participants, regardless of antiretroviral use. There were no significant associations between peripheral neuropathy and mtDNA content in any tissue.
Figure 1.
Mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) ratio in muscle, adipose tissue, and peripheral blood mononuclear cells (PBMCs) by patient group. A, mtDNA/nDNA in adipose tissue is significantly higher in patients infected with human immunodeficiency virus (HIV) not on therapy and significantly lower in patients receiving antiretroviral therapy (ART) compared with HIV-negative controls, whereas muscle and PBMC mtDNA/nDNA were not different among the groups. PBMC and tissue samples available for analysis for all HIV-negative participants (n = 15). PBMCs were available for all HIV-positive participants (n = 46). Muscle samples were available for 45 HIV-positive participants (28 on ART, 17 no ART). Adipose tissue samples were available for 40 HIV-positive participants (25 on ART, 15 no ART). B, mtDNA/nDNA in adipose tissue is decreased in patients with clinical lipodystrophy. C, mtDNA/nDNA is decreased in the adipose tissue of HIV-infected patients receiving thymidine analogues (zidovudine or stavudine) and/or D-drugs (didanosine or stavudine). Individuals receiving stavudine were included in both the thymidine analogue and D-drug groups. Box plots indicate median, interquartile range (box) and the 5%–95% range (whiskers). Outliers indicated by solid circles. Comparisons between groups are nonsignificant except where indicated.
Correlation Analysis
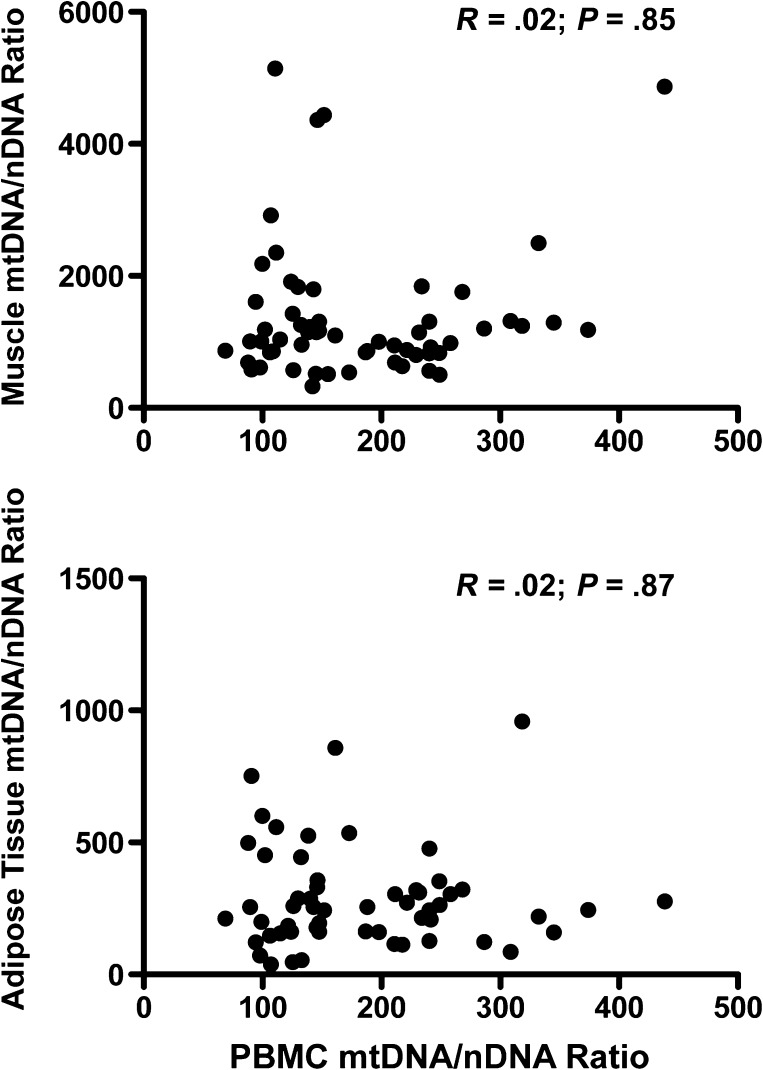
A secondary aim of the study was to determine whether mtDNA levels in PBMCs could serve as a surrogate marker for tissue mtDNA. However, no correlation was found between mtDNA content in PBMCs and muscle or adipose tissue in any study group (Figure 2). Similarly, no correlation was found between mtDNA content in muscle and adipose tissue.
Figure 2.
Relationship between mitochondrial DNA (mtDNA) to nuclear DNA (mtDNA/nDNA) in peripheral blood mononuclear cells (PBMCs) and tissues. There is no correlation between mtDNA/nDNA in PBMCs and mtDNA content in muscle (top) or adipose tissue (bottom). Each dot represents results for a single individual. Nonparametric correlation coefficients (Spearman) and P values are shown within the graphs.
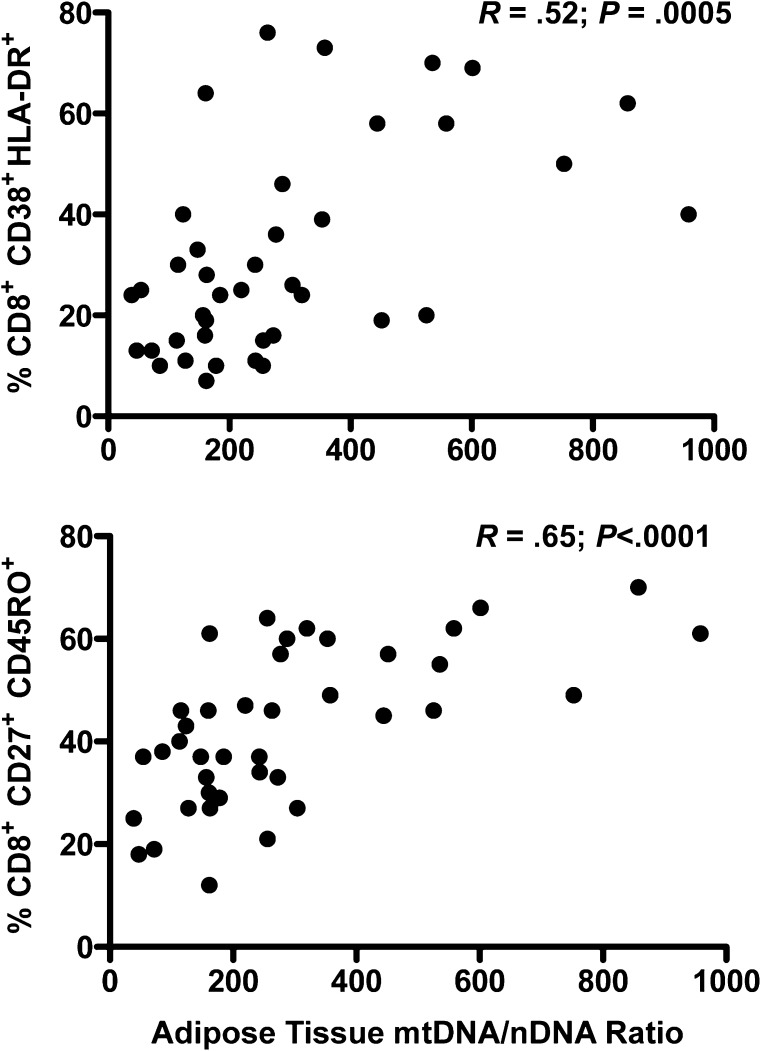
We then examined correlations between mtDNA content in the various compartments and other measured parameters. In HIV-infected participants, multiple phenotypic markers on lymphocytes correlated positively with mtDNA content in adipose tissue, but not with mtDNA content in PBMCs or muscle. The correlation was strongest with the percent of CD8+ cells that were activated (HLA-DR+/CD38+, r = 0.52, P = .0005) or had a central memory phenotype (CD27+/CD45RO+, r = 0.65, P < .0001) (Figure 3). A significant relationship was also seen with activated CD4+ cells (P = .005). These relationships remained significant after adjustment for HIV viral load. No correlation was seen between markers and mtDNA in HIV-negative participants.
Figure 3.
Relationship between adipose tissue mitochondrial DNA (mtDNA) to nuclear DNA (mtDNA/nDNA) and selected CD8+ T-lymphocyte subsets in patients infected with human immunodeficiency virus (HIV). A strong correlation was seen between adipose tissue mtDNA/nDNA and the percentage of total CD8+ cells that were activated (HLA-DR+/CD38+, top) or had a central memory phenotype (CD27+/CD45RO+, bottom). Nonparametric correlation coefficients (Spearman) and P values are shown within the graphs. A significant correlation was also seen between adipose tissue mtDNA/nDNA and the percentage of total CD4+ cells that were activated (HLA-DR+/CD38+; R = 0.44, P = .005; data not shown).
No correlation was found between mtDNA content at any site and fatigue score, as measured by validated questionnaires. Additionally, for all tissue types, mtDNA content did not correlate with serum lactate levels.
Histopathology of Muscle
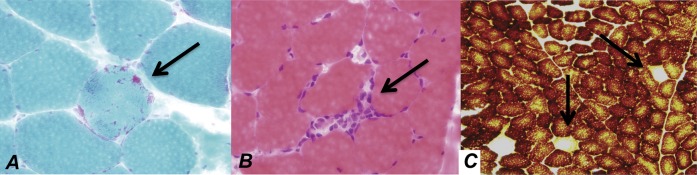
Detailed histopathologic analysis of muscle tissue was available for 54 biopsies. Samples were graded as normal, minimal change, or pathologic change. Pathologic changes were further categorized as mitochondrial, myopathic, or neuropathic in origin. At the light microscopy level, blind review of the biopsies demonstrated no disease-specific histologic abnormalities. However, pathologic changes were found in the muscle samples from 1 (7%) HIV-negative participant, 8 (27%) HIV-positive participants receiving ART, and 3 (18%) HIV-positive participants not receiving ART. Abnormalities included subtle variations in muscle fiber size, rare COX-negative fibers, rare ragged red fibers, and, in 2 HIV-positive patients not receiving ART, sparse endomysial infiltrates with MHC-1–expressing nonnecrotic muscle fibers (Figure 4). The frequency of pathologic abnormalities was not statistically significantly different between the groups. No pathologic changes were noted on light microscopic examination of adipose tissue or PBMCs.
Figure 4.
Histopathologic abnormalities noted on muscle biopsy of patients infected with human immunodeficiency virus (HIV) included (A) ragged red fibers (arrow, Gomori trichrome stain; red indicates mitochondrial proliferation), (B) inflammatory infiltrate (arrow, hematoxylin-eosin stain), and (C) cytochrome oxidase (COX) negative fibers (arrows, cytochrome oxidase stain, brown indicates cytochrome oxidase activity).
A subset of muscle (n = 33), adipose tissue (n = 19), and PBMC samples (n = 32) was evaluated by electron microscopy. Except for a rare vacuole, mitochondria in all muscle samples appeared normal. Minimal ultrastructural changes seen in muscle included rare specimens with lipid droplets in muscle fibers and lipofuscin with lipid in the perinuclear cytoplasm. Additionally, mitochondria appeared normal in adipocytes and endothelial cells of adipose tissue and in all lymphocytes and monocytes.
Mitochondrial Genome Sequencing
Eighteen SNPs occurred at higher frequencies in HIV-infected participants receiving ART compared with the HIV-positive participants not receiving ART and the HIV-negative participants. These SNPs had consistent call patterns and frequencies in the 3 tissue types. However, these differences were linked to race and therefore consistent with inherited haplotypes.
Differential Gene Expression Screened by Microarray
Because commercially available microarray chips did not include mitochondrial-encoded genes, we used a previously validated customized microarray chip to examine gene expression in tissue and PBMCs (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30310) [30]. Principal component analysis demonstrated that the first principal component grouped the same tissue types together, regardless of whether the source was an HIV-infected patient or HIV-negative control, highlighting that muscle, adipose tissue, and PBMCs each have highly related tissue-specific expression patterns. This also demonstrated that our tissue samples were primarily composed of a single tissue type.
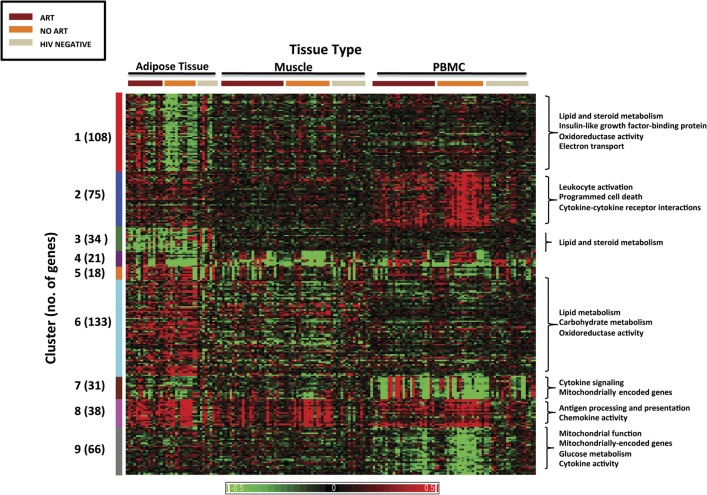
Using the criteria described in the methods, differential gene expression was identified in 524 of 4825 genes (11%) included in the custom microarray. Hierarchical clustering classified the genes into 9 distinct clusters based on differential expression patterns between the groups (Figure 5).
Figure 5.
Heat map of gene expression profile by a custom microarray. The findings for all 3 tissue types—fat, muscle, and peripheral blood mononuclear cells (PBMCs)—are shown. Each column represents an individual patient sample, and each row represents an individual gene. Differences in relative levels of gene expression (z score) are represented in color, where red indicates upregulation and green indicates downregulation relative to that of corresponding median gene expression in HIV-negative controls. The hierarchical analyses classified the genes into 9 distinct clusters based on differential expression pattern between the groups, indicated on the left. The numbers in parenthesis indicate the number of genes in each cluster. Functional annotation analysis identified biologically relevant pathways by cluster, indicated on the right for selected clusters.
The Supplementary Table summarizes the genes in each cluster. Differential regulation of genes related to immune activation and inflammation was found in HIV-infected adults compared with HIV-negative adults. Cluster 2 includes genes upregulated in the PBMCs of HIV-infected patients not receiving ART compared with HIV-infected patients receiving ART and HIV-negative controls. These genes are involved in leukocyte activation, cytokine–cytokine receptor interactions, and programmed cell death and include interferon γ, interleukins 1 and 15, chemokine receptors CCR1, CCR3, and CCR5, and tumor necrosis factor receptor superfamily and ligand. Cluster 8 consists of genes upregulated in all 3 tissue types of HIV-positive compared with HIV-negative participants and includes genes of the major histocompatibility complex and a number of chemokines. These findings confirm previous observations and help validate the accuracy of the microarray results, because inflammation and immune activation have been well described in HIV-infected patients, which are diminished but still increased in patients receiving ART compared with HIV-negative controls [35].
Cluster 1 consists of genes that are differentially expressed primarily in adipose tissue, where they are upregulated in HIV-infected patients receiving ART compared with the other 2 groups. This cluster includes genes related to lipid and steroid metabolism, and thus appear to represent effects of ART on fat metabolism. Pathways of significance in this cluster include genes related to insulinlike growth factor 1, growth hormone, and leptin signaling.
Cluster 4 includes genes that are downregulated in both adipose and muscle tissue of HIV patients not receiving ART. This cluster includes 10 mitochondrial-specific genes, including cytochrome b and NADH dehydrogenase 6, part of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that functions in the transfer of electrons from NADH to the respiratory chain. In cluster 9, downregulation of genes was seen in PBMCs of HIV-positive participants not receiving ART. This cluster is saturated with genes related to glucose metabolic processes and cytokine activity and includes mitochondrial-encoded genes, including ATP synthase FO subunit 6, involved in ATP synthesis; 3 mitochondrially encoded NADH dehydrogenases, part of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I); and nuclear encoded mitochondrial genes, including cytochrome c oxidase subunits, components of the terminal oxidase in mitochondrial electron transport, and ATP synthase. These changes thus appear to be a result of either direct or indirect effects (eg, via HIV-related immune activation or inflammation) of HIV itself.
Clusters 5 and 6 include genes upregulated in adipose tissue of HIV-positive participants (ART and no ART) compared with HIV-negative participants. These include genes involved in lipid metabolism, including lipoprotein lipase and hydroxysteroid 11-β dehydrogenase 1, as well as mitochondrial processes, including oxidoreductase activity. Similarly, cluster 7 consists of genes upregulated in the PBMCs of HIV-infected (ART and no ART) compared with HIV-negative participants. This cluster contains genes involved in cytokine signaling and mitochondrial function and is saturated with mitochondrial-encoded transfer RNAs. Again, this appears to reflect an HIV effect, which does not appear to be modulated by ART.
DISCUSSION
This study, which is the largest to date to comprehensively examine muscle, adipose tissue, and PBMCs in the same patient population, has demonstrated that mtDNA content in adipose tissue, but not muscle or PBMCs, is increased in individuals with untreated HIV infection and decreased in patients on a stable ART regimen. Furthermore, patients with clinical lipoatrophy, and those receiving zidovudine, stavudine, or didanosine, had significantly lower mtDNA content in adipose tissue, in keeping with previous studies [10–12, 36, 37].
These findings suggest that HIV, either directly or indirectly, leads to increases in mitochondrial content in adipose tissue, and further, these findings support the role of NRTI-related adipose tissue toxicity in the development of peripheral fat wasting. Although increases in mitochondrial mass and mtDNA content have been described in response to oxidative stress in vitro [38], this has not to our knowledge been previously described in HIV-infected adults. Mitochondrial DNA proliferation in adipose tissue may exist as a direct compensatory response to HIV-related mitochondrial toxicity or in response to the immune activation/inflammatory milieu induced by HIV. NRTI therapy may potentiate this initial damage, leading to increased risk of mitochondrial toxicity syndromes, including lipoatrophy.
Notably, we did not find evidence of mtDNA depletion in muscle, although we did identify subclinical pathologic changes, including COX negative fibers, in HIV-infected patients. Reversible mtDNA depletion in muscle has been described in patients with symptomatic AZT-associated myopathy [6]. Our requirement for patients to have been on stable ART regimens likely selected those tolerant of NRTI therapy without myopathic symptoms; other studies found similar results in patients on stable ART [39, 40].
No difference in mtDNA content was seen in PBMCs of HIV-infected adults, regardless of ART, compared with HIV-negative controls. This is consistent with the findings of some groups [10, 41–44] but in contrast to those of others that have identified depletion of mtDNA in PBMCs in treatment naive and antiretroviral-treated HIV-infected cohorts [8, 20, 21, 45–47]. These differences likely result from differences in patient populations, antiretroviral regimens, and duration of therapy.
One of the goals of the study was to identify an easily measured surrogate marker for mtDNA damage in less accessible tissues. Given the lack of correlation between the mtDNA content in PBMCs and that in muscle or adipose tissue, it is clear that PBMCs do not serve this function. Interestingly, however, expression of activation markers on CD4+ and CD8+ T cells, especially HLA-DR and CD38, were found to correlate with mtDNA levels in adipose tissue, suggesting that the same process that is responsible for expression of these markers, HIV-related immune activation or inflammation, may be accounting for the changes in mtDNA.
Our study found no evidence of significant deletions or insertions in the mitochondrial genome in individuals receiving ART with or without lipoatrophy. This suggests that decrease in mtDNA content seen in individuals receiving ART is not induced by deletions or duplications in the mitochondrial gene structure.
Exploratory microarray analysis identified significant changes in gene expression in adipose tissue. These included changes in genes regulating lipid and steroid metabolic processes and electron carrier activity in HIV-infected patients receiving ART. Additional genes that were involved in metabolic processes and mitochondrial function were found to be upregulated in the adipose tissue of HIV-positive patients compared with HIV-negative controls. Further confirmation of these results for the most biologically plausible genes is needed in other cohorts.
Although the use of microarray analysis in the study of HIV-associated lipodystrophy is limited, studies using other methods have investigated mitochondrial and nuclear gene expression in HIV-infected adults receiving ART, with or without lipodystrophy. Studies of adipogenic gene expression have found reduced expression of the differentiation factors sterol regulatory element binding protein-1 (SREBP-1) and peroxisome proliferator-activated receptor γ (PPAR-γ) in HIV-infected adults with lipoatrophy [48]. However, our microarray analysis did not identify significant differences in SREBP-1 or PPAR-γ between patient groups.
Our study has several limitations. Because it was cross-sectional, we can identify correlations but not demonstrate causality. The small sample size may have limited statistical power to detect differences in some analyses. Participants in the HIV-infected/ART cohort were receiving a variety of antiretroviral regimens for differing durations, likely limiting our ability to detect antiretroviral-specific effects. The evaluation of the effect of ART on mtDNA content may, because of survivor bias, represent a conservative estimate of the relationship; we likely excluded patients with early or severe ART toxicity by selecting patients tolerant of therapy. Functional assays, including respiratory chain enzyme or activity levels, were not performed. Prior studies have been contradictory on the correlation of functional assays and mtDNA depletion in HIV infection.
We have shown that mtDNA content in adipose tissue is decreased in HIV-infected adults receiving ART, with the most profound reductions seen in patients with lipodystrophy. We also demonstrated for the first time to our knowledge an increase in the mtDNA content in adipose tissue of HIV-infected patients not receiving ART, results that are consistent with our microarray data showing upregulation of expression of a subset of genes in adipose tissue. Further research should explore the role of gene pathways identified in this study in the pathogenesis of HIV-related mitochondrial toxicity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
We thank the patients for their willingness to participate in these studies, and the medical, nursing, and support staff of the National Institutes of Health (NIH), especially Catherine Rehm, Margaret Lloyd, and Brandie Freemire, for their help, which facilitated the completion of this study.
Disclaimers.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the NIH or the US Department of Health and Human Services.
Financial support.
This research was supported by the Intramural Research Programs of the NIH Clinical Center, the NIAID, and the National Institute of Arthritis and Musculoskeletal and Skin Diseases; by a Bench-to-Bedside award from the NIH, supported by the Office of AIDS Research; and by the K-22 National Institute of Nursing Research (grant number NR008672-01). This project has been funded in part with federal funds from the National Cancer Institute, NIH (contract number HHSN261200800001E).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; 2011. pp. 1–166. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 13 January 2011. [Google Scholar]
- 2.Chen CH, Cheng YC. Delayed cytotoxicity and selective loss of mitochondrial DNA in cells treated with the anti-human immunodeficiency virus compound 2′,3′-dideoxycytidine. J Biol Chem. 1989;264:11934–7. [PubMed] [Google Scholar]
- 3.Chen CH, Vazquez-Padua M, Cheng YC. Effect of anti-human immunodeficiency virus nucleoside analogs on mitochondrial DNA and its implication for delayed toxicity. Mol Pharmacol. 1991;39:625–8. [PubMed] [Google Scholar]
- 4.Brinkman K, ter Hofstede HJ, Burger DM, Smeitink JA, Koopmans PP. Adverse effects of reverse transcriptase inhibitors: mitochondrial toxicity as common pathway. AIDS. 1998;12:1735–44. doi: 10.1097/00002030-199814000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Lewis W, Dalakas MC. Mitochondrial toxicity of antiviral drugs. Nat Med. 1995;1:417–22. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- 6.Arnaudo E, Dalakas M, Shanske S, Moraes CT, DiMauro S, Schon EA. Depletion of muscle mitochondrial DNA in AIDS patients with zidovudine-induced myopathy. Lancet. 1991;337:508–10. doi: 10.1016/0140-6736(91)91294-5. [DOI] [PubMed] [Google Scholar]
- 7.Brinkman K, Smeitink JA, Romijn JA, Reiss P. Mitochondrial toxicity induced by nucleoside-analogue reverse-transcriptase inhibitors is a key factor in the pathogenesis of antiretroviral-therapy-related lipodystrophy. Lancet. 1999;354:1112–5. doi: 10.1016/S0140-6736(99)06102-4. [DOI] [PubMed] [Google Scholar]
- 8.Cote HC, Brumme ZL, Craib KJ, et al. Changes in mitochondrial DNA as a marker of nucleoside toxicity in HIV-infected patients. N Engl J Med. 2002;346:811–20. doi: 10.1056/NEJMoa012035. [DOI] [PubMed] [Google Scholar]
- 9.Cote HC, Yip B, Asselin JJ, et al. Mitochondrial:nuclear DNA ratios in peripheral blood cells from human immunodeficiency virus (HIV)-infected patients who received selected HIV antiretroviral drug regimens. J Infect Dis. 2003;187:1972–6. doi: 10.1086/375353. [DOI] [PubMed] [Google Scholar]
- 10.Cherry CL, Gahan ME, McArthur JC, Lewin SR, Hoy JF, Wesselingh SL. Exposure to dideoxynucleosides is reflected in lowered mitochondrial DNA in subcutaneous fat. J Acquir Immune Defic Syndr. 2002;30:271–7. doi: 10.1097/00126334-200207010-00002. [DOI] [PubMed] [Google Scholar]
- 11.Nolan D, Hammond E, Martin A, et al. Mitochondrial DNA depletion and morphologic changes in adipocytes associated with nucleoside reverse transcriptase inhibitor therapy. AIDS. 2003;17:1329–38. doi: 10.1097/00002030-200306130-00007. [DOI] [PubMed] [Google Scholar]
- 12.Walker UA, Bickel M, Lutke Volksbeck SI, et al. Evidence of nucleoside analogue reverse transcriptase inhibitor–associated genetic and structural defects of mitochondria in adipose tissue of HIV-infected patients. J Acquir Immune Defic Syndr. 2002;29:117–21. doi: 10.1097/00042560-200202010-00002. [DOI] [PubMed] [Google Scholar]
- 13.Mallal SA, John M, Moore CB, James IR, McKinnon EJ. Contribution of nucleoside analogue reverse transcriptase inhibitors to subcutaneous fat wasting in patients with HIV infection. AIDS. 2000;14:1309–16. doi: 10.1097/00002030-200007070-00002. [DOI] [PubMed] [Google Scholar]
- 14.Saint-Marc T, Partisani M, Poizot-Martin I, et al. A syndrome of peripheral fat wasting (lipodystrophy) in patients receiving long-term nucleoside analogue therapy. AIDS. 1999;13:1659–67. doi: 10.1097/00002030-199909100-00009. [DOI] [PubMed] [Google Scholar]
- 15.Barret B, Tardieu M, Rustin P, et al. Persistent mitochondrial dysfunction in HIV-1-exposed but uninfected infants: clinical screening in a large prospective cohort. AIDS. 2003;17:1769–85. doi: 10.1097/00002030-200308150-00006. [DOI] [PubMed] [Google Scholar]
- 16.Lund KC, Peterson LL, Wallace KB. Absence of a universal mechanism of mitochondrial toxicity by nucleoside analogs. Antimicrob Agents Chemother. 2007;51:2531–9. doi: 10.1128/AAC.00039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casademont J, Barrientos A, Grau JM, et al. The effect of zidovudine on skeletal muscle mtDNA in HIV-1 infected patients with mild or no muscle dysfunction. Brain. 1996;119:1357–64. doi: 10.1093/brain/119.4.1357. [DOI] [PubMed] [Google Scholar]
- 18.Lopez S, Miro O, Martinez E, et al. Mitochondrial effects of antiretroviral therapies in asymptomatic patients. Antivir Ther. 2004;9:47–55. [PubMed] [Google Scholar]
- 19.Mallon PW, Unemori P, Sedwell R, et al. In vivo, nucleoside reverse-transcriptase inhibitors alter expression of both mitochondrial and lipid metabolism genes in the absence of depletion of mitochondrial DNA. J Infect Dis. 2005;191:1686–96. doi: 10.1086/429697. [DOI] [PubMed] [Google Scholar]
- 20.Miura T, Goto M, Hosoya N, et al. Depletion of mitochondrial DNA in HIV-1-infected patients and its amelioration by antiretroviral therapy. J Med Virol. 2003;70:497–505. doi: 10.1002/jmv.10423. [DOI] [PubMed] [Google Scholar]
- 21.Miro O, Lopez S, Martinez E, et al. Mitochondrial effects of HIV infection on the peripheral blood mononuclear cells of HIV-infected patients who were never treated with antiretrovirals. Clin Infect Dis. 2004;39:710–16. doi: 10.1086/423176. [DOI] [PubMed] [Google Scholar]
- 22.Steer RA, Cavalieri TA, Leonard DM, Beck AT. Use of the Beck Depression Inventory for Primary Care to screen for major depression disorders. Gen Hosp Psychiatry. 1999;21:106–11. doi: 10.1016/s0163-8343(98)00070-x. [DOI] [PubMed] [Google Scholar]
- 23.Piper BF, Dibble SL, Dodd MJ, Weiss MC, Slaughter RE, Paul SM. The revised Piper Fatigue Scale: psychometric evaluation in women with breast cancer. Oncol Nurs Forum. 1998;25:677–84. [PubMed] [Google Scholar]
- 24.Grady C, Anderson R, Chase GA. Fatigue in HIV-infected men receiving investigational interleukin-2. Nurs Res. 1998;47:227–34. doi: 10.1097/00006199-199807000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Barroso J, Lynn MR. Psychometric properties of the HIV-Related Fatigue Scale. J Assoc Nurses AIDS Care. 2002;13:66–75. doi: 10.1016/S1055-3290(06)60242-2. [DOI] [PubMed] [Google Scholar]
- 26.Ware JE, Jr, Sherbourne CD. The MOS 36-item short-form health survey (SF-36). I. Conceptual framework and item selection. Med Care. 1992;30:473–83. [PubMed] [Google Scholar]
- 27.Brazier JE, Harper R, Jones NM, et al. Validating the SF-36 health survey questionnaire: new outcome measure for primary care. BMJ. 1992;305:160–4. doi: 10.1136/bmj.305.6846.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McHorney CA, Ware JE, Jr, Lu JF, Sherbourne CD. The MOS 36-item Short-Form Health Survey (SF-36): III. Tests of data quality, scaling assumptions, and reliability across diverse patient groups. Med Care. 1994;32:40–66. doi: 10.1097/00005650-199401000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Graham L, Orenstein JM. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat Protoc. 2007;2:2439–50. doi: 10.1038/nprot.2007.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voss JG, Raju R, Logun C, et al. A focused microarray to study human mitochondrial and nuclear gene expression. Biol Res Nurs. 2008;9:272–9. doi: 10.1177/1099800408315160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Coon KD, Valla J, Szelinger S, et al. Quantitation of heteroplasmy of mtDNA sequence variants identified in a population of AD patients and controls by array-based resequencing. Mitochondrion. 2006;6:194–210. doi: 10.1016/j.mito.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Lichtenstein KA, Ward DJ, Moorman AC, et al. Clinical assessment of HIV-associated lipodystrophy in an ambulatory population. AIDS. 2001;15:1389–98. doi: 10.1097/00002030-200107270-00008. [DOI] [PubMed] [Google Scholar]
- 35.Lange CG, Lederman MM, Madero JS, et al. Impact of suppression of viral replication by highly active antiretroviral therapy on immune function and phenotype in chronic HIV-1 infection. J Acquir Immune Defic Syndr. 2002;30:33–40. doi: 10.1097/00042560-200205010-00005. [DOI] [PubMed] [Google Scholar]
- 36.Cherry CL, Nolan D, James IR, et al. Tissue-specific associations between mitochondrial DNA levels and current treatment status in HIV-infected individuals. J Acquir Immune Defic Syndr. 2006;42:435–40. doi: 10.1097/01.qai.0000224974.67962.ce. [DOI] [PubMed] [Google Scholar]
- 37.Sievers M, Walker UA, Sevastianova K, et al. Gene expression and immunohistochemistry in adipose tissue of HIV type 1-infected patients with nucleoside analogue reverse-transcriptase inhibitor-associated lipoatrophy. J Infect Dis. 2009;200:252–62. doi: 10.1086/599986. [DOI] [PubMed] [Google Scholar]
- 38.Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348:425–32. [PMC free article] [PubMed] [Google Scholar]
- 39.Fleischman A, Johnsen S, Systrom DM, et al. Effects of a nucleoside reverse transcriptase inhibitor, stavudine, on glucose disposal and mitochondrial function in muscle of healthy adults. Am J Physiol Endocrinol Metab. 2007;292:E1666–73. doi: 10.1152/ajpendo.00550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haugaard SB, Andersen O, Pedersen SB, et al. Depleted skeletal muscle mitochondrial DNA, hyperlactatemia, and decreased oxidative capacity in HIV-infected patients on highly active antiretroviral therapy. J Med Virol. 2005;77:29–38. doi: 10.1002/jmv.20410. [DOI] [PubMed] [Google Scholar]
- 41.Cossarizza A, Riva A, Pinti M, et al. Increased mitochondrial DNA content in peripheral blood lymphocytes from HIV-infected patients with lipodystrophy. Antivir Ther. 2003;8:315–21. [PubMed] [Google Scholar]
- 42.Gerschenson M, Shiramizu B, LiButti DE, Shikuma CM. Mitochondrial DNA levels of peripheral blood mononuclear cells and subcutaneous adipose tissue from thigh, fat and abdomen of HIV-1 seropositive and negative individuals. Antivir Ther. 2005;10(Suppl 2):M83–9. [PubMed] [Google Scholar]
- 43.McComsey G, Tan DJ, Lederman M, Wilson E, Wong LJ. Analysis of the mitochondrial DNA genome in the peripheral blood leukocytes of HIV-infected patients with or without lipoatrophy. AIDS. 2002;16:513–8. doi: 10.1097/00002030-200203080-00001. [DOI] [PubMed] [Google Scholar]
- 44.Vittecoq D, Jardel C, Barthelemy C, et al. Mitochondrial damage associated with long-term antiretroviral treatment: associated alteration or causal disorder? J Acquir Immune Defic Syndr. 2002;31:299–308. doi: 10.1097/00126334-200211010-00005. [DOI] [PubMed] [Google Scholar]
- 45.Casula M, Weverling GJ, Wit FW, et al. Mitochondrial DNA and RNA increase in peripheral blood mononuclear cells from HIV-1-infected patients randomized to receive stavudine-containing or stavudine-sparing combination therapy. J Infect Dis. 2005;192:1794–800. doi: 10.1086/497140. [DOI] [PubMed] [Google Scholar]
- 46.Montaner JS, Cote HC, Harris M, et al. Nucleoside-related mitochondrial toxicity among HIV-infected patients receiving antiretroviral therapy: insights from the evaluation of venous lactic acid and peripheral blood mitochondrial DNA. Clin Infect Dis. 2004;38(Suppl 2):S73–9. doi: 10.1086/381449. [DOI] [PubMed] [Google Scholar]
- 47.Maagaard A, Holberg-Petersen M, Kollberg G, Oldfors A, Sandvik L, Bruun JN. Mitochondrial (mt)DNA changes in tissue may not be reflected by depletion of mtDNA in peripheral blood mononuclear cells in HIV-infected patients. Antivir Ther. 2006;11:601–8. [PubMed] [Google Scholar]
- 48.Bastard JP, Caron M, Vidal H, et al. Association between altered expression of adipogenic factor SREBP1 in lipoatrophic adipose tissue from HIV-1-infected patients and abnormal adipocyte differentiation and insulin resistance. Lancet. 2002;359:1026–31. doi: 10.1016/S0140-6736(02)08094-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.