Abstract
The skin functions as more than a physical barrier to infection. Epithelial cells of the skin can synthesize antimicrobial peptides, including defensins, which exhibit direct antimicrobial activity. Here we characterize the expression pattern, genetic variation and activity of the major β-defensin expressed in canine skin, canine β-defensin 103 (CBD103). The gene encoding CBD103 exhibits two forms of polymorphism: a common 3-basepair deletion allele and a gene copy-number variation. Golden retrievers and Labrador retrievers were the only breeds that encoded the variant allele of CBD103, termed CBD103ΔG23. Both these breeds also exhibited a CBD103 gene copy-number polymorphism that ranged from 2 to 4 gene-copies per diploid genome. Recombinant CBD103 and CBD103ΔG23, as well as the human ortholog human β-defensin 3 (hBD3) and hBD3ΔG23, showed potent and comparable antimicrobial killing against both methicillin-susceptible and methicillin-resistant Staphylococcus pseudintermedius. Skin biopsy specimens from dogs with atopic dermatitis revealed CBD103 expression levels similar to those in healthy controls and comparable at lesional and nonlesional sites. This expression pattern in dogs differs from the previously reported reduced expression of the human ortholog in atopic dermatitis. Overall, the similarities of CBD103 and its human ortholog reported here support the notion that the domestic dog may serve as a valuable model for studying β-defensin biology in the skin.
Key Words: Canine beta-defensin, Skin, Antimicrobial peptides, Epidermis, Atopic dermatitis
Introduction
Epithelial surfaces, including skin, are frequently confronted with pathogens, yet the incidence of infection is relatively low. Physical barriers to infection, circulating leukocytes and the endogenous microflora play important roles in host protection. In addition to these protective mechanisms, epithelial cells synthesize and secrete soluble effector molecules called antimicrobial peptides that serve as endogenous antibiotics to help thwart infection [1,2]. These epithelial antimicrobial peptides are expressed by many diverse species ranging from invertebrates to higher-level species, including mammals [3].
In mammals, defensins are a major family of antimicrobial peptides and this group can be classified into subfamilies: α-, β- and θ-defensins [4]. The α-defensins have been described in many mammals including glires, horses, mice, rats, primates and humans, while the θ-defensins are only present in nonhuman primates [5]. In contrast, the β-defensins represent the oldest evolutionary subfamily, found in diverse species ranging from horseshoe crabs and birds to hominids. The genomes of some mammals, such as ruminants and carnivores, appear to exclusively encode β-defensins.
β-defensins are expressed in many epithelial tissues, including a high expression in skin. They have many functions in innate immunity including host protection as endogenous antibiotics, signaling to promote chemotaxis and wound healing, and other intercellular communication activities [3,5,6,7,8]. The expression and biological role of β-defensins in various skin diseases have been the focus of much investigation [1,9,10,11,12,13]. However, most of our knowledge of β-defensins in skin stems from studies of human peptides, because animal models have not been fully developed. In this regard, the domestic dog is emerging as a potentially valuable model to study β-defensins [8,14,15,16]. A computational analysis of the canine genome, performed by Patil et al. [14], demonstrated that the canine genome encodes 43 β-defensin genes and pseudogenes. The genomic organization of these genes is similar to that for β-defensin genes in humans and mice. For example, the major human β-defensin cluster on chromosome 8 is syntenic to the canine cluster on chromosome 16 [14]. Two of the human β-defensins (hBD) encoded in this cluster, hBD2 and hBD3, are inducibly expressed to high levels in human skin, and both have potent antimicrobial activity [1,17,18]. Based on sequence comparisons, hBD3 and canine β-defensin 103 (CBD103) seem to be orthologs. CBD103 is expressed in canine skin [8,15] and respiratory tract [16], a tissue expression pattern similar to its human counterpart. Whether CBD103 shares the potent antibacterial activity of hBD3 has not yet been rigorously examined and is tested in this study. At present, no ortholog of hBD2 is evident in dogs. As is characteristic of the defensin family, there is often much species-to-species variability, so such differences are commonly identified [3] and important to consider when developing useful animal models.
Interestingly, CBD103 exhibits two types of genetic polymorphisms: (1) a variant allele with a 3-basepair deletion in its coding sequence, and (2) gene copy-number variation. The 3-basepair deletion in CBD103, termed CBD103ΔG23, was identified as the allelic variant responsible for a dominant black coat color in dogs [8]. CBD103ΔG23 lacks a glycine residue at the N-terminus of the mature peptide, and can bind with higher affinity than CBD103 to the melanocortin 1 receptor (Mc1r) on the surface of melanocytes. Due to its binding affinity and high expression levels, CBD103ΔG23 is thought to displace Agouti, an Mc1r signaling antagonist, allowing Mc1r to signal unabated, leading to the synthesis of the black pigment eumelanin and resulting in a black coat color.
In addition to the ΔG23 sequence variant, CBD103 appears to exhibit another form of genetic plasticity, the gene copy-number variation [19]. Whereas most genes are present in two copies per diploid genome, copy-number variation describes a phenomenon whereby the genes located at a single locus are present in the population at a variable copy number, compared to a reference genome. In humans, the gene encoding hBD3 resides in a β-defensin gene cluster that exhibits a copy-number polymorphism, which varies from 1 to 12 copies per diploid genome in human populations [20]. The gene copy-number polymorphism for this β-defensin cluster, including the gene encoding hBD3 and adjacent β-defensin genes, has been extensively investigated, including its association with skin disease susceptibility [11]. However, knowledge regarding gene copy-number variation of the putative canine ortholog, CBD103, is sparse.
Atopic dermatitis (AD) is a chronic inflammatory dermatopathy that is frequently complicated by secondary bacterial infection in both dogs and humans [21]. In human AD patients, it is thought that the inflammatory cytokines in the skin, such as IL-4 and IL-13, reduce the expression of hBD3 [22]. In addition, it has been noted that human AD skin is defective in killing Staphylococcus aureus due to aberrations in mobilization of hBD3 [23]. A recent study in dogs with AD indicates that CBD103 expression is equal between lesional and nonlesional skin, yet reduced when compared to healthy controls [24].
In this investigation, we sought to give insight on antimicrobial activity, tissue expression in healthy and disease specimens, and breed variation of CBD103 and its common genetic polymorphisms. As domestic dogs emerge as a model to study the biology of β-defensins in the skin, the data reported here highlight many similarities of CBD103 with its human counterpart, although some differences in expression are observed.
Materials and Methods
Tissue
Canine tissue was procured from client-owned dogs which, for reasons unrelated to this study, had been euthanized, processed for necropsy and deemed unrestricted for use in research. This took place at the University of California (UC) Davis School of Veterinary Medicine William R. Pritchard Veterinary Medical Teaching Hospital. Tissues (epididymis and skin) were placed in RNAlater (Ambion, Applied Biosystems, Foster City, Calif., USA) and rocked at room temperature overnight. Preserved specimens were stored at −80°C until further processing. Additional RNA from the gastrointestinal tract (tongue, stomach, duodenum, jejunum, ileum, colon and rectum), skin, lungs, kidneys, pancreas and bone marrow was procured from a commercial source (Zyagen, San Diego, Calif., USA).
Canine Genomic DNA
Blood samples were acquired from patients of the UC Davis William R. Pritchard Veterinary Medical Teaching Hospital. All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies, and all animal work was approved by the Institutional Animal Care and Use Committee-approved protocol for Animal Care and Use #15356. Genomic DNA was isolated using the QiaAmp DNA Blood Mini Kit (Qiagen Inc.).
PCR of Canine Defensins
RNA isolation and cDNA synthesis was performed as previously described [25,26]. Qualitative PCR was used as a broad screening tool to determine the expression patterns of defensins encoded by the canine genome. PCR primers were designed using MacVector software (MacVector Inc., Cary, N.C., USA) to specifically amplify canine defensin sequences deposited on the National Center for Biotechnology Information (NCBI) [14], and the housekeeping transcript, canine ribosomal protein S5 (RPS5) (online suppl. Table 1; for all online suppl. material, see www.karger.com?doi=10.1159/000334566) [27]. FastStart Taq (Roche Applied Sciences, Indianapolis, Ind., USA) was used to amplify cDNA representing 100 ng of total RNA with the following parameters: 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final extension at 72°C for 7 min. The epididymis and testis were used as positive controls for defensin expression as these tissues have been shown to express most of the defensins in other species [14]. Nontemplate controls substituted water for the DNA solution. PCR products were resolved by agarose gel electrophoresis (1.5% w/v) and detected using ethidium bromide staining. Positive PCR products for each defensin transcript were excised from the agarose gel, purified with QIAquick PCR purification kit (Qiagen) and directly sequenced to ensure identification of the amplified cDNA. Quantitative RT-PCR (qRT-PCR) was performed on cDNA from tissue from the initial screening using primers for CBD1 (2)s, CBD1 (118)a, CBD103 (12)s, CBD103 (117)a, CBD108 (23)s, CBD108 (126)a, RPS5 (54)s and RPS5 (235)a (online suppl. Table 1) [26].
Genetic Variation
To determine the prevalence of the ΔG23 genetic variation in CBD103, PCR primers were designed (MacVector) to flank the CBD103 genomic sequence surrounding the 3-base pair deletion, with a 6FAM fluorescently labeled sense primer (online suppl. Table 1). Approximately 10 ng of genomic DNA from different breeds (Doberman Pinscher (n = 15), German Shepherd (n = 15), Golden retriever (n = 13), Labrador retriever (n = 16) and Rottweiler (n = 15)) was PCR amplified with CBD103 (1662)s and CBD103 (1889)a (online suppl. Table 1) using the following parameters: 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final extension at 72°C for 7 min. A 1:20 dilution was made with GeneScan 400D Rox in Hi-Di formamide (Life Technologies Corporation, Carlsbad, Calif., USA). Samples were detected on an ABI 3100 Genetic Anaylyzer (Life Technologies Corporation) using STRand software developed by the UC Davis Veterinary Genetics Laboratory (http://www.vgl.ucdavis.edu/ informatics/strand.php) [28].
To determine GCN variation in CBD103, polyacrylamide gel electrophoresis-purified PCR primers were designed (MacVector) to amplify regions of CBD103 and a reference gene, TATA-binding protein (TBP), thought to be diploid in the genome [online suppl. Table 1: CBD103 CN (421)s, CBD103 CN (580)a, CTBP CN (17889)s, and CTBP CN (17999)a]. Quantitative PCR (qPCR) was performed using FastStart Taq (Roche) using 10 ng of genomic DNA from different breeds with the following parameters: 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final extension at 72°C for 7 min. GCN was calculated using the method detailed by Linzmeier and Ganz [29].
Immunohistochemistry
Canine skin samples were formalin-fixed and embedded in paraffin. Five-µm paraffin sections were mounted on charged slides and deparaffinized in xylene twice for 3 min. The slides were then incubated in 100% ethanol twice for 2 min each and endogenous peroxidases were quenched with a 0.1% hydrogen peroxide in methanol for 5 min. After rehydration in 95% ethanol for 2 min, then in 70% for 2 min, slides were placed in a humidified chamber at room temperature. A blocking step with 0.02% goat serum in phosphate-buffered saline (PBS) for 20 min was followed by application of the primary polyclonal antibody raised against hBD3 [24] (Allele Biotech, San Diego, Calif., USA) at a dilution of 1:500 in 0.02% goat serum in PBS. Negative control slides were incubated with 0.02% goat serum in PBS at 4°C overnight and then washed with PBS twice for 10 min. Biotinylated α-rabbit secondary antibody (Vector Elite Kit, Vector Laboratories, Burlingame, Calif., USA) was applied for 20 min. After two PBS washes for 2 min, ABC reagent (Vector Elite Kit) was applied to the slides and incubated for 20 min at room temperature. Following three washes with Tris-buffered saline (TBS) 3 times for 2 min, the chromagen 3,3′-diaminobenzidine (Vector Elite Kit) was added to each slide for 5 min. The final counterstaining with Light Green (Sigma-Aldrich, St. Louis, Mo., USA) for 5 min was performed after a wash with TBS for 4 min and a wash with deionized water for 4 min. After drying the slides, coverslips were mounted with Permount (ThermoFisher Scientific, Waltham, Mass., USA). Evaluation of the tissue and image recording was performed using an Olympus DP72 microscope (Olympus America Corporation, Center Valley, Pa., USA).
Recombinant Peptide
The cDNA encoding the predicted mature CBD103 (45 amino acids), CBD103ΔG23 (44 amino acids), hBD3 (45 amino acids) and hBD3ΔG23 (44 amino acids) was PCR amplified (online suppl. Table 1). A codon encoding a methionine residue was engineered into the sense PCR primer immediately adjacent to the N-terminal codon of the defensin peptides, so as to provide a cleavage site for cyanogen bromide that would allow cleavage of the mature peptide from the remainder of the expressed fusion protein. Each PCR product was restriction-digested with EcoRI and SalI and inserted (in frame) into a pET32a(+) expression vector (Novagen, EMD Chemicals, Darmstadt, Germany), containing an N-terminal His-tag and thioredoxin. The four (sequence-verified) expression constructs were subcloned into BL-21 (DE3) chemically competent cells (Invitrogen, Carlsbad, Calif., USA), propagated in lysogeny broth (LB) containing carbenicillin (50 µg/ml) at 37°C shaking overnight and glycerol stocks were frozen at −80°C. To obtain individual colonies for expression, each glycerol stock was streaked onto an LB plate containing carbenicillin (50 µg/ml) and incubated overnight at 37°C. An individual colony was used to inoculate a starter culture (LB and carbenicillin 50 µg/ml) and incubated at 37°C shaking overnight. The following morning, the entire starter culture was added to 1 liter of LB containing carbenicillin (50 µg/ml) and sterile phosphate buffer (36 mM K2HPO4, 8.5 mM KH2PO4). Peptide expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (final concentration 0.5 mM) at an OD600 equal to 0.5–0.6. The culture was allowed to induce for 4–5 h, at which point BL21 cells were pelleted at 5,000 g for 15 min. Pelleted BL21 cells were lysed with 6.0 M guanidine-HCl and 100 mM Tris pH 8.0, sonicated and centrifuged at 5,000 g for 30 min. The resulting clarified supernatant was incubated with Ni-NTA agarose beads (Novogen, 5 ml/l of induction) overnight at 4°C. The beads were then packed into a column, washed with 6 M guanidine/100 mM Tris (pH 8.0) and eluted with 6 M guanidine/100 mM Tris/1 M imidazole (pH 6.0). The eluent was dialyzed against 5% glacial acetic acid and lyophilized. To chemically cleave the His-tag and thioredoxin from the recombinant defensin peptide, the lyophilized product was reconstituted in 80% formic acid, purged with argon gas and cut with CNBr (20 mg/ml) overnight at room temperature. The cut reaction product was diluted 10-fold with Milli-Q H2O, dialyzed against 5% glacial acetic acid, lyophilized and reconstituted in 0.05% glacial acetic acid. The cleaved reaction products were then initially purified using C18 semipreparative reverse-phase high-performance liquid chromatography (RP-HPLC) with a 15–50% gradient buffer B (99.9% acetonitrile, 0.1% trifluoroacetic acid), monitoring absorbance at 230 nm, with fractions collected every minute at a flow rate of 1 ml/min. Approximately 10% of each fraction was added to 3× protein sample buffer (5% acetic acid, 3 M urea, 0.01% methyl green) and loaded into a 12.5% acid urea polyacrylamide gel electrophoresis (AU-PAGE) and run at 130 V in reverse polarity for 1.5 h. The resulting gels were stained with SimplyBlue (Invitrogen) and destained with water. Fractions that appeared to contain the cleaved mature peptide based on gel migration were combined and subjected to further rounds of RP-HPLC purification with a shallower gradient of buffer B (26–38% for CBD103, CBD103ΔG23, hBD3ΔG23 and 40–46% for hBD3) using a smaller bore C18 column. In the final purification step, each peptide was hand-collected upon elution. The overall yield was approximately 1 mg of peptide per 6 liters of bacterial culture. The isolated peptides were subject to RP-HPLC on a C18 analytical column with a gradient of acetonitrile at a flow rate of 1 ml/min (online suppl. fig. 3). Purified peptides were also analyzed by MALDI-TOF mass spectroscopy (online suppl. fig. 4). To further analyze the recombinant peptides by AU-PAGE and Western blotting, each of the peptides was reduced with 100 mM TCEP at 55°C for 25 min. Approximately 1 µg of reduced peptide was added to 3× protein sample buffer, loaded into a 12.5% AU-PAGE and run at 130 V for 1.5 h. The resulting gel was stained with SimplyBlue (Invitrogen) and destained with water. For Western blotting, 250 ng of each reduced peptide was added to 3× protein sample buffer, loaded onto a 12.5% AU-PAGE and run at 130 V in reverse polarity for 1.5 h. Peptides were transferred to a PVDF membrane (Millipore, Billerica, Mass., USA) in reverse polarity with 2 mA/cm2 current for 20 min. The blot was fixed with 0.05% glutaraldehyde, and probed with a polyclonal antibody raised against hBD3 (Allele Biotech) at a 1:3,000 dilution in TBS-Tween 20 and milk (0.05%). After subsequent washes, the blot was incubated with a goat α-rabbit secondary antibody (KPL, Gaithersburg, Md., USA) diluted in TBS-Tween 20 and milk (0.05%). Chemiluminescent signal was detected with a Biospectrum AC imaging system (UVP, Upland, Calif., USA).
Antimicrobial Assay
Clinical isolates of Staphylococcus pseudintermedius, both methicillin-susceptible and methicillin-resistant, were acquired, as well as bacteria isolated from canine skin. To confirm the identity of the clinical isolates, bacterial DNA was isolated using the QIAamp DNA Stool mini kit (Qiagen, Valencia, Calif., USA), universal bacterial primers [Bac16s (27)s and Bac16s (1391)a] were used to amplify the 16S rRNA gene, subcloned into pBluescript and sequenced. A radial diffusion antimicrobial assay was used to determine the antibacterial activity of unreduced CBD103, CBD103ΔG23, hBD3 and hBD3ΔG23 at various concentrations (250, 83, 28, 9 and 3 µg/ml) of each peptide [26,30].
Atopic Dermatitis Samples
Dogs included in the study had been presented for intradermal testing (IDT). All dogs had clinical signs and a history supportive of AD. Prior to presentation for intradermal testing, the possibility of pruritus attributable to flea allergy dermatitis had been ruled out with stringent flea control with an appropriate adulticide and dogs were still receiving either a topical or an oral adulticide as flea control at the time that skin biopsies and IDT were performed. Cutaneous adverse food reaction had been ruled out prior to presentation for IDT confirmed by completion of an elimination diet trial and subsequent rechallenge. All secondary infections with bacteria or yeast had been effectively treated. Dogs were not receiving either antihistamines or corticosteroids orally or topically for 4 weeks prior to presentation for IDT. Some were receiving cyclosporine for the management of pruritus at the time of skin testing.
A 6-mm biopsy punch was used to procure skin samples. Lesional skin refers to biopsies sampled from the axilla, an area that is typically affected in dogs with AD. Nonlesional samples were from the interscapular region, an area that is not typically affected in AD. Biopsy specimens were bisected sagitally, with one half formalin-fixed and the remaining half preserved in RNAlater (Ambion) and processed like other skin specimens described above. All dogs were examined by a Board-certified veterinary dermatologist (C.A.O.) and there were no clinical lesions other than mild erythema in the axilla of some dogs. Histologic examination of all skin biopsies was performed by a Board-certified veterinary dermatopathologist (V.K.A.) and no lesions were seen. All samples were collected in accordance with the UC Davis Clinical Trial Review Board and Institutional Animal Care and Use Committee, and there was written client consent.
Statistical Analysis
All statistical comparisons were performed using paired, 2-tailed, Student t tests in Excel (Microsoft Corporation, Redmond, Wash. USA).
Results
Canine β-Defensin Expression
In an effort to determine if CBD103 was the sole highly expressed β-defensin in skin, we embarked on the most comprehensive screen of canine β-defensin expression to date. PCR primers (online suppl. Table 1) were designed to specifically amplify each of the 41 putative β-defensin gene transcripts [14] from skin, as well as 13 other tissues. The male epididymis expressed nearly every β-defensin encoded by the canine genome, which conveniently served as a positive control for all but one PCR reaction (online suppl. fig. 1). CBD121 was the only canine β-defensin not detected in any tissue. The PCR product from each of the other β-defensin PCR reactions was directly sequenced to verify its identity. CBD1, CBD108 and CBD123 exhibited a generalized pattern of expression, as mRNA transcripts were identified in most of the tissue analyzed (online suppl. fig. 1). In contrast, a more specific pattern of expression was noted for CBD103, CBD109, CBD112, CBD119, CBD122, CBD139 and CBD140. The only two β-defensin genes with readily detectable expression in skin were CBD1 and CBD103.
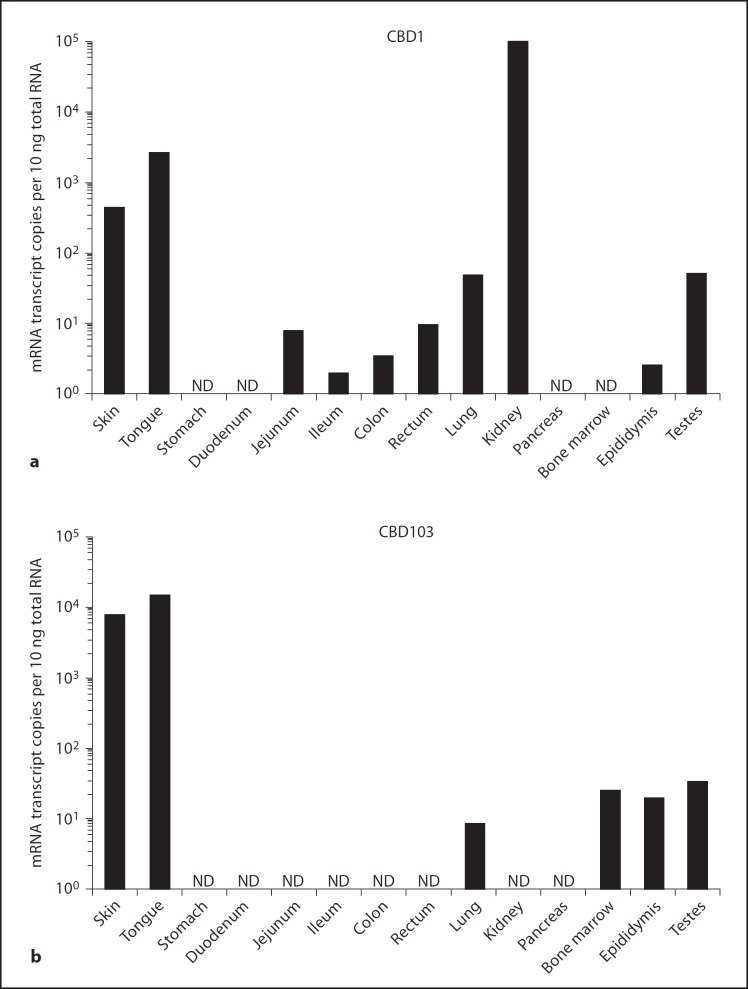
qRT-PCR assays were then designed for the three most highly expressed canine β-defensin transcripts (CBD1, CBD103 and CBD108) based on this initial qualitative PCR screening. Using this technique, CBD1 also showed a generalized pattern of expression with appreciable levels in the skin, tongue and kidneys (fig. 1a). CBD108 was expressed at low levels in any tissues tested, with higher levels in the kidneys and testes (online suppl. fig. 2A). In contrast, a more tissue-specific expression pattern was seen with CBD103, with expression at high levels detected in the skin (8.2E+03 copies/10 ng total RNA) and tongue (1.5E+04 copies/10 ng total RNA) (fig. 1b). RPS5, used as a housekeeping gene [27], exhibited relatively consistent and high levels of expression across all of the different tissues examined (online suppl. fig. 2B). These data indicated that CBD103 was the most highly expressed β-defensin in canine skin.
Fig. 1.
Expression of canine β-defensins. qRT-PCR analysis of absolute copy number of mRNA transcripts encoding CBD1 (a) and CBD103 (b) in RNA isolated from various tissues including the skin, gastrointestinal tract (tongue, stomach, duodenum, jejunum, ileum, colon, rectum), lungs, kidneys, pancreas, bone marrow, testes and epididymis. Each bar represents an average of replicate assays (standard deviation <10%) from a single specimen tested (n = 1). ND = Not detected.
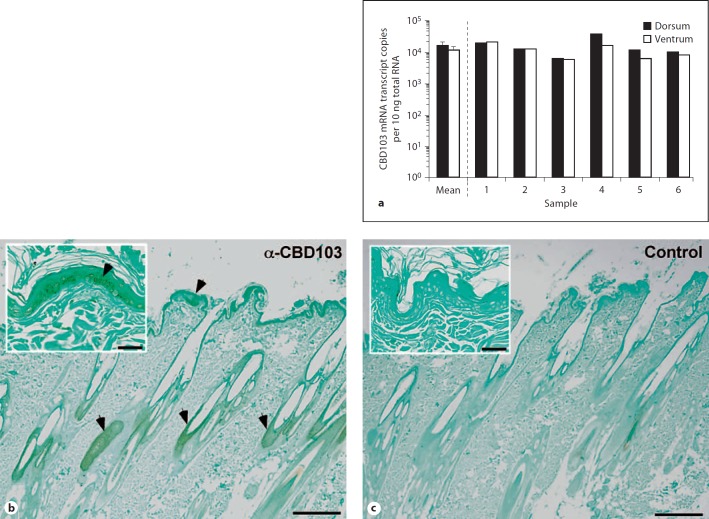
To extend our analysis of epidermal CBD103 expression, skin specimens were obtained from the ventrum and dorsum of 6 dogs of varying breeds and age. RNA was extracted from each tissue sample, and CBD103 expression was evaluated using qRT-PCR (fig. 2a). The mean CBD103 mRNA expression level was essentially the same when comparing skin from either the dorsal (mean: 1.7E+4 copies/10 ng total RNA) or ventral surface (mean: 1.3E+4 copies/10 ng total RNA). In addition, CBD103 mRNA was expressed to similar levels in the skin in each of the 6 different dogs examined (fig. 2a). To identify the site of CBD103 expression in skin, we employed immunohistochemistry using a polyclonal antibody raised against the human ortholog of CBD103, hBD3. Positive staining was detected in keratinocytes of the epidermis, as well as in follicular epithelial cells (fig. 2b).
Fig. 2.
Epidermal mRNA and peptide expression of CBD103. a qRT-PCR analysis of absolute copy number of mRNA transcripts encoding CBD103 in RNA isolated from the caudodorsal (dorsal) and inguinal (ventral) skin of dogs. Calculated mean of the 6 dogs, dorsal and ventral skin. Error bars represent standard error. b Immunostaining of CBD103 in canine skin (2×) using a polyclonal antibody (1:500) raised against hBD3. CBD103 localizes to keratinocytes and follicular epithelial cells. Inset is taken at 20×. c Negative control for staining with secondary antibody alone (2×). Inset is taken at 20×. Arrowheads indicate positive staining. Size bar in 2× view: 300 µm, and in 20× view: 30 µm.
CBD103 Genetic Variation
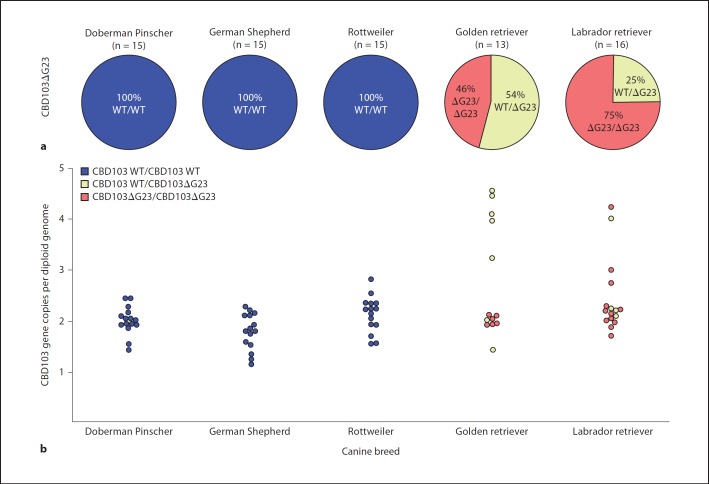
A common variant of CBD103, CBD103ΔG23, has a 3-basepair deletion encoding the amino terminal glycine of the mature peptide. This variant has previously been identified as the allele responsible for dominant black coat color in dogs [8]. A PCR-based genotyping assay was developed to evaluate the presence of this variant sequence in different dog breeds. Fluorescently labeled PCR products were separated using capillary gel electrophoresis, and alleles were discriminated based on product size. Of the 5 breeds evaluated, Rottweilers, Doberman Pinschers and German Shepherds did not possess a variant allele; only the wild-type CBD103 allele was detected (fig. 3a). Interestingly, the variant allele was found in all of the Golden retrievers (54% WT/ΔG23; 46% ΔG23/ΔG23) and Labrador retrievers (75% WT/ΔG23; 25% ΔG23/ΔG23).
Fig. 3.
Prevalence of CBD103 common variant and gene copy-number variation. a Pie graphs represent individuals of a given breed, divided into canine genotypes: CBD103/CBD103 (blue), CBD103/CBD103ΔG23 (yellow) and CBD103ΔG23/CBD103ΔG23 (red). Doberman Pinschers (n = 15), German Shepherds (n = 15) and Rottweilers (n = 15) do not appear to have the CBD103ΔG23 allele, whereas Golden retrievers (n = 13) and Labrador retrievers (n = 16) possess the CBD103ΔG23 allele as a heterozygote or homozygote variant. b Plot of CBD103 gene copy-number variation with colors indicating the CBD103 genotype. Doberman Pinschers, German Shepherds and Rottweilers encode 2 copies of CBD103 per diploid genome, whereas Golden retrievers and Labrador retrievers have variation in their total CBD103 gene copy number.
In addition to the CBD103ΔG23 allele, another form of genetic variation identified at the β-defensin locus on chromosome 16 was the gene copy-number polymorphism [19]. The CBD103 gene is located within a cluster of β-defensins that is syntenic with a cluster of orthologs on human chromosome 8, many of which are thought to be orthologous between the two species [14]. In humans, this cluster exhibits gene copy-number polymorphism, and a given individual can encode between 1 and 12 copies of this cluster per diploid genome [20,29]. To investigate similar gene copy-number polymorphism in dogs, we designed a qPCR assay with specific primers for the CBD103 gene and a diploid reference gene, TBP. Doberman Pinschers, German Shepherds and Rottweilers did not appear to exhibit CBD103 gene copy-number polymorphism, as each individual tested had a total CBD103 gene copy number of approximately two per diploid genome (fig. 3b). In contrast, Golden retrievers and Labrador retrievers exhibited gene copy-number polymorphism, with CBD103 gene copy numbers of 2–4 copies per diploid genome (fig. 3b).
Antimicrobial Activity
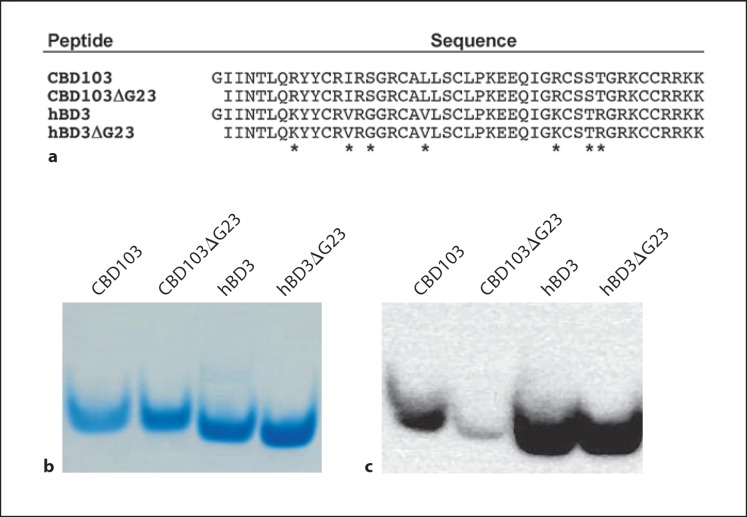
To investigate the possible differential antimicrobial activity of the wild-type CBD103 and its ΔG23 variant, recombinant peptides were expressed (fig. 4a), purified using RP-HPLC (online suppl. fig. 3), and analyzed using mass spectrometry (online suppl. fig. 4). For comparison, hBD3 and a corresponding variant, hBD3ΔG23, were similarly expressed, purified and analyzed (online suppl. fig. 3, 4). The purified peptides were subject to AU-PAGE under reducing conditions, and analyzed to assess immunoreactivity (fig. 4b, c). The Western blot of the recombinant peptides indicated that hBD3 and hBD3ΔG23 are equally recognized by the α-hBD3 polyclonal antibody, whereas the CBD103ΔG23 peptide was differentially recognized compared to CBD103 (fig. 4c). The result supports that N-terminal residues of CBD103 are accessible for protein-protein interactions, consistent with its proposed role in coat color [8].
Fig. 4.
CBD103 and hBD3 peptide sequence comparison and recombinant expression. a Predicted mature peptide sequences for CBD103 and CBD103ΔG23, as well as hBD3 and hBD3ΔG23. Asterisks designate amino acid differences, all of which are conservative except S37G and T58R. b AU-PAGE of TCEP reduced recombinant CBD103, CBD103ΔG23, hBD3 and hBD3ΔG23 showing peptide homogeneity by SimplyBlue staining. c Western blot of TCEP reduced recombinant CBD103, CBD103ΔG23, hBD3 and hBD3ΔG23 using a polyclonal antibody (1:3,000) raised against hBD3.
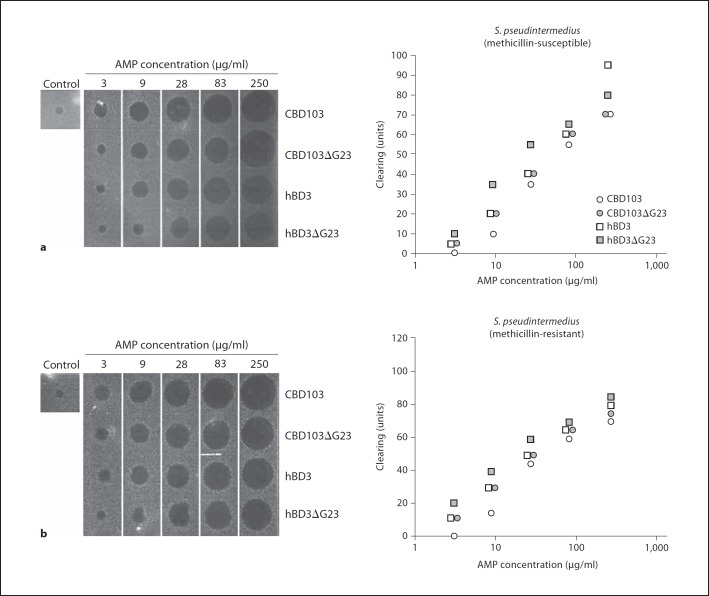
The antibacterial activity of the recombinant defensins was tested using a radial diffusion assay with both Gram-positive and Gram-negative bacteria. Clinical isolates of S. pseudintermedius, a common pathogen of dog skin, particularly canine patients with AD, were procured for antimicrobial testing. In addition to these clinical isolates, endogenous bacteria from canine skin were cultured for comparison with S. pseudintermedius. Both CBD103 and CBD103Δ23 exhibited relatively equal activity against methicillin-susceptible and methicillin-resistant S. pseudintermedius (fig. 5a, b). Similarly, hBD3 and hBD3ΔG23 were equally antimicrobial against S. pseudintermedius. Both sets of canine and human peptides exhibited potent antimicrobial activity against endogenous bacteria found on canine skin (Bacillus licheniformis, Micrococcus sp. and Bacillus cereus), as well as a laboratory strain of Escherichia coli D31 (table 1).
Fig. 5.
Antibacterial activity of recombinant CBD103. Determination of antibacterial activity of CBD103, CBD103ΔG23, hBD3 and hBD3ΔG23 using a radial diffusion antibacterial assay with clinical isolates of methicillin-susceptible (a) and methicillin-resistant (b) S. pseudintermedius. Graphical representation of the antibacterial clearing by CBD103, CBD103ΔG23, hBD3 and hBD3ΔG23 [30].
Table 1.
Antibacterial activity
| Bacteria | MIC ((µg/ml) |
|||
|---|---|---|---|---|
| CBD103WT | CBD103∆G23 | hBD3 WT | hBD3∆G23 | |
| Escherichia coli D31 | 1 | 1 | 1 | 1 |
| Bacillus licheniformis | 2 | 2 | 2 | 1 |
| Micrococcus sp. | 1 | 2 | 2 | 1 |
| Bacillus cereus | 2 | 1 | 1 | 1 |
| Staphylococcus pseudintermedius (susceptible) | 5 | 2 | 3 | 1 |
| Staphylococcus pseudintermedius (resistant) | 2 | 2 | 1 | 1 |
Minimum inhibitory concentration (MIC) of CBD103, CBD103ΔG23, hBD3 and hBD3ΔG23 using a radial diffusion antimicrobial assay.
Association with Atopic Dermatitis
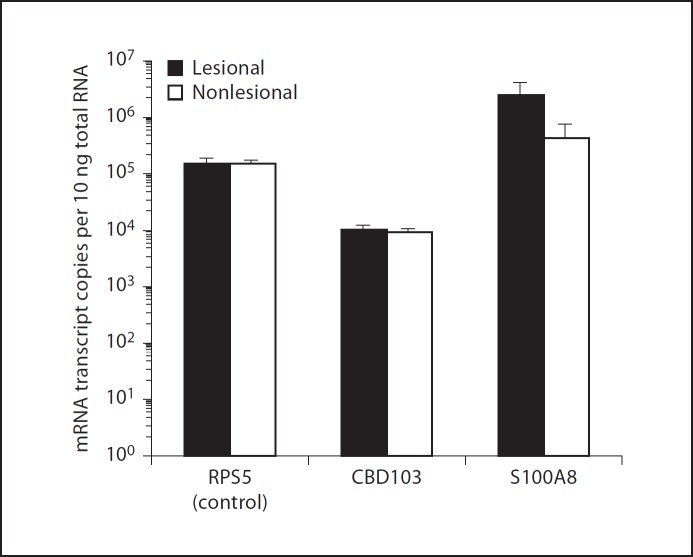
In human patients with AD, hBD3 mRNA levels and peptide mobilization is impaired [9,10,23]. We therefore hypothesized that canine patients with AD may also have a reduced defensin expression, predisposing them to secondary infection. To test this hypothesis, punch biopsy specimens of the skin were procured from both the lesional (axilla) and nonlesional (interscapular) skin of canine patients diagnosed with AD. We did not detect a statistically significant difference in CBD103 mRNA expression between lesional (mean: 1.1E+04 copies/10 ng total RNA) and nonlesional (mean: 9.4E+03 copies/10 ng total RNA) skin (p = 0.42, fig. 6). Interestingly, S100A8, an inflammatory protein known to be upregulated in the skin of atopic dogs [31], exhibited a 6-fold higher mRNA expression in lesional (mean: 2.5E+06 copies/10 ng total RNA) when compared with nonlesional (mean: 4.4E+05 copies/10 ng total RNA) skin. As a control in these experiments, the housekeeping gene, RPS5, was expressed to equal levels between the two skin sites (fig. 6).
Fig. 6.
CBD103 expression in AD. Quantitative RT-PCR analysis of absolute copy-number of mRNA transcripts encoding RPS5, CBD103 and S100A8 in RNA isolated from lesional and nonlesional skin biopsy specimens from dogs (n = 18) diagnosed with AD. Error bars represent standard error.
Discussion
β-defensins are key effector molecules of innate immunity expressed at many body surfaces, including skin. Studies on β-defensins in skin have almost exclusively focused on human peptides [32], including investigation on expression [17,18], regulation [33,34,35] and associations with skin infection [12,13,36] and other diseases, such as psoriasis and AD [10,11]. Few studies of skin β-defensins in animal models have been reported, except for the dog model [8,15,16,24,37,38]. The current study sought to extend our knowledge of canine β-defensins by quantifying CBD103 expression in healthy skin, determine genetic variation of CBD103 amongst and within different breeds, characterize the antibacterial activity of CBD103 and CBD103ΔG23 and evaluate expression of CBD103 from lesional and nonlesional skin of dogs with AD.
Of the canine β-defensins, CBD103 is expressed to the highest levels in skin. Consistent with the finding reported here, localized expression of CBD103 to keratinocytes and follicular epithelial cells has been reported previously [24]. To determine possible variation in CBD103 expression in canine skin, we procured dorsal and ventral skin specimens from dogs lacking skin disease. CBD103 mRNA expression was approximately equal between the sites of expression, both within the same dog and between others tested. These quantitative data corroborate the finding in a study by Wingate et al [15], where qualitative PCR demonstrated similar intensity PCR products by gel electrophoresis. The human ortholog of CBD103, hBD3, exhibits low-level constitutive expression; however, in the presence of inflammatory stimuli such as heat-inactivated bacteria and TNF-α, human keratinocyte cells responded with 20- to 30-fold increased hBD3 expression [18]. In contrast to the low baseline expression in humans, CBD103 mRNA constitutive expression is remarkably high in canine skin. In canine keratinocyte cell culture models, we were unable to induce CBD103 over constitutive levels with various bacterial or proinflammatory stimuli, including IL-1β, hemolytic Streptococcus, and S. pseudintermedius (unpubl. results). Further investigations will be necessary to determine if CBD103 expression levels deviate under any circumstances from the constitutive ones we report here.
The dog represents an interesting species to study β-defensin genetic variation, because there are two common polymorphisms: a 3-basepair deletion in CBD103, and the gene copy-number polymorphism. We found that the CBD103ΔG23 variant sequence is common in Golden retrievers and Labrador retrievers, in accordance with another study, which also reported this variant in the Great Dane, Basenji and Poodle [8]. While CBD103ΔG23 is responsible for a dominant black coat color in some breeds, coat color in Golden retrievers and in yellow Labrador retrievers is independent of their CBD103 genotype, due to genetic mutations in Mc1r [39]. A second aspect of genetic heterogeneity is gene copy-number polymorphism, which provides genetic plasticity and is observed with both canine and human β-defensins. Both Golden retrievers and Labrador retrievers had gene copy numbers of CBD103 that varied from approximately 2–4 copies per diploid genome. Two studies using human biopsy specimens and cell culture demonstrated a direct correlation between β-defensin mRNA expression and gene copy numbers [40,41]. Currently, it is not known if an increased gene copy number in the dog similarly results in increased β-defensin expression.
One goal of this study was to characterize the antimicrobial activity of CBD103 and the variant, CBD103ΔG23. Given the high allele frequency of CBD103ΔG23 in some breeds of dog, we questioned if altered antimicrobial activity of the variant peptide should be considered when developing models to explain the curiously high frequency of this polymorphism. To test the antibacterial activity, we procured clinical isolates of methicillin-susceptible and methicillin-resistant S. pseudintermedius, a common pathogen of canine skin. In addition, we cultured and isolated endogenous microflora from the skin of a healthy dog, including: B. licheniformis, Micrococcus sp. and B. cereus. Interestingly, CBD103 and CBD103ΔG23 exhibited equal in vitro antibacterial activity against all bacteria tested. This observation is especially important due to the prevalence of the CBD103ΔG23 genotype in some of the more common breeds of dog, including Labrador retrievers and Golden retrievers. CBD103 and CBD103ΔG23 also displayed equally potent antibacterial activity when compared to hBD3 and the corresponding variant, hBD3ΔG23. The similar activities of CBD103 and CBD103ΔG23 suggest that antimicrobial function may not be an important factor to explain changes in allele frequency in dog populations.
In both human and canine AD, patients are frequently complicated by recurrent secondary bacterial infections, S. aureus in humans [9,42] and S. pseudintermedius in dogs [43]. Previous studies have demonstrated the reduced killing capacity of the epidermis in human AD patients due to reduced hBD3 expression [9,10,22] and mobilization to the skin surface [23]. Interestingly, we did not find a difference in CBD103 mRNA transcript levels between lesional and nonlesional specimens. Our data are in agreement with another study reporting equal CBD103 mRNA expression from lesional and nonlesional skin in AD [24]. However, we did not appreciate any differences in CBD103 mRNA expression between healthy dogs and dogs with AD, in contrast to what was previously reported [24]. Genetic differences, in terms of the variant CBD103ΔG23 allele and/or gene copy-number variation, could potentially contribute to a selection bias with respect to environmental pathogens and/or disease associations, notably in those breeds where both variations are observed. However, we found that CBD103 and CBD103ΔG23 are both potent antibacterial peptides with virtually equivalent antibacterial activity, particularly against common pathogens of canine skin. Therefore, our data indicate that altered CBD103 expression in atopic skin does not account for recurrent secondary bacterial infections.
Overall, the expression in skin, potent antimicrobial activity, and gene copy-number polymorphism are properties shared between CBD103 and its human ortholog. On the other hand, apparent constitutive expression in healthy skin and AD distinguishes the canine β-defensin from its human counterpart. The findings of this investigation will help shape how the domestic dog can be further developed as a valuable model of β-defensin biology in skin.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
We thank Dr. Denise M. Imai for her assistance with imaging of immunohistochemical detection of CBD103 and Dr. Nicole Eckholm, Dr. Casey Stepnik, Monica Kratochvil, Judy Schettler and Nina Toebe for help with clinical sample procurement. We also thank Hiutung Chu for critical reading of the manuscript. The investigation was supported in part by grants from the UC Davis School of Veterinary Medicine Center for Companion Animal Health and from the NIH (AI32738, AI50843, and AI76246). B.C.L. received support from NIH T32AI60555 and T32RR021312, and the UC Davis School of Veterinary Medicine, Veterinary Scientist Training Program.
References
- 1.Schroder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci. 2006;63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard JJ, Gallo RL. Protecting the boundary: the sentinel role of host defense peptides in the skin. Cell Mol Life Sci. 2011;68:2189–2199. doi: 10.1007/s00018-011-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 4.Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. 2004;327:539–549. doi: 10.1016/j.crvi.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 6.Varoga D, Pufe T, Harder J, Schroder JM, Mentlein R, Meyer-Hoffert U, Goldring MB, Tillmann B, Hassenpflug J, Paulsen F. Human beta-defensin 3 mediates tissue remodeling processes in articular cartilage by increasing levels of metalloproteinases and reducing levels of their endogenous inhibitors. Arthritis Rheum. 2005;52:1736–1745. doi: 10.1002/art.21090. [DOI] [PubMed] [Google Scholar]
- 7.Bowdish DM, Davidson DJ, Hancock RE. Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol. 2006;306:27–66. doi: 10.1007/3-540-29916-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, Kerns JA, Schmutz SM, Millhauser GL, Barsh GS. A β-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, Gallo RL, Leung DY. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 10.Howell MD, Boguniewicz M, Pastore S, Novak N, Bieber T, Girolomoni G, Leung DY. Mechanism of hBD-3 deficiency in atopic dermatitis. Clin Immunol. 2006;121:332–338. doi: 10.1016/j.clim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, Reis A, Armour JA, Schalkwijk J. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer-Hoffert U, Schwarz T, Schroder JM, Glaser R. Expression of human beta-defensin-2 and −3 in verrucae vulgares and condylomata acuminata. J Eur Acad Dermatol Venereol. 2008;22:1050–1054. doi: 10.1111/j.1468-3083.2008.02675.x. [DOI] [PubMed] [Google Scholar]
- 13.Meyer-Hoffert U, Schwarz T, Schroder JM, Glaser R. Increased expression of human beta-defensin 3 in mollusca contagiosum. Clin Exp Dermatol. 2010;35:190–192. doi: 10.1111/j.1365-2230.2009.03640.x. [DOI] [PubMed] [Google Scholar]
- 14.Patil AA, Cai Y, Sang Y, Blecha F, Zhang G. Cross-species analysis of the mammalian β-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol Genomics. 2005;23:5–17. doi: 10.1152/physiolgenomics.00104.2005. [DOI] [PubMed] [Google Scholar]
- 15.Wingate KV, Torres SM, Silverstein KA, Hendrickson JA, Rutherford MS. Expression of endogenous antimicrobial peptides in normal canine skin. Vet Dermatol. 2009;20:19–26. doi: 10.1111/j.1365-3164.2008.00707.x. [DOI] [PubMed] [Google Scholar]
- 16.Erles K, Brownlie J. Expression of beta-defensins in the canine respiratory tract and antimicrobial activity against bordetella bronchiseptica. Vet Immunol Immunopathol. 2010;135:12–19. doi: 10.1016/j.vetimm.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 18.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 19.Chen WK, Swartz JD, Rush LJ, Alvarez CE. Mapping DNA structural variation in dogs. Genome Res. 2009;19:500–509. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardwick RJ, Machado LR, Zuccherato LW, Antolinos S, Xue Y, Shawa N, Gilman RH, Cabrera L, Berg DE, Tyler-Smith C, Kelly P, Tarazona-Santos E, Hollox EJ. A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum Mutat. 2011;32:743–750. doi: 10.1002/humu.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsella R, Girolomoni G. Canine models of atopic dermatitis: a useful tool with untapped potential. J Invest Dermatol. 2009;129:2351–2357. doi: 10.1038/jid.2009.98. [DOI] [PubMed] [Google Scholar]
- 22.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, Darst MA, Gao B, Boguniewicz M, Travers JB, Leung DY. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 23.Kisich KO, Carspecken CW, Fieve S, Boguniewicz M, Leung DY. Defective killing of Staphylococcus aureus in atopic dermatitis is associated with reduced mobilization of human beta-defensin-3. J Allergy Clin Immunol. 2008;122:62–68. doi: 10.1016/j.jaci.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 24.van Damme CM, Willemse T, van Dijk A, Haagsman HP, Veldhuizen EJ. Altered cutaneous expression of beta-defensins in dogs with atopic dermatitis. Mol Immunol. 2009;46:2449–2455. doi: 10.1016/j.molimm.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Wehkamp J, Chu H, Shen B, Feathers RW, Kays RJ, Lee SK, Bevins CL. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Letters. 2006;580:5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 26.Leonard BC, Chu H, Johns JL, Gallo RL, Moore PF, Marks SL, Bevins CL. Expression and activity of a novel cathelicidin from domestic cats. PLoS One. 2011;6:e18756. doi: 10.1371/journal.pone.0018756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkhof B, Spee B, Rothuizen J, Penning LC. Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal Biochem. 2006;356:36–43. doi: 10.1016/j.ab.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Toonen RJ, Hughes S. Increased throughput for fragment analysis on an ABI prism 377 automated sequencer using a membrane comb and STRand software. Biotechniques. 2001;31:1320–1324. [PubMed] [Google Scholar]
- 29.Linzmeier RM, Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22-p23. Genomics. 2005;86:423–430. doi: 10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 31.Merryman-Simpson AE, Wood SH, Fretwell N, Jones PG, McLaren WM, McEwan NA, Clements DN, Carter SD, Ollier WE, Nuttall T. Gene (mRNA) expression in canine atopic dermatitis: microarray analysis. Vet Dermatol. 2008;19:59–66. doi: 10.1111/j.1365-3164.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 32.Harder J, Schroder JM. Antimicrobial peptides in human skin. Chem Immunol Allergy. 2005;86:22–41. doi: 10.1159/000086650. [DOI] [PubMed] [Google Scholar]
- 33.Harder J, Meyer-Hoffert U, Wehkamp K, Schwichtenberg L, Schroder JM. Differential gene induction of human beta-defensins (hBD-1, −2, −3, and −4) in keratinocytes is inhibited by retinoic acid. J Invest Dermatol. 2004;123:522–529. doi: 10.1111/j.0022-202X.2004.23234.x. [DOI] [PubMed] [Google Scholar]
- 34.Sorensen OE, Cowland JB, Theilgaard-Monch K, Liu L, Ganz T, Borregaard N. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J Immunol. 2003;170:5583–5589. doi: 10.4049/jimmunol.170.11.5583. [DOI] [PubMed] [Google Scholar]
- 35.Sorensen OE, Thapa DR, Rosenthal A, Liu L, Roberts AA, Ganz T. Differential regulation of beta-defensin expression in human skin by microbial stimuli. J Immunol. 2005;174:4870–4879. doi: 10.4049/jimmunol.174.8.4870. [DOI] [PubMed] [Google Scholar]
- 36.Kisich KO, Howell MD, Boguniewicz M, Heizer HR, Watson NU, Leung DY. The constitutive capacity of human keratinocytes to kill Staphylococcus aureus is dependent on beta-defensin 3. J Invest Dermatol. 2007;127:2368–2380. doi: 10.1038/sj.jid.5700861. [DOI] [PubMed] [Google Scholar]
- 37.Fazakerley J, Crossley J, McEwan N, Carter S, Nuttall T. In vitro antimicrobial efficacy of beta-defensin 3 against Staphylococcus pseudintermedius isolates from healthy and atopic canine skin. Vet Dermatol. 2010;21:463–468. doi: 10.1111/j.1365-3164.2010.00893.x. [DOI] [PubMed] [Google Scholar]
- 38.Santoro D, Bunick D, Graves TK, Campbell KL. Expression and distribution of antimicrobial peptides in the skin of healthy beagles. Vet Dermatol. 2011;22:61–67. doi: 10.1111/j.1365-3164.2010.00911.x. [DOI] [PubMed] [Google Scholar]
- 39.Everts RE, Rothuizen J, van Oost BA. Identification of a premature stop codon in the melanocyte-stimulating hormone receptor gene (Mc1r) in Labrador and Golden retrievers with yellow coat color. Anim Genet. 2000;31:194–199. doi: 10.1046/j.1365-2052.2000.00639.x. [DOI] [PubMed] [Google Scholar]
- 40.Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, Radlwimmer B, Stange EF. A chromosome 8 gene cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn's disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groth M, Wiegand C, Szafranski K, Huse K, Kramer M, Rosenstiel P, Schreiber S, Norgauer J, Platzer M. Both copy number and sequence variations affect expression of human DEFB4. Genes Immun. 2010;11:458–466. doi: 10.1038/gene.2010.19. [DOI] [PubMed] [Google Scholar]
- 42.Boguniewicz M, Leung DY. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev. 2011;242:233–246. doi: 10.1111/j.1600-065X.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeBoer DJ, Marsella R. The ACVD task force on canine atopic dermatitis (XII): the relationship of cutaneous infections to the pathogenesis and clinical course of canine atopic dermatitis. Vet Immunol Immunopathol. 2001;81:239–249. doi: 10.1016/s0165-2427(01)00345-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data