Abstract
BACKGROUND
Inactivation of the maternally or paternally derived X chromosome (XCI) initially occurs in a random manner in early development; however as tissues form, a ‘patchiness’ will occur in terms of which X is inactivated if cells positioned near each other are clonally descended from a common precursor. Determining the relationship between skewed XCI in different tissues and in different samples from the same tissue provides a molecular assessment of the developmental history of a particular tissue that can then be used to understand how genetic and epigenetic variation arises in development.
METHODS
XCI skewing was evaluated in and compared between amnion, chorion, trophoblast and mesenchyme using multiple sampling sites from 14 term placentae. XCI was also evaluated in chorionic villus samples obtained at multiple sites and depths from four additional term placentae. The pattern of variation was then compared with methylation variation associated with the H19/IGF2 imprinting control region (ICR); promoter regions of KISS1, PTPN6, CASP8 and APC; and LINE-1 elements.
RESULTS
Mean placental level of skewing for amnion and chorion are correlated, consistent with a common developmental origin of at least a component of these membranes from inner cell mass derivatives subsequent to XCI, while trophoblast appears to be derived independently, consistent with its origin from the trophectoderm. Villus samples taken from different depths spanning the fetal to maternal side of the placenta were highly clonally related. Comparing patterns of clonal growth identified through XCI to the distribution of epigenetic variation in other genomic regions suggests that some variation arises early in development (e.g. LINE-1 methylation), whereas other variation arises predominantly after villus tree formation (e.g. methylation at H19/IGF2 ICR).
CONCLUSIONS
The patterns of XCI skewing are consistent with a model whereby each biopsied site of chorionic villi represents one or a few individual villus trees, each of which is clonally derived from only one or a few precursor cells. Sampling of placentae to evaluate changes associated with clinical pathology should be done with consideration of the tree-to-tree differences. A limitation of this study is the small number of placentas used and therefore placental-specific differences in variation could not be assessed.
Keywords: placenta, X chromosome inactivation, epigenetics, DNA methylation
Introduction
The human placenta is a transient organ that regulates fetal growth and development. Abnormal placental development is associated with a variety of poor pregnancy outcomes, including maternal hypertension and fetal growth restriction (Chelbi and Vaiman, 2008; Kinzler and Vintzileos, 2008). Understanding the normal developmental patterns of the extra-embryonic tissues is needed to clarify the etiology of placental abnormalities; however, the origin of each human extra-embryonic tissue sampled from a term placenta has not been clearly established.
One tool for evaluating the developmental relationships between cells is to measure X chromosome inactivation (XCI) skewing, which refers to the portion of cells inactivating either the maternally or paternally derived X chromosome (50% being random inactivation). XCI is the transcriptional silencing of one of the two X chromosomes in somatic cells of female mammals (Lyon, 1961, 1962). There is conflicting data concerning the exact timing of XCI in human embryos (van den Berg et al., 2009; Okamoto et al., 2011), though it is generally presumed to occur in a random manner in the embryonic precursors within the inner cell mass (ICM). While preferential inactivation of the paternal X is observed in the trophectoderm and primitive endoderm of the mouse (Takagi and Sasaki, 1975), this process is random in human chorionic villi (Looijenga et al., 1999; Uehara et al., 2000; Moriera de Mello et al., 2010). However, it has not been established when in development XCI occurs in the trophectoderm, nor has the relationship between skewing in different extra-embryonic tissues been previously assessed.
In a human placenta, the fetal vessels enter through the chorionic plate and branch into a network that is covered by two main layers of cells, an outer trophoblast layer and inner mesenchyme layer, making up the chorionic villus tree structures (Castellucci et al., 2000). Placental sampling strategies rarely consider this underlying tree structure. In particular, there may be a clustering of cellular and gene expression changes within the placenta if cells located near each other are derived from common progenitor cells. Evaluating XCI skewing within a tissue can identify such clonal patterns of growth, as a ‘patchiness’ will occur during development in terms of which X is inactivated if cells positioned near each other descended from a common precursor cell subsequent to XCI. While extreme variation from site to site within a placenta has been noted previously and attributed to the presence of clonal patches of chorionic villi (Moriera de Mello et al., 2010), the three-dimensional patterns of XCI skewing in the human placenta have not been previously evaluated.
To further clarify the pattern of XCI skewing in human placenta, the methylation-based HUMARA assay was used to assess skewing in different extra-embryonic tissues in multiple sampled sites from a series of term human placentae. The main objectives were to use a molecular approach to (i) evaluate the developmental relationships of different extra-embryonic tissues at term, (ii) determine the three-dimensional developmental relationship of chorionic villus samples. These results can be used to improve placental sampling strategies. Furthermore, using our understanding of this pattern of clonal growth in the placenta, we can then determine whether genetic and epigenetic variation follows a similar developmental pattern or instead exhibits variation that arises after cellular differentiation in a clonally independent fashion.
Materials and Methods
Sample ascertainment
All placentae were obtained from term deliveries of healthy female newborns of normal birthweight at the BC Children's and Women's Hospital, between 1996 and 2006. Prior to sampling, the placental diameter, thickness, weight, cord insertion, number of cord vessels and general appearance were recorded. Placental samples encoded as ‘PX’ (n = 14) were used for comparing different extra-embryonic tissues (trophoblast, mesenchyme, amnion and chorion). Placental samples encoded as ‘PM’ (n = 4) were obtained through a separate study for which clinical information has been reported previously (Avila et al., 2010). The collection of all samples was approved by the University of British Columbia and the BC Children's and Women's Hospital Ethics Board (H04-70488). A summary of the samples, methods and results is given in Supplementary data, Fig. S1.
Placental sampling
The PX placentae each had 4–9 distinct sites sampled from the fetal side of the placenta to avoid any possibility of maternal contamination. At least one site was located near the umbilical cord and one near the periphery of the placental disk. Other sites sampled were located approximately halfway between the cord and placental edge and well-spaced across the placenta such that no two sites were within 2 cm of each other. The amnion and chorion (fetal membranes) were taken from the placental surface of each sample site and separated by tweezers. The remaining chorionic villi (placenta) from each sampled site were subjected to enzymatic digestion to separate the mesenchyme from the trophoblast (Henderson et al., 1996; Robinson et al., 2010). This method yields ∼95% purity of the trophoblast and 60–70% purity of the mesenchyme (Grigoriu et al., 2011). The four PM placentae were sampled as described previously (Avila et al., 2010), with 3–7 sites spanning the placental disk, including one or more near the periphery and one or more near the cord insertion point. Each site was then subdivided into four equal sections by depth. Prior to extraction, the tissue samples were homogenized and digested with tissue lysis buffer and Proteinase K (Invitrogen, CA, USA). DNA from all samples was extracted using standard protocols and the presence of any significant maternal contamination was excluded by microsatellite markers and androgen receptor (AR) polymorphism comparisons with maternal decidua (data not shown).
X chromosome inactivation
The degree of allelic bias in terms of which X chromosome is inactivated can range from 50% (completely random) to 100% (completely skewed). If the directionality of bias is accounted for, then the degree of skewing can range from 0 to 100%. The pattern of XCI skewing was determined by assaying the allelic ratio of methylated alleles at the AR locus (Allen et al., 1992). This protocol has been described in detail previously (Beever et al., 2003). The allelic ratios were expressed in terms of the percent inactivation of the smaller AR allele when a 0–100% scale for skewing was used.
As it was previously reported that only trophoblast cells and not the mesenchymal cells within the chorionic villi are methylated at the sites used for the XCI assay at AR (Looijenga et al., 1999), we quantified the level of methylation at this site using a pyrosequencing assay (AR_HpaII-D) (Hanna et al., unpublished data). On average, methylation levels were 28% for trophoblast; 21% for mesenchyme, 19% for chorion and ∼35% for amnion. Thus, methylation appears to be present in all tissues but is lower in extra-embryonic samples than what we have observed in female blood, which was closer to the expected 50% (46%; Hanna et al., unpublished data). This observation is generally true for methylation at X-gene promoters in females (Cotton et al., 2009).
Methylation quantification
Several genes exhibiting intra- and inter-placental variability in DNA methylation based on our previous studies (Avila et al., 2010; Bourque et al., 2010) were assessed in this study. These included promoter regions of KISS1, APC, PTPN6 and CASP8, as well as the imprinted control region 1 (ICR1) of H19/IGF2. Primer information and partial data for these loci was previously published (Avila et al., 2010). Primers and conditions for the H19/IGF2 ICR1 were as previously published (Horike et al., 2009). An assay to assess methylation at four CpG sites from a consensus sequence found within LINE-1 retrotransposible elements was also performed according to the manufacturer's directions (PyroMark™ LINE-1 Kit, Biotage-Qiagen, Uppsala, Sweden). LINE-1 elements comprise ∼17% of the human genome and are often used as a surrogate measure for ‘genome-wide’ methylation. PCR was used to amplify the region of interest from bisulfite converted DNA. PCR products were sequenced using a PyroMark™ MD (Biotage AB).
Results
Sample-to-sample variability in XCI within a placenta
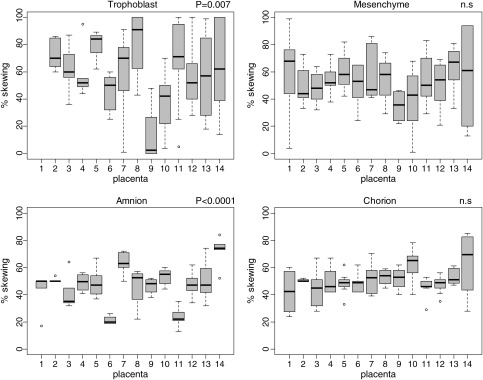
Sample-to-sample variability for a specific extra-embryonic tissue derived from the same pregnancy may reflect a compartmentalization of growth in development. Assessment of this variability is important for determining the degree to which one sample can be representative of the tissue as a whole. All tissues examined showed variability among the different (4–9) samples taken from each of the 14 placentae; however, trophoblast tended to show more extreme XCI skewing values than other tissues (Fig. 1). Using the standard deviation (SD) of the measurements for different samples taken from the same placenta to assess this variability, significantly more site-to-site variation was observed for mesenchyme or trophoblast (mean SD = 19.5 and 23.7) than for amnion and chorion (mean SD = 9.4 and 10.6) (Supplementary data, Table SI).
Figure 1.
Distribution of XCI skewing by placenta in different tissues. Box plot for each placenta (numbered on the x-axis) for amnion, chorion, trophoblast and chorion. Degree of skewing ranging from 0 to 100% is on the Y-axis. The P-value for the ANOVA to test for differences in means between placentae is given. The boxes represent the interquartile range (IQR) for XCI skewing values for the 4–9 sites analyzed per placenta. The whiskers demonstrate the last data point within ± 1.5 times the IQR. The bars indicate the medians and the dots show the outliers.
Between-placenta differences in mean XCI skewing
If XCI initially occurs at a point in which there is a large pool (e.g. >20) of progenitor cells then, even if there is localized sampling variability, the overall average level of skewing across different samples of the same tissue (in the same placenta) should be ∼50%. From examining Fig. 1, trophoblast and amnion show the greatest deviations in means from 50%, while a mean of close to 50% (random XCI) was observed for mesenchyme and chorion in most placentae. Using analysis of variance (ANOVA) to test for inter-placental differences in XCI skewing for each tissue, amnion (P < 0.0001) and trophoblast (P = 0.007) were significant.
Correlations in mean XCI skewing between different tissue samples from the same placenta
If different placental tissues are derived from a common pool of cells subsequent to XCI, then there may be a correlation in the direction of allelic bias of XCI between those two tissues. For example, if there is an initial bias towards inactivation of the maternal X chromosome, then different tissues derived from that same pool of cells should, on average, show that same tendency. We thus compared the average degree of skewing (from 0 to 100%) for different tissues from the same placenta. There was a modest correlation in mean placental XCI skewing for amnion and chorion (r = 0.64, P = 0.01) and for trophoblast and mesenchyme (r = 0.77, P = 0.002), but not for any other comparisons (Table I).
Table I.
Intra-placental correlation of means of XCI skewing measurements between different tissues.
| Amnion | Chorion | Trophoblast | Mesenchyme | |
|---|---|---|---|---|
| Amnion | — | 0.637 | 0.075 | 0.122 |
| Chorion | P = 0.014 | — | −0.23 | −0.195 |
| Trophoblast | n.s. | n.s. | — | 0.773 |
| Mesenchyme | n.s. | n.s. | P = 0.002 | — |
Correlation coefficients (r) are in upper quadrants, P-values are in lower quadrants.
Bold represents significant correlation values.
In addition to derivation from common precursor pools, a correlation in direction and degree of XCI skewing between tissues derived from the same sample might occur if there is cross-contamination of cells within a given placental sample. This could be a concern for amnion and chorion, which are adjacent membranes that are separated by teasing apart with tweezers, and for trophoblast and mesenchyme, which are separated using an enzymatic digestion procedure. To test for this possibility, we evaluated whether there was a correlation in skewing values from paired tissues sampled from within the same site. As shown in Supplementary data, Fig. S2a, there was no significant intra-site correlation in skewing values for amnion and chorion for any of the sampled placentas. Thus, the overall correlation in XCI skewing between these two tissues cannot be explained by sample cross-contamination. In contrast, the degree of skewing between paired trophoblast and mesenchyme samples derived from the same site is highly correlated (Supplementary data, Fig. S2b). Considering all placentae together, the correlation between trophoblast and mesenchyme for paired samples from the same site was 0.62 (P < 0.0001). No other pair-wise comparison of tissues showed a significant correlation.
Three-dimensional assessment of skewed XCI in chorionic villi
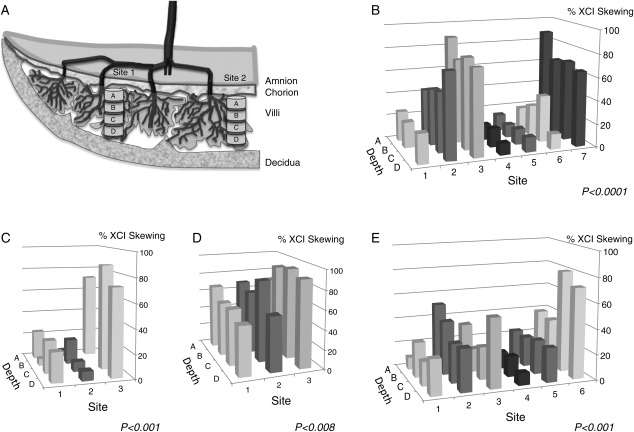
As the placenta is composed of ‘trees’ of villi that grow from the chorionic plate (fetal side of the placenta), samples taken vertically by depth (i.e. spanning the fetal to maternal planes) may show a more similar level of XCI skewing because of an increased chance of being derived from the same tree or set of trees. We thus analyzed samples of whole chorionic villi from four additional placentae for which we had previously separated each sampled site into four depths (Fig. 2a). As is apparent in Fig. 2, there is a striking concordance between XCI skewing values from samples taken from different depths within the same site, contrasting sharply to the considerable site-to-site variation observed when taking samples across the placental surface. Thus, cell division appears to occur in a clonal fashion by depth, consistent with the structure of the villus trees. This intra-site correlation is highly significant for each placenta as evaluated by ANOVA (Table II).
Figure 2.
Three-dimensional analysis of XCI skewing. (a) Schematic representation of the placental villus trees. (b–e) Degree of XCI skewing in different sites (1–7)/depths (A–D) in four control placentae. P-values correspond to ANOVA used to evaluate site-to-site differences within each placenta.
Table II.
ANOVA test for between-site differences (intra-site correlation) for each placenta.
| PM17, (n = 7 sites × 4 depths) | PM136, (n = 6 sites × 4 depths) | PM106, (n = 3 sites × 4 depths) | PM109, (n = 3 sites × 4 depths) | |
|---|---|---|---|---|
| XCI | <0.0001 | 0.001 | 0.008 | <0.0001 |
| ICR1 | n.s. | n.s. | n.s. | n.s. |
| LINE-1 | <0.0001 | 0.001 | n.s. | 0.01 |
| KISS1 | n.s. | 0.001 | 0.032 | n.s. |
| PTPN6 | n.s. | n.s. | n.s. | n.s. |
| APC | 0.024 | 0.007 | <0.0001 | <0.0001 |
| CASP8 | n.s. | <0.0001 | n.s. | n.s. |
| AR_54 (B) | n.s. | 0.001 | NA | NA |
| AR_34 (C) | n.s. | n.s. | n.s. | n.s. |
| AR_HpaII (D) | n.s. | 0.003 | n.s. | 0.009 |
Distribution of methylation variation at other genomic loci
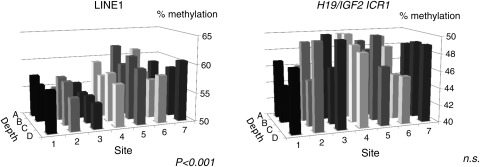
Knowing the patterns of clonal growth detected by XCI skewing, we can then ask whether variation in other genomic regions of the placenta also follow a trend to displaying more variation between sites, rather than within a given site. For example, methylation at the ICR associated with H19 and IGF2 has been reported to exhibit a high degree of intra-placental variation (Bourque et al., 2010; Turan et al., 2010). Such variation may arise in the blastocyst or early stages of tissue differentiation and then it may be stably maintained similar to what has been observed for XCI. If however, such variation arises due to either stochastic errors or environmental influences acting after development of the villus tree structure, there may not be a significant intra-site correlation. Using data on methylation within the promoters of APC, KISS1, PTPN6, CASP8 and AR as well as methylation at the H19/IGF2 ICR and at a consensus LINE-1 sequence, we used ANOVA to determine whether the variation in methylation was greater between samples taken from different sites than for those taken from different depths within the same site. (Fig. 3, Table II). While not as striking as we had observed for XCI skewing, methylation at APC and LINE-1 showed significant intra-site correlation, while methylation at the H19/IGF2 ICR did not. Results for the other loci were mixed (Table II).
Figure 3.
Three-dimensional analysis of DNA methylation variation for H19/IGF2 ICR1 and LINE-1. Distribution of methylation by site and depth for H19/IGF2 ICR1 and LINE-1 for control placenta PM17. Global (LINE-1) methylation follows a clonal pattern, while variation at H19/IGF2 ICR1appears to arise after XCI.
Discussion
Skewed XCI has long been used as a tool to examine the potential clonal origin of tumors in females and to make inferences about the expression of X-linked disease in females (Minks et al., 2008). However, past studies of skewed XCI in the placenta have largely been limited to demonstrating that there is no preferential inactivation of the paternal X (Migeon, 1979; Looijenga et al., 1999; Uehara et al., 2000; Moriera de Mello et al., 2010), as seen in mouse placenta (Takagi and Sasaki, 1975). In the present study, we used XCI skewing to better understand how patterns of development might influence the distribution of genetic and epigenetic variation in the placenta. Our data, along with previous data on XCI skewing (Looijenga et al., 1999; Moriera de Mello et al., 2010), trisomy mosaicism (Henderson et al., 1996; Robinson et al., 2010), gene expression and methylation (Avila et al., 2010; Turan et al., 2010), have all demonstrated a considerable degree of site-to-site heterogeneity in the placenta. Thus, a single sample may not be representative of the placenta as a whole (Yuen and Robinson, 2011). We further demonstrate that this variability is observed for both the trophoblast and mesenchyme components of the chorionic villi, while significantly less sampling variability is seen for amnion and chorion.
XCI patterns are consistent with developmental origins of extra-embryonic tissues
A correlation in skewing between different somatic tissues has been used to infer that all somatic tissues derive from 15 to 20 precursor cells present at the time an X is marked to be inactivated in each cell (Fialkow, 1973; Tonon et al., 1998; Amos-Landgraf et al., 2006). The marking of an X for inactivation in mouse is thought to occur in the ICM of the late blastocyst, while this process may occur later in humans (Okamoto et al., 2011). A correlation of skewing ratios is typically observed between different somatic tissues, but selection may additionally modify these ratios over time, particularly in lymphocytes (Azofeifa et al., 1996; Sharp et al., 2000). Our present data show a correlation in the placental-wide average skewing values between amnion and chorion (Table I), suggesting that these two tissues derive, at least in part, from the same pool of cells subsequent to the marking of an X for inactivation.
A question remains as to the timing of this common progenitor pool for amnion and chorion (sampled from a term placenta) relative to the origin of hypoblast and epiblast (before their derivation from the ICM or after). The chorionic membrane is initially derived from partial regression of chorionic villi and consists of several layers of undifferentiated trophoblast and stroma that is derived from extra-embryonic mesoderm (Ilancheran et al., 2009). Extra-embryonic mesoderm has been suggested to arise from the hypoblast of the ICM (Bianchi et al., 1993; Ilancheran et al., 2009), though an origin from epiblast is suggested by embryological studies in primates (Luckett, 1978; Enders et al., 1986) and molecular studies in mouse (Lawsen and Hage, 1994; Cross et al., 2003; Downs, 2011). While the amniotic mesoderm has also been cited as being derived from the hypoblast, we previously used data from cases of chromosomal mosaicism to demonstrate that in a term placenta this component of amnion is more likely derived from the subsequent contribution of mesodermal cells from the primitive streak rather than cells initially derived from the hypoblast (Robinson et al., 2002). Furthermore, we inferred that amniotic epithelium was the main contributor to the DNA derived from amnion and that this layer derives from the epiblast of the ICM of the embryo, prior to primitive streak formation, but after derivation of the extra-embryonic mesoderm (Robinson et al., 2002). The present data are consistent with the assumption that XCI for the major precursors of amnion and chorion either occurs in the ICM prior to the origin of hypoblast/epiblast or that there is a significant epiblast contribution to the chorionic membrane later in development. The greater deviation in mean skewing from 50% observed for amnion when compared with chorion would be consistent with amnion being derived from a smaller subset of this common cell pool.
There is no apparent relationship between XCI in trophoblast and amnion or chorion consistent with what we would expect if XCI occurs separately in the trophectoderm. While the chorion initially consists of an outer layer of trophoblast in early first trimester development and trophoblast stem cells appear to exist within the chorionic mesoderm (Genbacev et al., 2011), based on the present data, the trophoblast component may not contribute significantly to DNA extracted from this membrane (or at least the portion of it located on the placental disc) at term.
A clonal origin of the trophoblast within each villus
The formation of the chorionic villi begins with trophoblast budding at 12–18 days p.c., followed by mesenchymal invasion and then fetal capillary development (starting at 18–20 days p.c.) (Castellucci et al., 2000). Additional growth of villi occurs by repeated sprouting of branches off of each initial mesenchymal villus and an increase in the villous diameter. Thus, each villus tree is likely derived from the cells present in the initial villus bud without further input of cells. As the placenta is composed of roughly 60–70 individual chorionic villi which grow from the chorionic plate towards the basal plate (Bernischke and Kaufman, 2000), and as the surface area of a placenta is roughly 300–500 cm2, a single 2 cm biopsy should represent only one, or at most a few, of these villus trees.
As we found a strong concordance in degree and direction of XCI skewing between samples of chorionic villi (largely representing skewing in trophoblast) taken at intervals vertically from the fetal (chorionic plate) to maternal (basal plate) side of the placenta, samples taken at different depths from the same site likely represent samples from the same villus tree(s). Clonal development of trophoblast within each villus from a single precursor cell was previously proposed based on the expression of the FMRP protein in sections from placentae heterozygous for a mutation allele at the FMR1 locus encoding this gene (Willemsen et al., 2002). In females, depending on which X chromosome was inactivated, either all trophoblast cells of the villus would express FMRP (demonstrating that the normal allele was active) or no cells would express it (inferring that the normal allele was inactivated). As the staining was less clear in the mesenchymal core of the villi, no conclusions could be drawn for this component. Our skewing data would also be consistent with a model whereby the trophoblast cells within each villus tree represent a single clone in regard to XCI.
Application of three-dimensional studies to infer origin of epigenetic variation
A variety of recent studies demonstrate a significant degree of within-placenta variability in measurements of both DNA methylation and gene expression (Yuen and Robinson, 2011). Modulation of gene expression through epigenetic regulation may provide a mechanism to allow the placenta to buffer the consequences of a variety of adverse genetic and environmental conditions. The imprinting control region of H19 and IGF2 is an example of a region that shows considerable site-to-site variability within some placentae (Katari et al., 2009; Bourque et al., 2010; Ollikainen et al., 2010), and it was hypothesized that this variability might be a function of the number of stem cells from which the placental trophoblast derived, with placentae derived from fewer precursors having a greater variance (Katari et al., 2009). This hypothesis would then suggest that the methylation state is set in the preimplantation embryo and then stably maintained in subsequent divisions within the trophoblast, as occurs for XCI. However, our data at this ICR do not show a significant within-site correlation in methylation, suggesting that this variability predominantly arises after formation of the primary villi. If methylation at this site can change during pregnancy in response to environmental effects such as nutrient supply (Angiolini et al., 2006), the variation may not follow developmental patterns, but rather reflect more localized influences.
Interestingly, LINE-1 methylation, which has been reported to be altered in newborns in association with environmental factors such as homocysteine levels (Fryer et al., 2011) or maternal smoking (Breton et al., 2009), showed a significant between-site difference for three of four sampled placentae (Table II). Either LINE-1 methylation is more subject to preimplantation than post-implantation environmental influences, or those influences act in a manner such that all branches of a villus tend to be affected similarly. Nonetheless the within-site correlation was not as dramatic for LINE-1 methylation as was seen for XCI skewing and thus some variability at these repeated sequences must be acquired after cells are allocated into the primary villi.
While both LINE-1 and the H19/IGF2-ICR show similar levels of methylation across different placental tissues, the promoter region of KISS1 is unmethylated specifically in trophoblast, and this gene is expressed exclusively in proliferating trophoblast at the villus tips. We previously argued that within-placenta methylation variance at the KISS1 promoter could be due to sample-to-sample fluctuations in cell composition, specifically trophoblast (unmethylated): mesenchyme (methylated) ratios (Avila et al., 2010). As KISS1 showed significant within-site correlation in some placentae, it is possible that the proportion of trophoblast cells differ from tree-to-tree. In support of this explanation, electron microscopy of placental villi shows that different villi appear to be at various stages of maturation (Demir et al., 1995). Furthermore, some clusters of mesenchymal villi can be found with degenerative trophoblast and no evidence of proliferation, suggesting that their growth has been repressed (Castellucci et al., 2000). Thus, it is perhaps not surprising to see patterns of variation that align with such regional differences. CASP8 shows a significant intra-site correlation for the same placenta that shows the most significant intra-site correlation for KISS1. As this region of CASP8 is also methylated distinctly in trophoblast when compared with mesenchyme, it may similarly be affected by tree-to-tree variation in cell composition. On the other hand, PTPN6 methylation tends to be correlated with KISS1 methylation (Avila et al., 2010) but did not show the same effect.
Interestingly, APC shows a significant clonal pattern of variation in all placentae. We recently reported evidence that this gene is imprinted in placenta, possibly in a cell-specific or polymorphic fashion (Yuen et al., 2011). Although conjectural, this locus could show polymorphic imprinting within a placenta that varies from villus to villus.
Summary
In summary, the patterns of XCI skewing observed in this study are consistent with developmental patterns in the placenta inferred from early embryological studies. This knowledge is important in order to interpret how observations based on a single placental sample relate to the underlying placental biology. While it is known that there can be considerable within-placental variability in gene expression and methylation, it is important to consider that this may be due to (i) very early alterations that are maintained clonally in each villus tree; (ii) varying patterns of growth and maturation of individual villi and (iii) local environmental influences on different placental regions. For example, pre-eclampsia is associated with reduced placental perfusion, which is thought in turn to lead to secondary effects such as accelerated maturation of villi and increase in syncytial knots. However, villi in areas of the placenta with the lowest oxygen exposure may grow and behave differently from others for which maternal blood flow is more adequate. Clearly, multiple dispersed samples ensuring representation from different villi are needed to adequately assess the average level of function of the placenta as a whole.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors’ roles
M.S.P.: study design, data collection and analysis and manuscript writing; R.J.: technical assistance; L.A.: data collection and analysis; R.K.C.Y.: data collection; C.J.B.: study design and interpretation; W.P.R.: study design, data analysis and manuscript writing.
Funding
This work has been funded by the Canadian Institutes of Health Research operating grant (to W.P.R. and C.J.B.). M.S.P. was partially funded by a Graduate Degree fellowship from FUNDACYT at initial collection of the samples and XCI studies; R.K.C.Y. received a doctoral fellowship research award from the Child & Family Research Institute. Funding to pay the Open Access publication charges for this article was provided by a Canadian Institutes of Health Research Operating Grant (WPR).
Conflict of interest
The authors declare that they have no competing interests.
Supplementary Material
Acknowledgements
We thank Dr Angela Devlin for the use of the Biotage PyroMark_MD and the women who donated the placentae from their pregnancies for research. We also thank Drs Valerie Désilets and Doug Wilson for assisting in placental ascertainment.
References
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, Schwartz CE, Longshore J, Willard HF. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006;79:493–499. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiolini E, Fowden A, Coan P, Sandovici I, Smith P, Dean W, Burton G, Tycko B, Reik W, Sibley C, et al. Regulation of placental efficiency for nutrient transport by imprinted genes. Placenta. 2006;27(Suppl A):S98–S102. doi: 10.1016/j.placenta.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Avila L, Yuen RK, Diego-Alvarez D, Penaherrera MS, Jiang R, Robinson WP. Evaluating DNA methylation and gene expression variability in the human term placenta. Placenta. 2010;31:1070–1077. doi: 10.1016/j.placenta.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Azofeifa J, Waldherr R, Cremer M. X-chromosome methylation ratios as indicators of chromosomal activity: evidence of intraindividual divergencies among tissues of different embryonal origin. Hum Genet. 1996;97:330–333. doi: 10.1007/BF02185765. [DOI] [PubMed] [Google Scholar]
- Beever CL, Stephenson MD, Penaherrera MS, Jiang RH, Kalousek DK, Hayden M, Field L, Brown CJ, Robinson WP. Skewed X-chromosome inactivation is associated with trisomy in women ascertained on the basis of recurrent spontaneous abortion or chromosomally abnormal pregnancies. Am J Hum Genet. 2003;72:2. doi: 10.1086/346119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernischke K, Kaufman P. Pathology of the Human Placenta. New York: Springer-Verlag; 2000. [Google Scholar]
- Bianchi DW, Wilkins-Haug LE, Enders AC, Hay ED. Origin of extraembryonic mesoderm in experimental animals: relevance to chorionic mosaicism in humans. Am J Med Genet. 1993;46:542–550. doi: 10.1002/ajmg.1320460517. [DOI] [PubMed] [Google Scholar]
- Bourque DK, Avila L, Peñaherrera MS, von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci M, Kosanke G, Verdenelli F, Huppertz B, Kaufmann P. Villous sprouting: fundamental mechanisms of human placental development. Hum Reprod Update. 2000;6:485–494. doi: 10.1093/humupd/6.5.485. [DOI] [PubMed] [Google Scholar]
- Chelbi ST, Vaiman D. Genetic and epigenetic factors contribute to the onset of preeclampsia. Mol Cell Endocrinol. 2008;282:120–129. doi: 10.1016/j.mce.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Cotton AM, Avila L, Penaherrera MS, Affleck JG, Robinson WP, Brown CJ. Inactive X chromosome-specific reduction in placental DNA methylation. Hum Mol Genet. 2009;18:3544–3552. doi: 10.1093/hmg/ddp299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- Demir R, Demir N, Ustunel I, Erbengi T, Trak I, Kaufmann P. The fine structure of normal and ectopic (tubal) human placental villi as revealed by scanning and transmission electron microscopy. Zentralbl Pathol. 1995;140:427–442. [PubMed] [Google Scholar]
- Downs KM. Lineage commitments: emphasis on embryonic-extraembryonic interfaces. EMBO Rep. 2011;12:987–990. doi: 10.1038/embor.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders AC, Schlafke S, Hendrickx AG. Differentiation of the embryonic disc, amnion, and yolk sac in the rhesus monkey. Am J Anat. 1986;177:161–185. doi: 10.1002/aja.1001770205. [DOI] [PubMed] [Google Scholar]
- Fialkow PJ. Primordial cell pool size and lineage relationships of five human cell types. Ann Hum Genet. 1973;37:39–48. doi: 10.1111/j.1469-1809.1973.tb01813.x. [DOI] [PubMed] [Google Scholar]
- Fryer AA, Emes RD, Ismail KM, Haworth KE, Mein C, Carroll WD, Farrell WE. Quantitative, high-resolution epigenetic profiling of CpG loci identifies associations with cord blood plasma homocysteine and birth weight in humans. Epigenetics. 2011;6:86–94. doi: 10.4161/epi.6.1.13392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbacev O, Donne M, Kapidzic M, Gormley M, Lamb J, Gilmore J, Larocque N, Goldfien G, Zdravkovic T, McMaster MT, et al. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 2011;29:1427–1436. doi: 10.1002/stem.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. Cell specific patterns of methylation in the human placenta. Epigenetics. 2011;6:53–64. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KG, Shaw T, Barrett I, Telenius A, Wilson R, Kalousek D. Distribution of mosaicism in human placentae. Hum Genet. 1996;97:650–654. doi: 10.1007/BF02281877. [DOI] [PubMed] [Google Scholar]
- Horike S, Ferreira JC, Meguro-Horike M, Choufani S, Smith AC, Shuman C, Meschino W, Chitayat D, Zackai E, Scherer SW, et al. Screening of DNA methylation at the H19 promoter or the distal region of its ICR1 ensures efficient detection of chromosome 11p15 epimutations in Russell-Silver syndrome. Am J Med Genet A. 2009;149A:2415–2423. doi: 10.1002/ajmg.a.33065. [DOI] [PubMed] [Google Scholar]
- Ilancheran S, Moodley Y, Manuelpillai U. Human fetal membranes: a source of stem cells for tissue regeneration and repair? Placenta. 2009;30:2–10. doi: 10.1016/j.placenta.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler WL, Vintzileos AM. Fetal growth restriction: a modern approach. Curr Opin Obstet Gynecol. 2008;20:125–131. doi: 10.1097/GCO.0b013e3282f7320a. [DOI] [PubMed] [Google Scholar]
- Lawsen KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. In: Marsh J, Goode J, editors. Germline Development. London: John Wiley and Sons; 1994. pp. 68–91. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Verkerk AJ, van Putten WL, Oosterhuis JW. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet. 1999;64:1445–1452. doi: 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckett WP. Origin and differentiation of the yolk sac and extraembryonic mesoderm in presomite human and rhesus monkey embryos. Am J Anat. 1978;152:59–97. doi: 10.1002/aja.1001520106. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–145. [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Do TT. In search of non-random X inactivation: studies in the placenta from newborns heterozygous for G6PD. Am J Hum Genet. 1979;31:581–585. [PMC free article] [PubMed] [Google Scholar]
- Minks J, Robinson WP, Brown CJ. A skewed view of X chromosome inactivation. J Clin Invest. 2008;118:20–23. doi: 10.1172/JCI34470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriera de Mello JC, Souza de Aroujo ES, Stabellini R, Fraga AM, Santana de Souza JE, Sumita DR, Camargo AA, Periera LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, Abdul Aziz NK, Carlin JB, Morley R, Saffery R, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet. 2010;19:4176–4188. doi: 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Barrett IJ, Kuchinka B, Penaherrera MS, Bruyere H, Best R, Pediera D, McFadden DE, Langlois S, Kalousek DK. Origin of amnion and implications for evaluation of the fetal genotype in cases of mosaicism. Prenat Diagn. 2002;22:1078–1087. doi: 10.1002/pd.483. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Penaherrera MS, Jiang R, Avila L, Sloan J, McFadden DE, Langlois S, von Dadelszen P. Assessing the role of placental trisomy in preeclampsia and intrauterine growth restriction. Prenat Diagn. 2010;30:1–8. doi: 10.1002/pd.2409. [DOI] [PubMed] [Google Scholar]
- Sharp A, Robinson D, Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000;107:343–349. doi: 10.1007/s004390000382. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Tonon L, Bergamaschi G, Dellavecchia C, Rosti V, Lucotti C, Malabarba L, Novella A, Vercesi E, Frassoni F, Cazzola M. Unbalanced X-chromosome inactivation in haemopoietic cells from normal women. Br J Haematol. 1998;102:996–1003. doi: 10.1046/j.1365-2141.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, Coutifaris C, Sapienza C. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet. 2010;6:e1001033. doi: 10.1371/journal.pgen.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara S, Tamura M, Nata M, Ji G, Yaegashi N, Okamura K, Yajima A. X-chromosome inactivation in the human trophoblast of early pregnancy. J Hum Genet. 2000;45:119–126. doi: 10.1007/s100380050197. [DOI] [PubMed] [Google Scholar]
- van den Berg IM, Laven JS, Stevens M, Jonkers I, Galjaard RJ, Gribnau J, van Doorninck JH. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009;84:771–779. doi: 10.1016/j.ajhg.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Bontekoe CJ, Severijnen LA, Oostra BA. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110:601–605. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- Yuen R, Robinson W. Review: a high capacity of the human placenta for genetic and epigenetic variation: implications for assessing pregnancy outcome. Placenta. 2011;32:s136–s141. doi: 10.1016/j.placenta.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Yuen R, Jiang R, Peñaherrera M, McFadden D, Robinson W. Genome-wide mapping of imprinted genes by DNA methylation profiling of human placentas from triploidies. Epigenetics Chromatin. 2011;4:10. doi: 10.1186/1756-8935-4-10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.