Abstract
Increased morbidity and mortality associated with ischemic heart failure (HF) in type 2 diabetic patients requires a deeper understanding of the underpinning pathogenetic mechanisms. Given the implication of microRNAs (miRNAs) in HF, we investigated their regulation and potential role. miRNA expression profiles were measured in left ventricle biopsies from 10 diabetic HF (D-HF) and 19 nondiabetic HF (ND-HF) patients affected by non–end stage dilated ischemic cardiomyopathy. The HF groups were compared with each other and with 16 matched nondiabetic, non-HF control subjects. A total of 17 miRNAs were modulated in D-HF and/or ND-HF patients when compared with control subjects. miR-216a, strongly increased in both D-HF and ND-HF patients, negatively correlated with left ventricular ejection fraction. Six miRNAs were differently expressed when comparing D-HF and ND-HF patients: miR-34b, miR-34c, miR-199b, miR-210, miR-650, and miR-223. Bioinformatic analysis of their modulated targets showed the enrichment of cardiac dysfunctions and HF categories. Moreover, the hypoxia-inducible factor pathway was activated in the noninfarcted, vital myocardium of D-HF compared with ND-HF patients, indicating a dysregulation of the hypoxia response mechanisms. Accordingly, miR-199a, miR-199b, and miR-210 were modulated by hypoxia and high glucose in cardiomyocytes and endothelial cells cultured in vitro. In conclusion, these findings show a dysregulation of miRNAs in HF, shedding light on the specific disease mechanisms differentiating diabetic patients.
A variety of clinical conditions can trigger ventricle remodeling, leading to chamber dilatation and heart failure (HF). In particular, myocardial infarction results in a number of structural changes involving both the infarcted and noninfarcted zones (1). A link between hyperglycemia and HF is recognized, and diabetes is an HF risk factor independent of coronary artery disease or hypertension (2). After myocardial infarction, diabetic patients have an increased HF risk and, once HF develops, diabetic patients have a poorer prognosis (3,4). Moreover, diabetic patients display increased morbidity and mortality upon cardiac surgery (5).
Several findings support the notion that increased incidence of ischemic HF occurs in diabetic patients even in the absence of larger infarct size, lower left ventricular ejection fraction (LVEF), or reduced infarct artery patency (6,7). The mechanisms leading to a greater burden of HF in diabetic patients after infarction are likely multifactorial, including increased comorbidities (e.g., coronary artery disease and hypertension), more severe atherosclerosis, endothelial and microvascular dysfunction, and deregulation of the autonomic nervous system. Recent studies show that hyperglycemia can cause apoptotic cardiomyocyte loss, which in turn may play a key role in diabetic cardiac damage (8,9). However, the molecular mechanisms by which diabetes contributes to worsening ischemic HF remain unclear.
MicroRNAs (miRNAs) are 21–23 nucleotide RNAs that regulate the stability and/or the translational efficiency of target messenger RNAs (mRNAs) (10), affecting a variety of cell functions. Furthermore, there is emerging evidence that miRNAs may play an important role in the pathogenesis of HF, regulating the expression levels of genes governing the process of adaptive and maladaptive cardiac remodeling (11,12).
Evidence of miRNA dysregulation in HF derives from studies either in cell culture models of hypertrophy (13–15) or in animal models of heart pathologic remodeling (13,14,16). Several studies have also been performed in humans (17–21). However, analyzed samples mostly were derived from end-stage patients, potentially missing important determinants of HF progression. Moreover, many discrepancies can be observed among the different studies, possibly as a result of the lack of standardized protocols, the small numbers of patients analyzed, and the high degree of variability in expression levels among patients. Several miRNAs are dysregulated in association with diabetes complications (22); however, thus far, no data on miRNA modulation in D-HF patients are available.
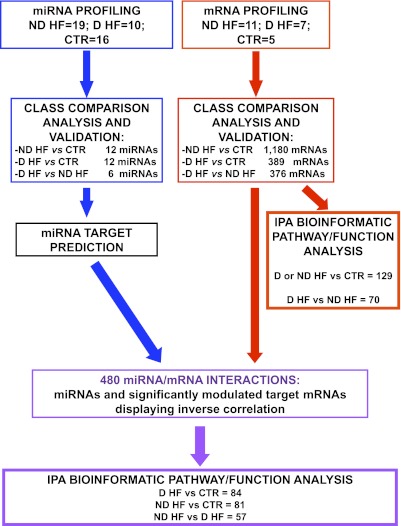
The goal of this study was to investigate miRNA regulation in non–end stage failing hearts, comparing diabetic HF (D-HF) and nondiabetic HF (ND-HF) patients with similar hemodynamic impairment. To this aim, we took advantage of the sampling opportunity represented by the ventricular restoration procedure (23) to analyze heart biopsies harvested from the nonischemic portion of the myocardium of both D-HF and ND-HF patients. The parallel analysis of miRNA and mRNA expression profiles allowed us to evaluate the impact of miRNA dysregulations and their potential pathogenetic role. The adopted experimental plan is summarized in Fig. 1.
FIG. 1.
Experimental plan. miRNA (blue) and mRNA (red) expression profiles were determined, and class comparison analysis identified significantly modulated miRNAs and mRNAs in the indicated classes of patients. Validation was performed by qRT-PCR for both miRNAs and mRNAs. IPA was used to analyze the molecular pathways and functions enriched among the differentially expressed mRNA (far right branch). For the miRNAs identified by class comparison, target mRNAs were predicted by dedicated software. Next, predicted target datasets were intersected with significantly modulated mRNAs deriving from class comparison analysis. Only miRNA/mRNA couples displaying inverse significant correlations were selected (purple) and further analyzed by IPA.
RESEARCH DESIGN AND METHODS
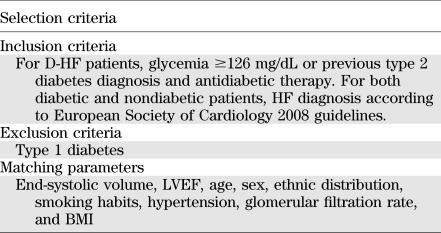
Patient selection and tissue collection.
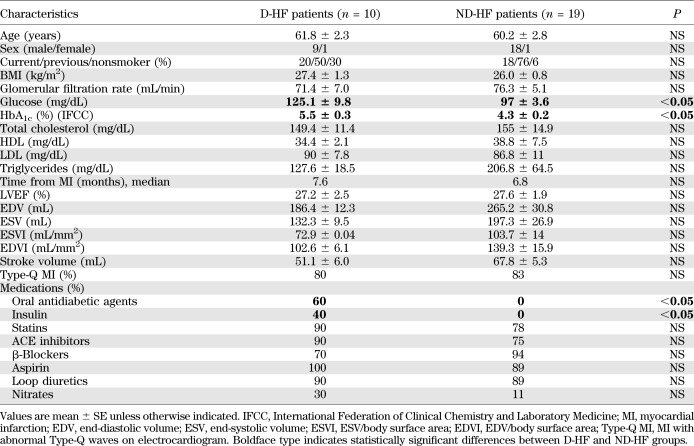
Left ventricle cardiac biopsies were derived from consecutive patients affected by dilated hypokinetic ischemic cardiomyopathy and obtained during surgical ventricular restoration procedures performed as described previously (23). For each patient, two biopsies were harvested: one from the peri-infarctual area (border zone) and one from the nonischemic, remote myocardium (remote zone). Samples were immediately immersed in RNAlater (QIAGEN GmbH) and stored at 4°C for <24 h before RNA extraction. D-HF (n = 10) and ND-HF (n = 19) patients were selected as described in Table 1. D-HF and ND-HF patients did not differ significantly for any clinical parameter other than type 2 diabetes (Table 1). In the validation procedure, the ND-HF group was extended to 4 more patients, with characteristics similar to those shown in Table 2. The control (CTR) cohort was constituted by age- and sex-matched subjects who died from causes other than stroke, ischemia, or cachexia for chronic diseases (n = 16, [7 females/9 males], aged 57 ± 3.2 years). Left ventricle samples were harvested <24 h after death and snap frozen at −80°C.
TABLE 1.
Selection criteria of HF patients
TABLE 2.
Clinical characteristics of study population
Bioptic specimens were taken after informed consent disclosing future use for research. The investigation conformed to the principles outlined in the Helsinki Declaration and to Italian laws and guidelines and was authorized by a local ethics committee.
RNA isolation and miRNA profiling.
Total RNA was extracted using TRIzol (Invitrogen) and the TissueLyser system (QIAGEN). TaqMan Low Density miRNA arrays (Applied Biosystems), containing quantitative real-time RT-PCR (qRT-PCR) assays for 365 human mature miRNAs, were performed using 100 ng total RNA as indicated by the manufacturer. Samples were normalized according to the median cycle threshold (Ct) value. Relative miRNA expression was calculated with the 2-ΔΔCt method. miRNAs with a Ct ≤33 and a ΔΔCt ≥ |1| in at least one of the three comparisons were further analyzed. Data distribution was evaluated by the Kolmogorov-Smirnov normality test. Permutational multivariate ANOVA was performed by the multivariate nonparametric PERMANOVA test, using 9,999 permutations, Bonferroni post hoc test, and Gower test for distance measurement. Adjusted P < 0.05 was considered statistically significant.
mRNA profiling.
Gene expression profiles were measured using Affymetrix GeneChips Human Gene 1.0ST arrays in 7 D-HF, 11 ND-HF, and 5 CTR subjects. Total RNA (100 ng) was analyzed according to the manufacturer’s instructions. Gene expression profiles were analyzed using the class comparison between-groups function of BRB-ArrayTools (Richard Simon and BRB-ArrayTools Development Team). Since our arrays have a small number of samples per class, differentially expressed genes were evaluated using the random variance version of the two-sample t test, as suggested by the BRB-ArrayTools user manual. The class comparison function also performs random permutations of the class labels, recomputing the random variance t tests of each gene for each random permutation. False discovery rate was computed per gene using the Benjamini and Hochberg (24) method. Differentially modulated genes were identified using the random variance version of the two-sample t test, with P < 0.001 and false discovery rate <0.05 as significance thresholds. The complete dataset of our study is available from the National Center for Biotechnology Information Gene Expression Omnibus database (GEO GSE26887).
qRT-PCR assays.
Individual mature miRNAs were measured using TaqMan MicroRNA single assays (Applied Biosystems) (25). Samples were normalized to miR-16 expression.
mRNA levels were measured using the SYBR Green qRT-PCR method (Applied Biosystems), and samples were normalized to the average level of glyceraldehyde-3-phosphate dehydrogenase and ubiquitin C. For both miRNAs and mRNAs, relative expression was calculated using the comparative Ct method (2-ΔΔCt).
To determine miRNA scores, the mean fold change of all the miRNAs belonging to each specific analysis (D-HF vs. CTR, ND-HF vs. CTR, and ND-HF vs. D-HF) was calculated for each patient and the minus sign was inverted before calculating the mean.
Bioinformatic analysis.
Bioinformatic prediction of miRNA target genes was performed using miRonTop (26,27). Predicted miRNA target genes were compared with differently expressed genes (P ≤ 0.01) displaying an opposite modulation with their corresponding miRNA. The correlation analysis between mRNA and miRNA expression was evaluated by the quantitative trait tool in BRB-ArrayTools (P ≤ 0.01).
Pathway analysis was performed using Ingenuity Pathways Knowledge Base version 8.8 (Ingenuity Systems) as reference set and assuming direct and indirect relationships. Fisher exact test P value <0.05 without multiple-testing correction was deemed statistically significant.
Cell cultures.
Mouse neonatal cardiomyocytes were prepared and cultured as previously described (28) from CD-1 mice (Harlan Laboratories, Inc.). All animal procedures complied with the Guidelines of the Italian National Institutes of Health and with the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Academy of Sciences, Bethesda, MD) and were approved by the institutional animal care and use committee. Primary human coronary artery endothelial cells (HCAECs), human umbilical vein endothelial cells (HUVECs) (Clonetics Corp.), and human HEK293T cell line (American Type Culture Collection) were cultured as previously described (25). For transforming growth factor (TGF)-β stimulations, HUVECs were cultured in 1% FBS endothelial basal medium (Lonza Group Ltd.) for 14 h and then stimulated for 48 h with 0–50 ng/mL TGF-β (R&D Systems). For hypoxia and high glucose stimulations, cells were cultured in the presence of either 25 mmol/L glucose or 5 mmol/L glucose plus 20 mmol/L mannitol (iso-osmotic control) for 72 h. Next, they were incubated in hypoxia (1.0% O2) or normoxia for a further 24 h, as previously described (25).
Statistics.
Continuous variables are expressed as mean ± SE. Gaussian distribution was tested by using the Kolmogorov-Smirnov test. For group-wise comparisons, Mann-Whitney test (two groups), ANOVA, Kruskal-Wallis test (n groups), or t test (two groups) were used as appropriate. For categorical variables, χ2 test was used. Spearman rank correlation was used to compare levels of miRNAs with clinical factors. All tests were performed two-sided and P ≤ 0.05 was considered statistically significant. For statistical analysis and heat map data visualization, GraphPad Prism version 4.03 (GraphPad Software Inc.) and Genesis 1.7.2 (Graz University of Technology, Institute for Genomics and Bioinformatics) software were used, respectively.
RESULTS
Identification of differentially expressed miRNAs in D-HF and ND-HF patients.
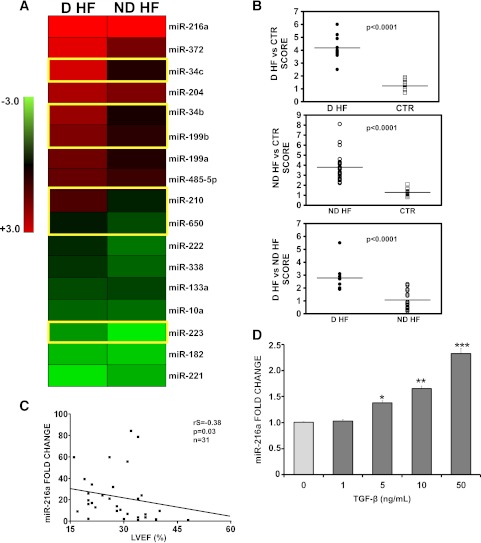
Patients were carefully selected so that D-HF and ND-HF patients displayed similar heart function impairment (Table 1). The aim was minimizing miRNA expression differences due to disease progression per se, highlighting differences resulting from the concomitant occurrence of diabetes. Myocardial biopsies were derived from the nonischemic, remote area of the left ventricle of 19 ND-HF and 10 D-HF patients. None of the clinical parameters other than the occurrence of type 2 diabetes and HbA1c concentration were significantly different between these patient groups (Table 2). A total of 16 age- and sex-matched nondiabetic individuals were used as CTR subjects. Total RNA was extracted, and miRNA profiling showed that out of 365 miRNAs measured, ∼250 were readily detectable in all groups. We found that 41 miRNAs were significantly modulated greater than or equal to twofold in D-HF and/or in ND-HF compared with CTR subjects. Moreover, 10 of these miRNAs were also expressed differently in D-HF compared with ND-HF patients (Fig. 2A).
FIG. 2.
miRNAs modulated in D-HF and ND-HF patients. Validation of the significantly modulated miRNAs by qRT-PCR. D-HF (n = 10) and ND-HF (n = 23) patients were compared with each other and with CTR subjects (n = 16). A: Average values are expressed using a log2 scale (-ΔΔCt) in a heat map where green and red colors indicate down- or upregulation, respectively; yellow boxes indicate those miRNAs statistically deregulated in D-HF vs. ND-HF patients. B: Scores summarizing miRNA differences observed in each patient were calculated. The fold change of all the miRNAs belonging to each specific analysis (D-HF vs. CTR, ND-HF vs. CTR, and ND-HF vs. D-HF) was calculated for each patient. Next, the sign of negative values was inverted and the mean of all values was calculated. Statistical difference between each group couple was identified by unpaired, two-tailed t test. C: Inverse correlation of miR-216a fold change compared with CTR subjects and LVEF values (Spearman analysis; P < 0.05). D: miR-216a modulation by TGF-β in vitro. HUVEC cells were growth-factor starved for 14 h and then stimulated with 1, 5, 10, or 50 ng/mL TGF-β or with vehicle only (CTR) for 48 h. miR-216a levels were measured by qRT-PCR and expressed as fold change compared with CTR subjects. n = 4. *P ≤ 0.05, **P ≤ 0.01, ***P < 0.001.
Significantly modulated miRNAs were validated by more sensitive and specific qRT-PCR assays. This validation step included the same patients and CTR subjects used for profiling and was extended to four more ND-HF patients. We found that 17 miRNAs were significantly modulated in D-HF and/or ND-HF patients compared with CTR subjects. Comparing D-HF with ND-HF, we observed that 6 miRNAs were differently modulated: miR-34b, miR-34c, miR-199b, miR-210, miR-650, and miR-223 (Fig. 2A and Supplementary Table 1).
In addition, the average fold changes of the differentially expressed miRNAs in each patient group were summarized in three scores: D-HF versus CTR, ND-HF versus CTR, and D-HF versus ND-HF. Figure 2B shows that each score exhibited highly significant differences in the corresponding patient cohorts.
miR-216a is induced in HF patients and by TGF-β.
miR-216a is the miRNA displaying the highest induction in both HF patient classes. Of interest, we found that miR-216a expression was inversely correlated with LVEF (Fig. 2C).
TGF-β is particularly expressed in the hypertrophic myocardium progressing toward HF (29) and can also induce miR-216a expression in diabetic nephropathy (30). Thus, we asked whether TGF-β may trigger miR-216a expression in the heart as well. While miR-216a is barely detectable in isolated mouse cardiac myocytes and fibroblasts (not shown), it was readily detectable in human endothelial cells. Upon TGF-β stimulation of these cells, we found that miR-216a expression was significantly induced in a dose-dependent (Fig. 2D) and time-dependent (not shown) fashion, suggesting that TGF-β may play a similar role in HF.
miRNA deregulation in the border zone.
We tested whether miRNA expression differences observed in the myocardial portion remote from the infarction region were also present in the border zone (i.e., in the myocardial portion around the scar tissue formed after the infarction). We found that all tested miRNAs were similarly dysregulated in the remote zone and in the border zone (not shown). It is intriguing that miR-216a increase was even higher in the border than in the remote zone (D-HF = 65 ± 3.8 and ND-HF = 49 ± 2.5 vs. CTR, P < 0.0001, for both). We also found that most expression differences between D-HF and ND-HF patients were attenuated in the border zone (Fig. 3 and Supplementary Table 2). For miR-34b, miR-34c, miR-210, and miR-199b, the expression differences were not due to less severe dysregulation in diabetic patients but, rather, to the opposite; indeed, expression levels for these miRNA were induced in ND-HF as well. Likewise, miR-223 decrease in the border zone was attenuated for both D-HF and ND-HF, in keeping with the less pronounced miR-223 downmodulation in D-HF than in ND-HF, as observed in the remote zone (Supplementary Table 1). Thus, at least for the analyzed miRNAs, the dysregulations observed only in diabetic patients when the remote myocardium was analyzed were found in the border zone of both patient classes.
FIG. 3.
miR-34b, miR-34c, miR-210, miR-199b, and miR-223 are deregulated in the border zone (BZ) of both D-HF and ND-HF patients. miRNAs deregulated comparing ND-HF with D-HF in the myocardial portion remote from the infarction region (REM) were measured in the BZ as well. The top panel shows a heat map representing the average values expressed using a log2 scale (-ΔΔCt), where green and red colors indicate down- or upregulation, respectively.
Impact of HF dysregulated miRNAs on the transcriptome.
To investigate the functional implications of miRNA dysregulations identified in both D-HF and ND-HF patients, we tested their impact on the transcriptome.
First, we measured gene expression profiles using high-density oligonucleotide microarrays. As expected, unsupervised hierarchical clustering analysis segregated the profiles of D-HF, ND-HF, and CTR subjects (Supplementary Fig. 1). Class comparison analysis versus the CTR group was performed and qRT-PCR validated (Supplementary Fig. 2), identifying 389 genes differentially expressed in D-HF and 1,180 genes differentially expressed in ND-HF. Moreover, the comparison between D-HF and ND-HF groups identified 376 modulated genes that could be attributed to diabetes (data not shown; see Gene Expression Omnibus database, GEO GSE26887 for a complete gene list).
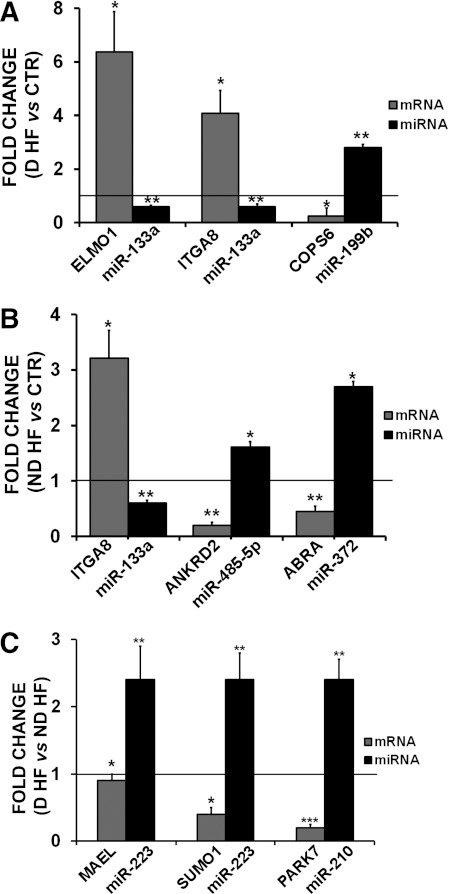
Next, experimentally validated and predicted targets of each miRNA were analyzed, and mRNA/miRNA couples displaying reciprocal modulation were selected. Statistical analysis allowed the identification of 140, 329, and 11 miRNA/mRNA interactions in the D-HF versus CTR, ND-HF versus CTR, and D-HF versus ND-HF groups, respectively (Supplementary Table 3). A subset of the identified miRNA-target modulations was further validated by qRT-PCR in cardiac biopsies (Fig. 4) and in a cell culture system (Supplementary Fig. 3).
FIG. 4.
A–C: Validation by qRT-PCR of miRNA predicted targets. We measured the mRNA expression of selected miRNA predicted targets (34 mRNAs), and modulation of the indicated miRNA predicted targets was validated in the relevant patient groups. The modulation of the corresponding targeting miRNA is also shown. ND-HF (n = 16), D-HF (n = 6), and CTR (n = 15). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Gene interaction and pathway analysis of global gene expression changes.
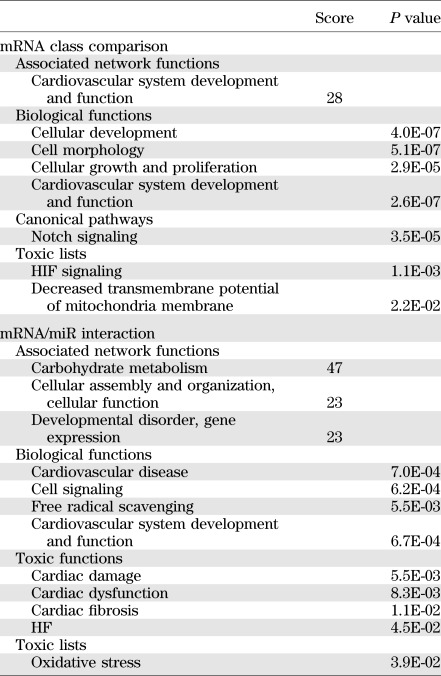
To facilitate the interpretation of the complex gene expression changes observed in the different experimental groups, we used Ingenuity Pathway Analysis software (IPA), exploring for enriched biological functions and pathways. We applied a low-stringency approach, prioritizing sensitivity to maximize the identification of the potentially involved categories.
To assess the potential functional relevance of the global gene expression changes observed in D-HF and ND-HF patients, class comparison genes were uploaded to the IPA database (Table 3). Several interesting network functions were enriched significantly in the D-HF group compared with the ND-HF group, such as the cardiovascular system development and function (Table 3). It is remarkable that IPA analysis also revealed a significant association of the hypoxia-inducible factor (HIF) signaling pathway to the mRNAs modulated in the D-HF group. In addition, a significant association of the transmembrane potential of mitochondria decrease and notch signaling pathway to the D-HF mRNAs was found (Table 3).
TABLE 3.
IPA analysis of mRNA class comparison and mRNA/miR interaction
Gene interaction and pathway analysis of miRNA target genes.
To investigate the specific functional impact of miRNA-dependent gene expression deregulation, IPA analysis was performed considering only predicted miRNA target genes displaying an inverse correlation with their corresponding miRNA (Supplementary Table 3). This analysis identified several interesting functions, canonical pathways, toxic functions, and toxic lists significantly enriched for the ND-HF or D-HF versus CTR groups (Table 3). Most of these categories were enriched in both HF groups. However, other functions were associated with the D-HF versus CTR group only (e.g., free radical scavenging, oxidative stress, cardiac damage, cardiac dysfunction, cardiac fibrosis, and HF) (Table 3), suggesting the involvement of the identified miRNAs in these events. The low number of targets correlating with D-HF versus ND-HF miRNAs precluded IPA analysis.
Activation of the HIF signaling pathway in D-HF patients.
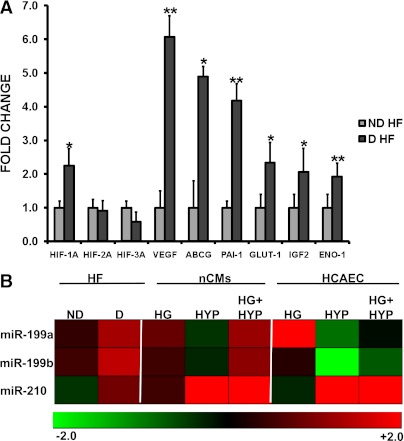
HIF signaling pathway enrichment among genes modulated comparing D-HF with ND-HF patients appeared particularly significant, since one of the modulated miRNAs, miR-210, is a known HIF-1α target (26). Thus, the activation of the HIF pathway was investigated further in HF biopsies by qRT-PCR. We found that HIF-1α mRNA levels were increased in the remote myocardium of D-HF patients compared with ND-HF patients, while the other members of the HIF family, HIF-2α and HIF-3α, were not modulated (Fig. 5A). This HIF-1α increase appeared functionally relevant since it was associated with higher levels of HIF target genes (Fig. 5A).
FIG. 5.
HIF pathway activation in D-HF patients and miR-199a, miR-199b, and miR-210 regulation by hypoxia (HYP) and high glucose (HG) in vitro. A: mRNA expression level of the HIF family and HIF target genes were assayed by qRT-PCR in D-HF (n = 6) and ND-HF (n = 16) samples. *P ≤ 0.05, **P ≤ 0.01. B: Neonatal mouse cardiomyocytes (nCMs) and HCAECs were cultured in HYP (1% O2), in HG (25 mmol/L), in HG followed by HYP (HG+HYP), or in normoxic and iso-osmotic control conditions. Next, total RNA was extracted and miR-199a, miR-199b, and miR-210 levels were assayed. Data are expressed using a log2 scale (-ΔΔCt) by a heat map, where green and red colors indicate down- or upregulation, respectively. As a reference, miR-199a, miR-199b, and miR-210 levels observed in D-HF and ND-HF heart biopsies are indicated as well.
miR-199a, miR-199b, and miR-210 modulation by hypoxia and high glucose.
These findings prompted us to investigate whether hypoxia was a significant stimulus regulating HF miRNA expression. The experiments were conducted in the presence or absence of high glucose, a crucial pathogenic component of diabetes, in both cardiomyocytes and endothelial cells.
Hypoxia stimulated miR-210 expression in both cell types (Fig. 5B). In endothelial cells, the combination of hypoxia with high glucose slightly but significantly increased this induction (Supplementary Table 4); when miR-199a and miR-199b levels were measured, we found that hypoxia decreased their expression in both cell types. This is in apparent contradiction with our observation in D-HF patients. However, we also found that high glucose strongly modified hypoxia response, since miR-199a and miR-199b downmodulation was prevented in endothelial cells (changes not significant) (Supplementary Table 4) and inverted in cardiomyocytes: Fig. 5B shows that the combination of hypoxia and high glucose led to miR-199a and miR-199b induction.
DISCUSSION
As a result of its high morbidity and mortality, HF has been the objective of many gene expression studies in the past few years. For the first time, we compared D-HF and ND-HF patients with similar functional impairment, identifying a subset of miRNAs that were specifically dysregulated in D-HF, in ND-HF, or in both. It is interesting that some of these miRNAs were previously identified as dysregulated in a variety of heart disease studies performed in humans, as summarized in Supplementary Table 5. One limitation of this study is the low number of female patients recruited, which did not allow us to determine the possible effects of sex.
Of interest, miR-216a significantly increased in both D-HF and ND-HF patients compared with CTR subjects, and although miR-216a has been found to be induced in senescent-cultured endothelial cells (31), this is the first time that miR-216a overexpression has been related to human HF. Accordingly, several miR-216a–predicted targets (Supplementary Fig. 3) are involved in heart diseases. PDE4D, an miR-216a–predicted target downmodulated in HF patients, is one of the isoforms of the enzyme that hydrolyzes and inactivates cAMP and finely tunes cAMP levels in different cardiomyocyte subcellular compartments. Inactivation of PDE4D potentiates epinephrine-evoked (32) or exercise-induced arrhythmias in mice and the development of cardiomyopathy, increasing mortality after cardiac infarction (33). Another miR-216a target is CAV2, a member of the caveolin family of scaffolding proteins, which regulates signal transduction pathways and lipid metabolism. Loss of caveolin expression results in the activation of a program of progressive hypertrophy in cardiac myocytes, and their deletion results in severe cardiomyopathy (34). Thus, the inhibitory activity of miR-216a on the expression of these transcripts could be deleterious for the cardiac activity. Moreover, ventricular myocytes from the infarcted myocardium overexpress TGF-β protein and mRNA (35), and TGF-β is particularly overexpressed in myocardium during transition from stable hypertrophy to HF (29). We found that TGF-β induced miR-216a expression increase in endothelial cells. In view of these findings, the release of TGF-β from myocytes after infarction and/or during transition to HF may be the cause for the miR-216a expression increase in HF patients.
We found that the expression of six miRNAs (miR-34b, miR-34c, miR-199b, miR-210, miR-223, and miR-650) changed specifically in the failing myocardium of diabetic patients. It is intriguing that most of these miRNAs also were similarly deregulated in ND-HF patients when the border zone was analyzed, suggesting a link with tissue damage.
Indeed, we previously showed that miR-34c was induced after acute ischemic damage (36), and the upregulation of miR-34b was also found in end-stage HF patients by Thum et al. (18), who, however, did not stratify patients for diabetes. miR-34b and miR-34c belong to an evolutionary conserved miRNA family that plays a fundamental role in the p53 tumor suppressor network (37). Since hyperglycemia promotes p53-dependent apoptosis (38), it is tempting to speculate that the p53/miR-34 axis may be involved in a hyperglycemia-induced death pathway.
We also found that miR-223 levels were sharply decreased in the failing myocardium. While further studies are needed, miR-223 downmodulation likely represents a compensatory adaptive mechanism, since its decrease is less evident both in the remote zone of D-HF patients and in the border zone of ND-HF patients. In keeping with our data, Lu et al. (39) found that miR-223 levels were increased in the myocardium of diabetic patients in the absence of concomitant myocardial infarction, indicating that diabetes-associated miR-223 induction might, at least in part, counteract miR-223 decrease triggered by other stimuli. Likewise, van Rooij et al. (40) reported that miR-223 is increased in end-stage ischemic cardiomyopathy, suggesting that miR-233 downmodulation may represent an adaptive mechanism lost in end-stage patients.
We also demonstrated that miR-199 is upregulated in the diabetic failing heart. Of interest, da Costa Martins et al. (41) found that miR-199b was activated by a calcineurin/nuclear factor of activated T cells pathway and that its overexpression in mice caused a significant left ventricular fibrosis and HF (41). When miR-199a and miR-199b levels were examined in cultured cardiomyocytes and endothelial cells, the levels were decreased by hypoxia as expected (26,42). However, the concomitant stimulus of high glucose prevented this decrease, and miR-199a and miR-199b levels were actually induced in cardiomyocytes exposed to hypoxia and high glucose. Since nuclear factor of activated T cells acts as a glucose sensor in smooth muscle cells (43), one can hypothesize that the same mechanism also may be at work in D-HF patients.
There is evidence that diabetic hearts have a compromised hypoxia signaling pathway (44). Indeed, in the healthy heart, responses to reductions in cellular O2 concentrations are conveyed mostly by the transcription factor family HIF (45). The HIF pathway is inactive in the presence of physiological levels of O2 and is activated in hypoxia, thereby upregulating a gene expression program facilitating O2-independent ATP synthesis.
IPA analysis of the transcriptomic changes observed in D-HF patients identified the HIF signaling pathway as significantly deregulated. Indeed, HIF-dependent genes and miRNAs were induced in the remote, noninfarcted myocardium of D-HF patients. These data are in keeping with previous evidence of HIF mRNA and HIF-pathway induction in the nonischemic myocardium observed in rat diabetic models of myocardial infarction (46) and, more general, with a pseudohypoxia metabolic state induced by hyperglycemia (47). It is also worth noting that while steady-state HIF levels are increased in diabetic subjects, several reports exist of impaired HIF induction by hypoxia, leading to an overall inhibition of HIF-dependent hypoxic response in diabetic patients (44,48).
Among different potential HIF activation mechanisms in D-HF patients, increased oxidative stress is a likely candidate. Indeed, reactive oxygen species can induce the HIF/HIF pathway (45), and increased oxidative stress levels are a hallmark of diabetes (49).
Accordingly, we found that oxidative stress and free radical scavenging functions were associated with the genes targeted by the miRNA deregulated in D-HF. Paradoxically, increased oxidative stress may also contribute to the dampening of HIF activation after ischemia (50). In this respect, PARK7, a predicted miR-210 target downregulated in HF diabetic patients and a ubiquitous redox-responsive cytoprotective protein (51), may be particularly important.
Among the multitude of genes modulated by HIF-1α, miR-210 can be considered a “master” miRNA of hypoxic response (26). It is not clear whether miR-210 activation in the noninfarcted myocardial region represents an adaptive or a maladaptive mechanism. PARK7 targeting suggests that miR-210 induction may be detrimental. However, we and others have found that miR-210 protected endothelial cells and cardiomyocytes from apoptosis (25,52,53), highlighting a possible protective role of miR-210 in D-HF.
Thus, while the role played by the HIF/HIF pathway in diabetic patients needs further investigation, it seems clear that HIF-dependent response to hypoxia is disrupted in the failing heart of diabetic patients.
In conclusion, HF in patients with or without diabetes is a result of the concomitant cross-talk between various deleterious and compensatory signaling pathways. The balance and the interaction between these two dynamic forces ultimately determine how the pathology will evolve. We identified a subset of miRNAs that are specifically dysregulated in D-HF. Gene expression analysis strongly implicated them in the pathogenetic disease mechanisms. Future investigations of the deregulated miRNA actions and functions will substantially improve our understanding of diabetic cardiac and vascular complications.
ACKNOWLEDGMENTS
This work was supported by Ministero della Salute.
No potential conflicts of interest relevant to this article were reported.
S.G. designed and performed most experiments and wrote the manuscript. P.F. and D.A. contributed to bioinformatic analysis. S.C., M.D.D., A.M., and L.M. enrolled and characterized patients and harvested biopsies. Y.D. and M.C.C. contributed to discussion. F.M. designed the experiments and wrote the manuscript. All authors reviewed the manuscript. F.M. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Part of this study was presented as an oral communication at the European Society of Cardiology Congress, Paris, France, 27–31 August 2011.
The authors thank Dr. Daniele Catalucci, Institute of Biomedical Technologies, National Research Council, Milan, Italy, for his help in cardiomyocyte isolation and Mauro Helmer-Citterich, Istituto Dermopatico dell’Immacolata IRCCS, and Lucia Cicchillitti, IRCCS Policlinico San Donato, for their contribution to Affymetrix gene expression analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0952/-/DC1.
REFERENCES
- 1.Pfeffer MA, Braunwald E. Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation 1990;81:1161–1172 [DOI] [PubMed] [Google Scholar]
- 2.Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation 2007;115:3213–3223 [DOI] [PubMed] [Google Scholar]
- 3.Mukamal KJ, Nesto RW, Cohen MC, et al. Impact of diabetes on long-term survival after acute myocardial infarction: comparability of risk with prior myocardial infarction. Diabetes Care 2001;24:1422–1427 [DOI] [PubMed] [Google Scholar]
- 4.Estep JD, Aguilar D. Diabetes and heart failure in the post-myocardial infarction patient. Curr Heart Fail Rep 2006;3:164–169 [DOI] [PubMed] [Google Scholar]
- 5.Castelvecchio S, Menicanti L, Baryshnikova E, de Vincentiis C, Frigiola A, Ranucci M, Surgical and Clinical Outcome Research (SCORE) Group Comparison of morbidity and mortality in diabetics versus nondiabetics having isolated coronary bypass versus coronary bypass plus valve operations versus isolated valve operations. Am J Cardiol 2011;107:535–539 [DOI] [PubMed] [Google Scholar]
- 6.Stone PH, Muller JE, Hartwell T, et al. The MILIS Study Group The effect of diabetes mellitus on prognosis and serial left ventricular function after acute myocardial infarction: contribution of both coronary disease and diastolic left ventricular dysfunction to the adverse prognosis. J Am Coll Cardiol 1989;14:49–57 [DOI] [PubMed] [Google Scholar]
- 7.Mak KH, Moliterno DJ, Granger CB, et al. Influence of diabetes mellitus on clinical outcome in the thrombolytic era of acute myocardial infarction. GUSTO-I Investigators. Global Utilization of Streptokinase and Tissue Plasminogen Activator for Occluded Coronary Arteries. J Am Coll Cardiol 1997;30:171–179 [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Li Y, Feng Q, Arnold M, Peng T. Calpain activation contributes to hyperglycaemia-induced apoptosis in cardiomyocytes. Cardiovasc Res 2009;84:100–110 [DOI] [PubMed] [Google Scholar]
- 9.Shen E, Li Y, Li Y, et al. Rac1 is required for cardiomyocyte apoptosis during hyperglycemia. Diabetes 2009;58:2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 2008;9:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res 2008;103:1072–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang G, Tang X, Mendu V, et al. The art of microRNA: various strategies leading to gene silencing via an ancient pathway. Biochim Biophys Acta 2008;1779:655–662 [DOI] [PubMed] [Google Scholar]
- 13.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618 [DOI] [PubMed] [Google Scholar]
- 14.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Ji R, Yue J, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol 2007;170:1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007;100:416–424 [DOI] [PubMed] [Google Scholar]
- 17.Ikeda S, Kong SW, Lu J, et al. Altered microRNA expression in human heart disease. Physiol Genomics 2007;31:367–373 [DOI] [PubMed] [Google Scholar]
- 18.Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258–267 [DOI] [PubMed] [Google Scholar]
- 19.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol 2008;45:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naga Prasad SV, Duan ZH, Gupta MK, et al. Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J Biol Chem 2009;284:27487–27499 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Matkovich SJ, Van Booven DJ, Youker KA, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation 2009;119:1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes 2011;60:1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castelvecchio S, Menicanti L, Donato MD. Surgical ventricular restoration to reverse left ventricular remodeling. Curr Cardiol Rev 2010;6:15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 1995;57:289–300 [Google Scholar]
- 25.Fasanaro P, Greco S, Lorenzi M, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem 2009;284:35134–35143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life 2011;63:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Brigand K, Robbe-Sermesant K, Mari B, Barbry P. MiRonTop: mining microRNAs targets across large scale gene expression studies. Bioinformatics 2010;26:3131–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Stefano V, Giacca M, Capogrossi MC, Crescenzi M, Martelli F. Knockdown of cyclin-dependent kinase inhibitors induces cardiomyocyte re-entry in the cell cycle. J Biol Chem 2011;286:8644–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boluyt MO, O’Neill L, Meredith AL, et al. Alterations in cardiac gene expression during the transition from stable hypertrophy to heart failure. Marked upregulation of genes encoding extracellular matrix components. Circ Res 1994;75:23–32 [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menghini R, Casagrande V, Cardellini M, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 2009;120:1524–1532 [DOI] [PubMed] [Google Scholar]
- 32.Galindo-Tovar A, Kaumann AJ. Phosphodiesterase-4 blunts inotropism and arrhythmias but not sinoatrial tachycardia of (-)-adrenaline mediated through mouse cardiac beta(1)-adrenoceptors. Br J Pharmacol 2008;153:710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehnart SE, Wehrens XH, Reiken S, et al. Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell 2005;123:25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park DS, Woodman SE, Schubert W, et al. Caveolin-1/3 double-knockout mice are viable, but lack both muscle and non-muscle caveolae, and develop a severe cardiomyopathic phenotype. Am J Pathol 2002;160:2207–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson NL, Bazoberry F, Speir EH, et al. Transforming growth factor beta-1 in acute myocardial infarction in rats. Growth Factors 1988;1:91–99 [DOI] [PubMed] [Google Scholar]
- 36.Greco S, De Simone M, Colussi C, et al. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J 2009;23:3335–3346 [DOI] [PubMed] [Google Scholar]
- 37.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer 2007;7:819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiordaliso F, Leri A, Cesselli D, et al. Hyperglycemia activates p53 and p53-regulated genes leading to myocyte cell death. Diabetes 2001;50:2363–2375 [DOI] [PubMed] [Google Scholar]
- 39.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 2010;86:410–420 [DOI] [PubMed] [Google Scholar]
- 40.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.da Costa Martins PA, Salic K, Gladka MM, et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol 2010;12:1220–1227 [DOI] [PubMed] [Google Scholar]
- 42.Rane S, He M, Sayed D, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res 2009;104:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson LM, Nilsson-Ohman J, Zetterqvist AV, Gomez MF. Nuclear factor of activated T-cells transcription factors in the vasculature: the good guys or the bad guys? Curr Opin Lipidol 2008;19:483–490 [DOI] [PubMed] [Google Scholar]
- 44.Heather LC, Clarke K. Metabolism, hypoxia and the diabetic heart. J Mol Cell Cardiol 2011;50:598–605 [DOI] [PubMed] [Google Scholar]
- 45.Semenza GL. Vascular responses to hypoxia and ischemia. Arterioscler Thromb Vasc Biol 2010;30:648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malfitano C, Alba Loureiro TC, Rodrigues B, et al. Hyperglycaemia protects the heart after myocardial infarction: aspects of programmed cell survival and cell death. Eur J Heart Fail 2010;12:659–667 [DOI] [PubMed] [Google Scholar]
- 47.Williamson JR, Chang K, Frangos M, et al. Hyperglycemic pseudohypoxia and diabetic complications. Diabetes 1993;42:801–813 [DOI] [PubMed] [Google Scholar]
- 48.Marfella R, Esposito K, Nappo F, et al. Expression of angiogenic factors during acute coronary syndromes in human type 2 diabetes. Diabetes 2004;53:2383–2391 [DOI] [PubMed] [Google Scholar]
- 49.Rains JL, Jain SK. Oxidative stress, insulin signaling, and diabetes. Free Radic Biol Med 2011;50:567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thangarajah H, Vial IN, Grogan RH, et al. HIF-1alpha dysfunction in diabetes. Cell Cycle 2010;9:75–79 [DOI] [PubMed] [Google Scholar]
- 51.Kahle PJ, Waak J, Gasser T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic Biol Med 2009;47:1354–1361 [DOI] [PubMed] [Google Scholar]
- 52.Hu S, Huang M, Li Z, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation 2010;122(Suppl.):S124–S131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fasanaro P, D’Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 2008;283:15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]