Abstract
Protein tyrosine phosphatase 1B (PTP1B) is postulated to modulate insulin action by dephosphorylating the insulin receptor signaling proteins and attenuating insulin signaling. We sought to determine the relationship of skeletal muscle PTP1B to whole-body insulin sensitivity. We studied 17 African Americans with type 2 diabetes mellitus (T2DM) and 16 without diabetes. PTP1B gene expression and protein abundance were determined in the biopsied skeletal muscles at the baseline of a hyperinsulinemic-euglycemic clamp. PTP1B gene expression was significantly higher in subjects with T2DM versus control (P < 0.0001) and remained significantly different after adjusting for age and insulin sensitivity (P = 0.05). PTP1B gene expression was positively related to protein abundance (rs = 0.39; P = 0.03; adjusted for age and insulin sensitivity) and negatively related to insulin sensitivity (rs = −0.52; P = 0.002; adjusted for age). Overexpression and interference RNA of PTP1B were performed in primary human skeletal muscle culture. PTP1B overexpression resulted in reduction of Akt phosphorylation in the control subjects. Moreover, interference RNA transfection downregulated PTP1B expression and enhanced Akt phosphorylation in subjects with T2DM. These data show that skeletal muscle PTP1B gene expression is increased in African American subjects with T2DM, is negatively associated with whole-body insulin sensitivity, and contributes to modulation of insulin signaling.
Insulin resistance in skeletal muscle is a key pathophysiologic feature of obesity and type 2 diabetes (1–3). In insulin signaling, tyrosine phosphorylation of the receptor is the key component in regulating the propagation of the insulin signal throughout the cell and activation of other biological processes (4). As such, insulin receptor signaling is a balance between phosphorylation of tyrosine residues by protein tyrosine kinases and dephosphorylation of tyrosine residues by protein tyrosine phosphatases (PTPases). PTPases within the cell are postulated to oppose the effects of the protein tyrosine kinases, and insulin sensitivity is observed clinically if the process is normally regulated. However, an excess of PTPase activities (i.e., hyperactivation of PTPase system) may alter the balance between the two enzymes, resulting in a relative reduction in tyrosine phosphorylation and attenuating insulin signaling on a cellular level. Thus, hyperactivation of PTPases is postulated to contribute to the development of insulin resistance on a whole-body level (5–7).
Protein tyrosine phosphatase 1B (PTP1B) is a specific nonreceptor-type PTPase that has unique structural features, which promotes its interaction with the insulin receptor and insulin receptor substrates (8–11). PTP1B has been shown to dephosphorylate these proteins, thus serving as a negative regulator of insulin receptor signaling. Several studies have postulated that elevation of PTP1B might be an etiologic factor in the tissue insulin resistance that occurs in human obesity and type 2 diabetes. Increased activities and abundance of PTPase and PTP1B were observed in adipose tissue and skeletal muscle of obese and insulin-resistant (i.e., no diabetes) humans (5,12,13), and rodents and humans with diabetes (6,7,14,15). However, conflicting data have been presented in humans with type 2 diabetes, thus suggesting PTPase and PTP1B activities and abundance may be decreased in skeletal muscle and adipose tissue (12,16,17). In addition, some studies have found that variants in the PTP1B gene have been linked to type 2 diabetes and other metabolic traits related to the metabolic syndrome, whereas other studies have found weak or no associations (18–24).
African Americans are an underrepresented population in most studies. However, observations suggest that they have an increased risk of developing insulin resistance and type 2 diabetes when compared with Caucasians (25). In addition, after developing type 2 diabetes, they are also more likely to develop complications from the disease and experience greater disability from the complications than Caucasians. Given the observation that there is a paucity of data regarding specific cellular mechanisms for this diabetes-susceptible group, insight into the etiology of disease presentation would be specifically relevant and would provide novel information. Thus, we sought to evaluate whether skeletal muscle PTP1B gene expression and protein abundance were altered in an African American population with type 2 diabetes when compared with nondiabetic control subjects. In addition, we evaluated and assessed the relationship of PTP1B gene expression and relative protein abundance in skeletal muscle to whole-body insulin sensitivity and other metabolic traits related to the metabolic syndrome. We hypothesized that PTP1B gene expression and protein abundance would be increased in African American subjects with type 2 diabetes and related to insulin sensitivity.
RESEARCH DESIGN AND METHODS
Subjects.
The study cohort consisted of 33 African Americans, 17 with type 2 diabetes and 16 without diabetes. The subjects evaluated were a subset from a larger cohort that evaluated both African Americans and Caucasians (26). To specifically focus on the more underrepresented and diabetes-susceptible population, we evaluated the 33 African Americans who had skeletal muscle biopsies in addition to having complete comprehensive phenotyping analyses (i.e., body composition and hyperinsulinemic-euglycemic clamp studies).
As previously described, if the subject had a history of type 2 diabetes, he/she was required to be on dietary therapy only (i.e., drug naive), with a fasting plasma glucose between 125 (6.9 mmol/L) and 175 mg/dL (9.7 mmol/L). Diabetes status was confirmed by an oral glucose tolerance test. Exclusions were: 1) medications known to affect glucose metabolism, including hormone replacement therapy; 2) untreated thyroid or chronic liver, renal, or cardiovascular disease; and 3) a history of drug and/or alcohol abuse or psychiatric disease prohibiting adherence to study protocol (26). All subjects signed a written informed consent before participation in the study. The study protocol was approved and conducted in strict compliance with Pennington Biomedical Research Center’s Institutional Review Board for human subjects.
Assessment of insulin sensitivity in vivo.
Whole-body insulin sensitivity was assessed in all subjects with use of the hyperinsulinemic-euglycemic clamps (27). After a 12-h overnight fast, insulin was administered at a primed-continuous infusion rate of 120 mU ⋅ m−2 ⋅ min−1. Blood samples were collected frequently during the 120-min clamp procedure. Arterialized serum glucose was measured periodically, and a variable infusion of dextrose (20% solution) was given to maintain serum glucose concentrations at ∼100 mg/dL (5.6 mmol/L). Whole-body insulin-mediated glucose disposal was calculated as described (27) and divided by lean mass to assess insulin sensitivity. In addition, a skeletal muscle biopsy from the vastus lateralis was obtained at the baseline of the clamp.
Isolation and culture of human skeletal muscle cells.
Human skeletal muscle cells (HSMC) were isolated from vastus lateralis muscle biopsies and grown as described previously (28). Briefly, ∼50 mg of muscle tissue was minced with surgical scissors and digested by 0.55% trypsin and 2.21 mmol/L ethylenediaminetetraacetic acid with constant shaking at 37°C. After centrifugation to remove fat and debris, myoblasts were grown in monolayer culture in Classic Liquid Media Hams Nutrient Mixture F12 (HyClone brands; Thermo Scientific) containing 10 μg/L human epidermal growth factor, 10% (v/v) fetal bovine serum, 1% (v/v) antibiotics (10,000 units/mL of penicillin G and 10 mg/mL streptomycin), and 2 mmol/L glutamine. Cells had been differentiated with 2% horse serum for 5 to 6 days after they reached to ∼85% confluence. Cells were used within five passages.
PTP1B protein abundance.
Skeletal muscle tissue lysates were prepared by dissection and homogenized in buffer A (25 mmol/L N-2-hydroxyethylpiperazine-N´-2-ethanesulfonic acid [pH 7.4], 1% Nonidet P-40, 137 mmol/L NaCl, 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, 1 μg/mL pepstatin, and 5 μg/mL leupeptin) using a PRO 200 homogenizer (PRO Scientific, Oxford, CT). The samples were centrifuged at 14,000 × g for 20 min at 4°C, and protein content of the supernatant was determined by Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Supernatants (50 µg) were resolved by SDS-PAGE and subjected to immunoblotting using chemiluminescence reagent (PerkinElmer Life Science, Boston, MA) and quantified as described (29,30). Antibodies for PTP1B were obtained from Millipore (Billerica, MA), and β-actin from Affinity Bioreagents (Golden, CO). Results of scanning for each gel were normalized by β-actin, and the data were presented as mean ± SEM of fold change in subjects with no diabetes versus type 2 diabetes.
PTP1B vectors transfection.
Primary HSMC from lean healthy subjects or subjects with type 2 diabetes were seeded in six-well plates. For each transfection sample, the PTP1B over expression vectors (pCMV6H-PTP1B vector; a gift from Dr. Jonathan Chernoff, Fox Chase Cancer Center, Philadelphia, PA) and PTP1B small interfering RNA (siRNA) vectors (Addgene, Cambridge, MA) were combined with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) at a ratio of PTP1B 1:2.5 (µg/µL). They were mixed gently and incubated for 20 min at room temperature. Five hundred microliters of transfection mixtures was added to cultured cells. The cells were incubated at 37°C in a CO2 incubator for 6 h and then changed to normal cell culture medium. The cultured cells were collected for Western blot 48–72 h after transfection.
PTP1B gene expression
RNA extraction.
Total RNA was extracted from skeletal muscle for real-time quantitative PCR assays as previously described (29). Frozen tissues were placed in a mortar in liquid nitrogen, and the tissue was pulverized into powder using a pestle on dry ice. Total RNA was isolated from the tissue powder using TRIzol reagent (Invitrogen). After DNase I (Invitrogen) digestion, RNA was further purified with the RNeasy Mini Kit (Qiagen, Germantown, MD). RNA concentration and quality were measured by RNA 6000 Nano LabChip kit (Agilent Technologies, Santa Clara, CA).
Real-time quantitative PCR.
The primer sequences of candidate genes were designed using Primer Express Software version 3.0 (Applied Biosystems, Foster, CA). The sequences of the specific primers obtained from IDT (San Diego, CA) were as follows: PTP1B gene expression, 5′-CGACCAGCTGCGCTTCTC-3′ (forward) and 5′-GTCCCCCATGATGAATTTGG-3′ (reverse); and RPLP0, 5′-TTCTCCTTTGGGCTGGTCAT-3′ (forward) and 5′-CAGGGAG CGAGAATGCAGAGT-3′ (reverse). RPLP0 gene encodes the human acidic ribosomal phosphoprotein large P0 subunit and is used as a special housekeeping gene for human skeletal muscle (31,32). A 1-µg aliquot of total RNA for each sample was reverse-transcribed in a 100-µl reaction volume with a commercial High-Capacity cDNA Archive Kit (Applied Biosystems PE) according to the manufacturer’s protocol. The quantitative PCR reaction was conducted in 384-well microtiter plates on the ABI Prism Sequence Detector 7900 (Applied Biosystems) with Bio-Rad iTaqTM SYBR Green Supermix with ROX Kits (Bio-Rad). For each sample of each gene, PCR amplification was performed in triplicate. The mRNA content of each gene was determined simultaneously in paired muscle samples assessed by DNA array analysis. The assay was performed in duplicate. Results for PTP1B gene expression were normalized by RPLP0, and the data were presented as mean ± SEM of fold change in subjects with no diabetes versus type 2 diabetes.
Statistical analysis.
Characteristics of the study subjects were summarized by health status as counts (number of subjects) for categorical data and means ± standard errors (mean ± SEM) for continuous data. Fisher exact test and Student two-sample t test were employed for counts and continuous data, respectively, to compare the subjects with no diabetes and type 2 diabetes. Spearman rank correlation coefficients were used to measure monotonic association between variable pairs because it is robust to the influence of outlying observations. Spearman partial correlation coefficients were used to evaluate the statistical associations between PTP1B gene expression and protein abundance and metabolic assessments while adjusting (controlling) for concomitant variations such as those attributable to age and insulin sensitivity. PTP1B gene expression and protein abundance differences between the groups were evaluated by ANOVA and adjusted for age and insulin sensitivity. Statistical significance was declared for statistical tests resulting in P ≤ 0.05. All statistical analyses were performed using the software package SAS Verson 9.1 (SAS Institute, Cary, NC).
RESULTS
The characteristics of the study cohort are listed in Table 1. Subjects with type 2 diabetes were older and had a higher fat distribution than the control subjects (P ≤ 0.05). Also, the subjects with type 2 diabetes had higher fasting glucose, insulin, and triglyceride concentrations when compared with the control subjects (P ≤ 0.05). The observations from the clamps demonstrated that the control subjects had significantly higher insulin sensitivity than the subjects with type 2 diabetes.
TABLE 1.
Characteristics of the study subjects
PTP1B gene expression
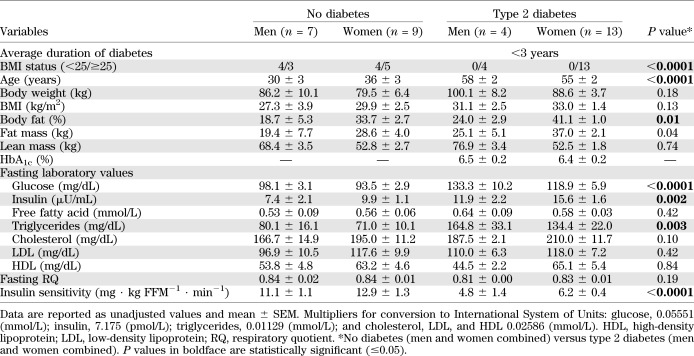
PTP1B gene expression in skeletal muscle was determined, and individual values of PTP1B mRNA for the 33 study participants are shown in Fig. 1A. The subjects with type 2 diabetes had a significantly higher average PTP1B gene expression level than the control subjects (unadjusted values; Fig. 1B). PTP1B gene expression remained higher in people with type 2 diabetes than control subjects even after adjusting for age and insulin sensitivity (control, 1.0 ± 2.6 versus type 2 diabetes, 10.6 ± 2.5; P = 0.05).
FIG. 1.
All study participants (n = 33) and their individual PTP1B gene expression values (A) (expressed as arbitrary units). Average PTP1B gene expression in both groups (mean ± SEM) (B). *No diabetes (control; n = 16) versus type 2 diabetes (n = 17); P < 0.0001 (unadjusted).
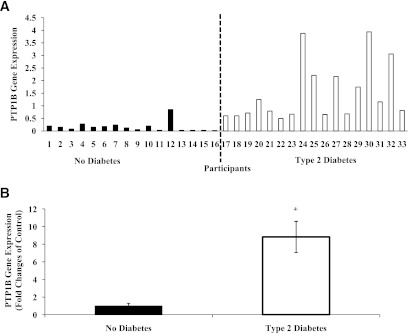
PTP1B gene expression in skeletal muscle for the entire cohort was positively and significantly (P ≤ 0.05) related to protein abundance, age, fasting glucose, and insulin concentrations and negatively correlated to insulin sensitivity. After adjustments, PTP1B gene expression remained positively related to protein abundance (rs = 0.39; P = 0.03; adjusted for age and insulin sensitivity; unadjusted, Fig. 2A) and negatively related to insulin sensitivity (rs = −0.52; P = 0.002; adjusted for age; unadjusted, Fig. 2B).
FIG. 2.
Relationship of PTP1B gene expression to PTP1B protein abundance (A) and insulin sensitivity (B). Relationship of PTP1B protein abundance to fasting glucose (C). PTP1B gene expression and protein abundance values are expressed as arbitrary units. Unadjusted values are presented. Black circles, no diabetes; white circles, type 2 diabetes.
PTP1B protein abundance.
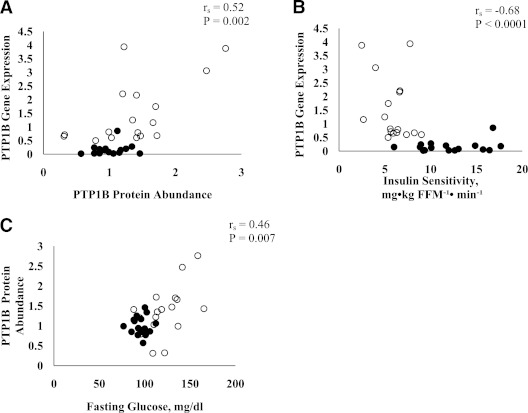
PTP1B protein abundance in skeletal muscle was determined, and individual values of PTP1B protein abundance for the 33 study participants are shown in Fig. 3A. On average, the subjects with type 2 diabetes had a higher PTP1B protein abundance in skeletal muscle than the control subjects (1.0 ± 0.1 versus 1.4 ± 0.2; fold changes of control; P = 0.04). However, the difference between the groups was not observed after adjusting for age and insulin sensitivity (1.0 ± 0.2 versus 1.1 ± 0.2; P = 0.68). Fig. 3B demonstrates a representative gel for skeletal muscle protein abundance for PTP1B as assessed in six subjects representing health status (three without and three with type 2 diabetes) and different phenotype as assessed by BMI (lean and obese) without (basal) and with insulin stimulation. As demonstrated, skeletal muscle PTP1B content was greater in obese subjects with type 2 diabetes than in lean nondiabetic individuals.
FIG. 3.
All study participants (n = 33) and their individual PTP1B protein abundance values (A) (expressed as arbitrary units). Protein abundance in lean (no diabetes; n = 3) and obese (type 2 diabetes; n = 3) subjects (B).
PTP1B protein abundance in skeletal muscle for the entire cohort was positively and significantly (P ≤ 0.05) related to PTP1B gene expression, age, and fasting glucose concentrations (Fig. 2C). After adjusting for age and insulin insensitivity, PTP1B protein abundance remained positively correlated to PTP1B gene expression (rs = 0.39; P = 0.03).
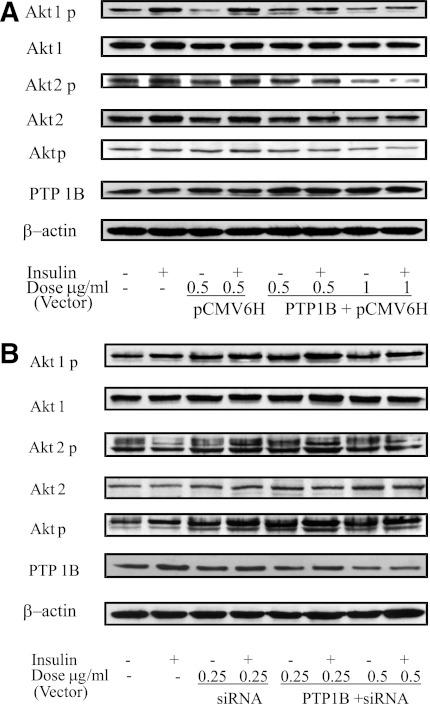
To confirm the role PTP1B plays in insulin signaling, we used PTP1B overexpression vector transfection in HSMC from a lean and healthy (i.e., no diabetes) subject. The results showed that PTP1B abundance was greatly increased and the insulin-stimulated Akt phosphorylation was reduced when compared with the empty vector transfected and control cell (Fig. 4A). Additionally, in the primary HSMC from an obese subject with type 2 diabetes, suppression of PTP1B expression by using a PTP1B siRNA vector transfection resulted in significantly reduced PTP1B protein abundance. In addition, insulin-stimulated Akt phosphorylation was increased in comparison with nontransfected muscle cells (Fig. 4B).
FIG. 4.
Effects of PTP1B overexpression in the skeletal muscle cells from a lean and healthy (no diabetes) subject (A) and interference in the skeletal muscle cells of an obese subject with type 2 diabetes (B) on PTP1B abundance and insulin-signaling protein Akt. The expression levels were normalized against β-actin. p, phosphorylation.
DISCUSSION
To our knowledge, this is the first study to evaluate skeletal muscle PTP1B gene expression and abundance in an African American population. Our data showed African American subjects with type 2 diabetes had a statistically significant increase in skeletal muscle PTP1B gene expression when compared with African American subjects without type 2 diabetes. In addition, increased PTP1B gene expression was significantly related to decreased insulin sensitivity (i.e., insulin resistance) and an increase in PTP1B protein abundance. These results were supported by preclinical and clinical data. Ahmad and colleagues (14) found an increase in skeletal muscle PTP1B protein abundance and expression of mRNA in diabetes-induced rats. Likewise, other studies with diabetes-induced rats observed an increase in PTP1B protein content and activity in skeletal muscle (6,7). In subjects with type 2 diabetes, Cheung et al. (15) reported higher adipose tissue PTP1B protein abundance when compared with lean nondiabetic subjects. Also, obese and insulin-resistant (i.e., no diabetes) subjects had increases in PTPase and PTP1B activities and protein abundance in skeletal muscle and adipose tissue (5,12,13). Based on our data and previous data, PTP1B in skeletal muscle and adipose tissue reflect in vivo insulin sensitivity and is increased in insulin-resistant states. Unlike the previous studies, we accessed PTP1B gene expression and protein abundance in the skeletal muscle of humans with type 2 diabetes because skeletal muscle accounts for >75% of insulin-dependent glucose uptake (33). An interesting comparison and a future direction would be to compare PTP1B gene and protein abundance in both skeletal muscle and adipose tissue from the same subjects.
In contrast to our study, previous studies (12,16,17) have suggested that the activity and protein abundance of PTPase and PTP1B are reduced in skeletal muscle of subjects with type 2 diabetes. There are many possible reasons for discrepancies in the data. The major differences may be the subject characteristics of the cohort evaluated. Specifically, subjects in the current study were much earlier in the course of their disease process [i.e., duration of the disease on average was <3 years versus 5 years reported in the study by Worm et al. (17)]. In the current study, diabetic subjects were naive to any antidiabetic medication, whereas medication use was reported in the other studies (12,16,17). The mean BMI differed in the current study participants (32.5 kg/m2) when compared with the mean value reported by Ahmad et al. (12) (51.5 kg/m2). In regards to glycemia, the current study had a lower HbA1c (6.4%) than Worm et al. (17) (7.7%). Also, the current study had a lower fasting glucose (122.3 mg/dL) compared with the other studies [141 (17), 146 (12), and 236 (16) mg/dL]. Thus, given the natural history of type 2 diabetes and the fact that different comorbidities exist depending on the stage of type 2 diabetes, the difference in values reported for PTP1B may be a reflection of the underlying comorbidities as observed in the cohort characteristics at the stage of the disease. Thus, our study clearly differed from the other reported studies (12,16,17) in regards to medication use, level of glycemia, BMI, duration of disease, and the fact that we evaluated the PTPase system with not only protein levels, but also specific gene expression for skeletal muscle PTP1B, which was not observed in the other studies (12,16,17).
Our data clearly demonstrate a significant positive relationship between PTP1B gene expression and protein abundance in the entire cohort. However, the relationship to insulin sensitivity appeared different for gene expression and protein abundance. Also, after adjusting for age and insulin sensitivity, gene expression remained significantly different between the nondiabetic and diabetic subjects, whereas the protein abundance difference disappeared between the groups. Given that it is the protein, not the mRNA, which exerts the biological activity, our observations showing no differences between groups for skeletal muscle PTP1B protein abundance would suggest a negative study in this regard. There are many possible reasons proposed to explain these differences between gene expression and protein abundance. It is possible that mRNA expression does not always reflect changes in protein levels. Tian et al. (34) observed differential up or down expression of mRNA only reflected ∼40% of the variation of protein level. They concluded that the differences between mRNA and proteins could be attributable to the underlying biological mechanisms. Clearly, the specific mechanisms underlying this observation are not precisely known, but could involve many complicated and varied posttranscriptional mechanisms involved in turning mRNA into proteins that are not yet sufficiently well defined to be able to compute protein concentrations from mRNA. Other reasons may involve different in vivo half-lives for the proteins of interest as the result of varied protein synthesis and degradation. In this regard, Di Paola et al. (35) identified a 3′ untranslated region of the PTP1B gene, a 1484insG variation, that was associated with several features of insulin resistance. Specifically, they reported that subjects carrying the 1484insG variant showed PTP1B mRNA overexpression in skeletal muscle.
Although there are limited data as to what factors specifically regulate PTP1B abundance, our study demonstrated that when PTP1B was overexpressed in primary HSMC from a nondiabetic lean subject, insulin-stimulated phosphorylation of the insulin- signaling protein Akt was reduced. However, when PTP1B expression was reduced with PTP1B siRNA vector transfection, the insulin-induced phosphorylation of Akt was increased in primary HSMC from a subject with type 2 diabetes. Although not evaluated in this study, increased phosphorylation of Akt could potentially restore insulin sensitivity in subjects with type 2 diabetes. Akt is a Ser/Thr kinase downstream from the insulin receptor substrate in the insulin-signaling pathway that has been implicated as an effector of insulin-stimulated glucose metabolism (36–39). Our findings are supported by previous studies that have observed similar effects on insulin signaling proteins (i.e., Akt, insulin receptor, and insulin receptor substrate) when PTP1B was overexpressed (9,40,41) and inhibited (42–44).
In vitro and in vivo data suggest that an acquired dysmetabolic state of an individual could potentially drive PTP1B gene expression at the transcriptional level. For example, elevated free fatty acids and inflammatory markers are considered as part of the dysregulated metabolic state of obesity and insulin resistance. The addition of a specific free fatty acid (i.e., palmitate) along with inflammatory markers (i.e., macrophages, tumor necrosis factor-α, and IL-6) to cell cultures enhanced PTP1B gene expression in skeletal muscle cells and impaired insulin signaling (45,46). In addition, when the proinflammatory cytokine (i.e., tumor necrosis factor-α) was administered to high fat–fed mice, PTP1B mRNA was increased in the adipose tissue, liver, skeletal muscle, and hypothalamic arcuate (47). However, when the PTP1B gene was disrupted, insulin sensitivity increased, blood glucose and insulin concentrations decreased, and phosphorylation of skeletal muscle proteins in the insulin-signaling pathway increased (43). Additional studies are needed to determine the mechanisms for PTP1B gene expression in the skeletal muscle.
Even though this study is the first to provide findings regarding PTP1B gene expression and its relationship to insulin sensitivity in an African American population, we realize type 2 diabetes is a growing health problem that is not only restricted to the African American community. We studied African Americans, but the results could possibly be applicable to Caucasians and other underrepresented minority populations that have type 2 diabetes and similar BMI as the African Americans. Future research would need to be conducted. Also, we recognize that our study has several limitations. First, our study reported on the major cytosolic PTPase (i.e., PTP1B), and other PTPases (i.e., leukocyte common antigen-related PTPase, PTPε) were not evaluated. Secondly, based on a preclinical study (47), it would have been of interest to measure inflammatory markers to determine if circulating levels were related to gene or protein expression. These limitations provide guidance and direction for future studies.
In conclusion, this study reported that PTP1B gene expression in skeletal muscle is higher in African Americans with type 2 diabetes and negatively related to insulin sensitivity. In addition, with PTP1B transfection studies, we conclusively demonstrated that PTP1B modulates insulin signaling in primary HSMC from this well-characterized minority population. This study was specifically designed to observe the expression of PTP1B in an underrepresented population and the unknown questions about the differences in gene expression of PTPases between races and the mechanism by which a negative regulator of cellular insulin receptor signaling induces this clinical effect is the subject of ongoing investigation.
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health (NIH) Grants RO1 DK060126 (to W.T.C.) and T32 AT004094 (to A.J.S.). This project used Genomics Core facilities that are supported in part by the Center of Biomedical Research Excellence (NIH P20-RR021945) and a Nutrition Obesity Research Center Grant (NIH 1P30-DK072476) from NIH.
No other potential conflicts of interest relevant to this article were reported.
A.J.S. analyzed data and wrote manuscript. Z.Q.W., X.H.Z., and Y.Y. performed experiments and reviewed and edited the manuscript. W.D.J. analyzed data and reviewed and edited the manuscript. W.T.C. directed the study and reviewed and edited the manuscript. W.T.C. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Parts of this study were presented in abstract form at the 20th World Diabetes Congress (International Diabetes Federation), Montreal, Quebec, Canada, 18–22 October 2009 and at the 71st Scientific Sessions of the American Diabetes Association, San Diego, California, 24–28 June 2011.
REFERENCES
- 1.DeFronzo RA. Insulin resistance, hyperinsulinemia, and coronary artery disease: a complex metabolic web. J Cardiovasc Pharmacol 1992;20(Suppl. 11):S1–S16 [DOI] [PubMed] [Google Scholar]
- 2.Haffner SM. The insulin resistance syndrome revisited. Diabetes Care 1996;19:275–277 [DOI] [PubMed] [Google Scholar]
- 3.Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 1988;37:1595–1607 [DOI] [PubMed] [Google Scholar]
- 4.White MF, Kahn CR. The insulin signaling system. J Biol Chem 1994;269:1–4 [PubMed] [Google Scholar]
- 5.Ahmad F, Considine RV, Goldstein BJ. Increased abundance of the receptor-type protein-tyrosine phosphatase LAR accounts for the elevated insulin receptor dephosphorylating activity in adipose tissue of obese human subjects. J Clin Invest 1995;95:2806–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dadke SS, Li HC, Kusari AB, Begum N, Kusari J. Elevated expression and activity of protein-tyrosine phosphatase 1B in skeletal muscle of insulin-resistant type II diabetic Goto-Kakizaki rats. Biochem Biophys Res Commun 2000;274:583–589 [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Ouyang JP, Wu K, Wang SS, Wen CY, Xia ZY. Rosiglitazone ameliorates abnormal expression and activity of protein tyrosine phosphatase 1B in the skeletal muscle of fat-fed, streptozotocin-treated diabetic rats. Br J Pharmacol 2005;146:234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandyopadhyay D, Kusari A, Kenner KA, et al. Protein-tyrosine phosphatase 1B complexes with the insulin receptor in vivo and is tyrosine-phosphorylated in the presence of insulin. J Biol Chem 1997;272:1639–1645 [DOI] [PubMed] [Google Scholar]
- 9.Goldstein BJ, Bittner-Kowalczyk A, White MF, Harbeck M. Tyrosine dephosphorylation and deactivation of insulin receptor substrate-1 by protein-tyrosine phosphatase 1B. Possible facilitation by the formation of a ternary complex with the Grb2 adaptor protein. J Biol Chem 2000;275:4283–4289 [DOI] [PubMed] [Google Scholar]
- 10.Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell 2000;6:1401–1412 [DOI] [PubMed] [Google Scholar]
- 11.Xue B, Kim YB, Lee A, et al. Protein-tyrosine phosphatase 1B deficiency reduces insulin resistance and the diabetic phenotype in mice with polygenic insulin resistance. J Biol Chem 2007;282:23829–23840 [DOI] [PubMed] [Google Scholar]
- 12.Ahmad F, Azevedo JL, Cortright R, Dohm GL, Goldstein BJ. Alterations in skeletal muscle protein-tyrosine phosphatase activity and expression in insulin-resistant human obesity and diabetes. J Clin Invest 1997;100:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuire MC, Fields RM, Nyomba BL, et al. Abnormal regulation of protein tyrosine phosphatase activities in skeletal muscle of insulin-resistant humans. Diabetes 1991;40:939–942 [DOI] [PubMed] [Google Scholar]
- 14.Ahmad F, Goldstein BJ. Alterations in specific protein-tyrosine phosphatases accompany insulin resistance of streptozotocin diabetes. Am J Physiol 1995;268:E932–E940 [DOI] [PubMed] [Google Scholar]
- 15.Cheung A, Kusari J, Jansen D, Bandyopadhyay D, Kusari A, Bryer-Ash M. Marked impairment of protein tyrosine phosphatase 1B activity in adipose tissue of obese subjects with and without type 2 diabetes mellitus. J Lab Clin Med 1999;134:115–123 [DOI] [PubMed] [Google Scholar]
- 16.Kusari J, Kenner KA, Suh KI, Hill DE, Henry RR. Skeletal muscle protein tyrosine phosphatase activity and tyrosine phosphatase 1B protein content are associated with insulin action and resistance. J Clin Invest 1994;93:1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worm D, Vinten J, Staehr P, Henriksen JE, Handberg A, Beck-Nielsen H. Altered basal and insulin-stimulated phosphotyrosine phosphatase (PTPase) activity in skeletal muscle from NIDDM patients compared with control subjects. Diabetologia 1996;39:1208–1214 [DOI] [PubMed] [Google Scholar]
- 18.Bento JL, Palmer ND, Mychaleckyj JC, et al. Association of protein tyrosine phosphatase 1B gene polymorphisms with type 2 diabetes. Diabetes 2004;53:3007–3012 [DOI] [PubMed] [Google Scholar]
- 19.Palmer ND, Bento JL, Mychaleckyj JC, et al. Insulin Resistance Atherosclerosis Study (IRAS) Family Study Association of protein tyrosine phosphatase 1B gene polymorphisms with measures of glucose homeostasis in Hispanic Americans: the Insulin Resistance Atherosclerosis Study (IRAS) Family Study. Diabetes 2004;53:3013–3019 [DOI] [PubMed] [Google Scholar]
- 20.Spencer-Jones NJ, Wang X, Snieder H, Spector TD, Carter ND, O’Dell SD. Protein tyrosine phosphatase-1B gene PTPN1: selection of tagging single nucleotide polymorphisms and association with body fat, insulin sensitivity, and the metabolic syndrome in a normal female population. Diabetes 2005;54:3296–3304 [DOI] [PubMed] [Google Scholar]
- 21.Cheyssac C, Lecoeur C, Dechaume A, et al. Analysis of common PTPN1 gene variants in type 2 diabetes, obesity and associated phenotypes in the French population. BMC Med Genet 2006;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florez JC, Agapakis CM, Burtt NP, et al. Association testing of the protein tyrosine phosphatase 1B gene (PTPN1) with type 2 diabetes in 7,883 people. Diabetes 2005;54:1884–1891 [DOI] [PubMed] [Google Scholar]
- 23.Meshkani R, Taghikhani M, Al-Kateb H, et al. Polymorphisms within the protein tyrosine phosphatase 1B (PTPN1) gene promoter: functional characterization and association with type 2 diabetes and related metabolic traits. Clin Chem 2007;53:1585–1592 [DOI] [PubMed] [Google Scholar]
- 24.Traurig M, Hanson RL, Kobes S, Bogardus C, Baier LJ. Protein tyrosine phosphatase 1B is not a major susceptibility gene for type 2 diabetes mellitus or obesity among Pima Indians. Diabetologia 2007;50:985–989 [DOI] [PubMed] [Google Scholar]
- 25.Cowie CC, Rust KF, Byrd-Holt DD, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health And Nutrition Examination Survey 1999-2002. Diabetes Care 2006;29:1263–1268 [DOI] [PubMed] [Google Scholar]
- 26.Stull AJ, Galgani JE, Johnson WD, Cefalu WT. The contribution of race and diabetes status to metabolic flexibility in humans. Metabolism 2010;59:1358–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol 1979;237:E214–E223 [DOI] [PubMed] [Google Scholar]
- 28.Wang ZQ, Ribnicky D, Zhang XH, Raskin I, Yu Y, Cefalu WT. Bioactives of Artemisia dracunculus L enhance cellular insulin signaling in primary human skeletal muscle culture. Metabolism 2008;57(Suppl. 1):S58–S64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ZQ, Floyd ZE, Qin J, et al. Modulation of skeletal muscle insulin signaling with chronic caloric restriction in cynomolgus monkeys. Diabetes 2009;58:1488–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang ZQ, Zhang XH, Russell JC, Hulver M, Cefalu WT. Chromium picolinate enhances skeletal muscle cellular insulin signaling in vivo in obese, insulin-resistant JCR:LA-cp rats. J Nutr 2006;136:415–420 [DOI] [PubMed] [Google Scholar]
- 31.Costford SR, Bajpeyi S, Pasarica M, et al. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol Endocrinol Metab 2010;298:E117–E126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haugen F, Norheim F, Lian H, et al. IL-7 is expressed and secreted by human skeletal muscle cells. Am J Physiol Cell Physiol 2010;298:C807–C816 [DOI] [PubMed] [Google Scholar]
- 33.Baron AD, Brechtel G, Wallace P, Edelman SV. Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am J Physiol 1988;255:E769–E774 [DOI] [PubMed] [Google Scholar]
- 34.Tian Q, Stepaniants SB, Mao M, et al. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics 2004;3:960–969 [DOI] [PubMed] [Google Scholar]
- 35.Di Paola R, Frittitta L, Miscio G, et al. A variation in 3′ UTR of hPTP1B increases specific gene expression and associates with insulin resistance. Am J Hum Genet 2002;70:806–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong LN, Chen H, Li Y, et al. Physiological role of Akt in insulin-stimulated translocation of GLUT4 in transfected rat adipose cells. Mol Endocrinol 1997;11:1881–1890 [DOI] [PubMed] [Google Scholar]
- 37.Esposito DL, Li Y, Cama A, Quon MJ. Tyr(612) and Tyr(632) in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology 2001;142:2833–2840 [DOI] [PubMed] [Google Scholar]
- 38.Hirata AE, Alvarez-Rojas F, Carvalheira JB, Carvalho CR, Dolnikoff MS, Abdalla Saad MJ. Modulation of IR/PTP1B interaction and downstream signaling in insulin sensitive tissues of MSG-rats. Life Sci 2003;73:1369–1381 [DOI] [PubMed] [Google Scholar]
- 39.Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem 1996;271:31372–31378 [DOI] [PubMed] [Google Scholar]
- 40.Kenner KA, Anyanwu E, Olefsky JM, Kusari J. Protein-tyrosine phosphatase 1B is a negative regulator of insulin- and insulin-like growth factor-I-stimulated signaling. J Biol Chem 1996;271:19810–19816 [DOI] [PubMed] [Google Scholar]
- 41.Zabolotny JM, Haj FG, Kim YB, et al. Transgenic overexpression of protein-tyrosine phosphatase 1B in muscle causes insulin resistance, but overexpression with leukocyte antigen-related phosphatase does not additively impair insulin action. J Biol Chem 2004;279:24844–24851 [DOI] [PubMed] [Google Scholar]
- 42.Bakhtiyari S, Meshkani R, Taghikhani M, Larijani B, Adeli K. Protein tyrosine phosphatase-1B (PTP-1B) knockdown improves palmitate-induced insulin resistance in C2C12 skeletal muscle cells. Lipids 2010;45:237–244 [DOI] [PubMed] [Google Scholar]
- 43.Elchebly M, Payette P, Michaliszyn E, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999;283:1544–1548 [DOI] [PubMed] [Google Scholar]
- 44.Swarbrick MM, Havel PJ, Levin AA, et al. Inhibition of protein tyrosine phosphatase-1B with antisense oligonucleotides improves insulin sensitivity and increases adiponectin concentrations in monkeys. Endocrinology 2009;150:1670–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohammadtaghvaei N, Meshkani R, Taghikhani M, Larijani B, Adeli K. Palmitate enhances protein tyrosine phosphatase 1B (PTP1B) gene expression at transcriptional level in C2C12 skeletal muscle cells. Inflammation 2011;34:43–48 [DOI] [PubMed] [Google Scholar]
- 46.Parvaneh L, Meshkani R, Bakhtiyari S, et al. Palmitate and inflammatory state additively induce the expression of PTP1B in muscle cells. Biochem Biophys Res Commun 2010;396:467–471 [DOI] [PubMed] [Google Scholar]
- 47.Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J Biol Chem 2008;283:14230–14241 [DOI] [PMC free article] [PubMed] [Google Scholar]