Abstract
Common genetic risk variants for type 2 diabetes (T2D) have primarily been identified in populations of European and Asian ancestry. We tested whether the direction of association with 20 T2D risk variants generalizes across six major racial/ethnic groups in the U.S. as part of the Population Architecture using Genomics and Epidemiology Consortium (16,235 diabetes case and 46,122 control subjects of European American, African American, Hispanic, East Asian, American Indian, and Native Hawaiian ancestry). The percentage of positive (odds ratio [OR] >1 for putative risk allele) associations ranged from 69% in American Indians to 100% in European Americans. Of the nine variants where we observed significant heterogeneity of effect by racial/ethnic group (Pheterogeneity < 0.05), eight were positively associated with risk (OR >1) in at least five groups. The marked directional consistency of association observed for most genetic variants across populations implies a shared functional common variant in each region. Fine-mapping of all loci will be required to reveal markers of risk that are important within and across populations.
Over the past decade, genome-wide association studies (GWAS) and candidate gene association studies have been successful in identifying common risk variants for type 2 diabetes (T2D) (1–15). The loci revealed have provided insight into the genetic basis of this common disease, as well as biological pathways important in its pathogenesis. Most of these previously reported risk variants were identified in very large studies or meta-analyses conducted among populations of European and Asian ancestry and have been associated with modest increases in T2D risk (per-allele odds ratios [ORs] between 1.1 and 1.4) (12). Subsequent testing of these well-established variants in other racial and ethnic groups has been limited (12,16–24), and most of the studies have been undersized and underpowered to provide reliable risk estimates and clarity regarding generalizability of the associations in non-European populations. Aggregating results from multiple studies conducted among racially and ethnically diverse populations is one approach to amass an adequate sample size for replicating these modest genetic associations and extend our understanding of T2D genetics to non-European populations. As part of the Population Architecture using Genomics and Epidemiology (PAGE) Consortium, we have tested 20 validated risk variants for association with T2D. These 20 variants represent 18 risk regions and were examined in as many as 16,235 diabetes case and 46,122 control subjects from six major U.S. population groups (European Americans, African Americans, Hispanics, East Asians, Native Hawaiians, and American Indians) from six large population-based studies.
RESEARCH DESIGN AND METHODS
The PAGE Consortium consists of large ongoing population-based studies or consortia (25). The following studies are included in the current study: from the CALiCo (Causal Variants Across the Life Course) consortium, ARIC (the Atherosclerosis Risk in Communities Study) (26), CHS (Cardiovascular Health Study) (27), and SHS (Strong Heart Study) (28,29); EAGLE (Epidemiologic Architecture of Genes Linked to Environment, based on three National Health and Nutrition Examination Surveys [NHANES]) (30–33); MEC (The Multiethnic Cohort) (34); and WHI (Women’s Health Initiative) (35,36). Detailed information about each study can be found in Supplementary Data.
Diabetes case and control definitions.
To facilitate harmonization of diabetes case definitions across studies, data-collection methods were reviewed and compared between studies. All studies collected self-reported information on previous diagnosis by a physician or medical professional and use of medication for treatment of diabetes; however, not all studies measured fasting blood glucose levels, which more specifically define uncontrolled or undiagnosed T2D. In order to incorporate the T2D information across studies, two case definitions were allowed: self-report and exam based. To be classified as a case subject according to the self-report definition, participants had to report both a previous diagnosis of diabetes and use of medication to treat diabetes. To be classified as a control subject (self-report), participants had to report neither previous diagnosis nor use of diabetes medications. To be classified as a case subject according to the exam-based definition, participants had to either meet the self-report case definition or have a fasting (≥8 h) blood glucose ≥126 mg/dL. To be classified as a control subject (exam based), participants had to be classified as a control subject per the self-report definition and have a fasting blood glucose <126 mg/dL. Both prevalent and incident cases were included. For both definitions, case subjects with reported diabetes diagnosis before age 30 years were excluded. Sensitivity analyses in the ARIC study suggested that the magnitude of association between candidate variants and T2D did not differ systematically according to the case definitions we applied (Supplementary Data). Additional study-specific details on the data-collection methods and case definitions can be found in the Supplementary Data.
A total of 16,235 diabetes case and 46,122 control subjects were included in this study (case and control subjects, respectively, by study: ARIC, 1,348/10,978; CHS, 859/4,488; SHS, 1,575/1,249; MEC, 6,298/9,980; EAGLE/NHANES, 1,029/4,502; and WHI, 5,126/14,925). None of these studies was involved in the initial discovery efforts of these T2D risk loci. The data from the MEC have previously been reported (37).
Genotyping.
The 20 variants evaluated in the current study were selected from 18 genomic regions found to be significantly associated with risk of T2D in studies published as of September 2009 (Supplementary Table 1). In the CDKN2A/CDKN2B and KCNQ1 regions, more than one variant was investigated, as many of the index signals identified in the initial GWAS populations are not perfectly correlated. An additional variant, rs8050136, at the FTO locus, was also examined but not associated with risk in any population after adjustment for BMI (data not shown).
Genotyping was conducted in study-specific laboratories using a number of different platforms. Cross-laboratory and cross-platform reproducibility was assessed by genotyping 360 HapMap samples from populations most relevant to PAGE samples in each laboratory. A description of the platforms and quality-control metrics from each study/laboratory is provided in Supplementary Data. The genotype concordance for single nucleotide polymorphisms (SNPs) evaluated in the HapMap samples in more than one laboratory was >98.5% per SNP, with an average concordance of 99.8%.
We excluded results for SNP rs13266634 (SLC30A8) in all populations except European Americans and Hispanics, as there is an adjacent SNP 1 bp away (rs16889462) that has a frequency of 10% in African Americans, 4% in Asians, and 2% in Native Hawaiians (<1% in Hispanics and Europeans) and interferes with genotyping assays, thus resulting in genotype misclassification.
Genetic markers that distinguish the major ancestral populations (African, European, and Asian) were available in three studies. For ARIC, principal components of ancestry were derived from 200,000 SNPs genotyped on a custom array. For WHI (all populations) and MEC (African Americans and Native Hawaiians), ∼100 ancestry-informative markers were used in a principal-components analysis to assess major axes of variation (38,39). For a subset of the MEC Latinos, principal components were derived from markers on the Illumina 2.5M array. Genetic ancestry information was not available for the majority of the American Indian (SHS) or East Asian (MEC) samples or samples in EAGLE.
Statistical analysis.
β values and SEs for each variant were obtained by unconditional logistic regression or Cox proportional hazards regression. For each variant, the allele tested was the allele that was associated with increased risk in previous studies. In each study, models were run separately for each racial/ethnic population and adjusted for sex, age (continuous), and BMI (continuous). Approximately 13% of the WHI cohort was selected for inclusion in PAGE. This selection was nonrandom; therefore, analyses in WHI incorporated inverse probability weighting to account for sampling. For SHS, models were also run separately for each center.
Information on genetic ancestry was available for a large number of European Americans (∼64%), African Americans (∼85%), Hispanics (65%), and Native Hawaiians (∼83%). Results were similar after adjustment for population structure in all populations except for five SNPs in Native Hawaiians and four SNPs in Hispanics, where log ORs changed by >20% and P values changed by more than one order of magnitude in either direction (Supplementary Table 2). For each ethnic group, a pooled estimate was calculated using a fixed-effects model in which the effect measures were weighted by the inverse of the variance of the log OR. A combined estimate across ethnic groups was calculated using a random-effects model. We tested also for heterogeneity by study and by race using the Q statistic. For Native Hawaiians (MEC), we used the results adjusted for genetic ancestry. Similarly, for Latinos results are presented for MEC and WHI, as no ancestry information was available in EAGLE. All reported P values were derived from two-sided statistical tests. A P value <0.05 was used to declare an association as statistically significant. For each SNP in each racial/ethnic population, we estimated the statistical power to detect the previously reported relative risks in discovery populations of European or Asian ancestry (40) (Supplementary Table 1).
RESULTS
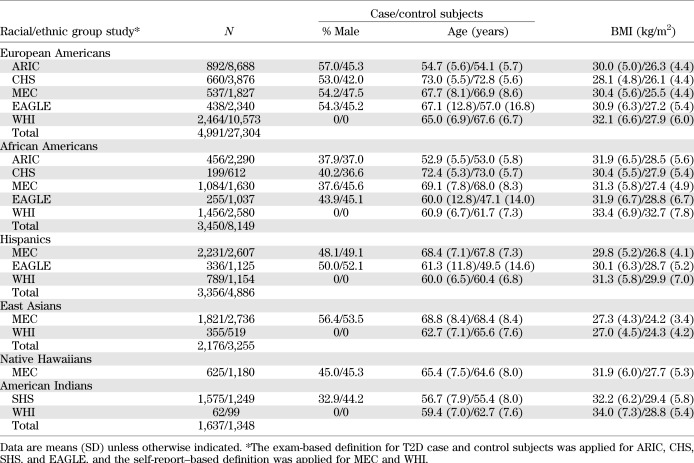
The descriptive characteristics of case and control subjects by racial/ethnic group and study are presented in Table 1. The mean age of case or control subjects ranged across studies from 47.1 (EAGLE, African American control subjects) to 73.0 (CHS, European American case subjects and African American control subjects). Both men and women were represented in each study except for WHI, which included only women. Case subjects were consistently heavier than control subjects in each study and population (Table 1).
TABLE 1.
Descriptive characteristics of diabetes case and control subjects in PAGE studies
We found no significant association with the first principal component (a measure of European admixture) and T2D risk in African Americans (in ARIC, MEC, or WHI). In Native Hawaiians, the first principal component is a measure of European admixture (and ancestry) and was significantly inversely associated with T2D risk (P = 3.2 × 10−8) (Supplementary Fig. 1). In Native Hawaiians, the significance of the association with three variants, which were all more common in Native Hawaiians than European Americans, diminished after adjustment for stratification (rs10010131, WFS1; rs7754840, CDKAL1; and rs864745, JAZF1). In contrast, the variants at TCF7L2 (rs7903146) and KCNQ1 (rs2237897) became nominally significant. The observation of larger β values for TCF7L2 and KCNQ1 variants after adjustment for stratification is consistent with negative confounding due to lower risk allele frequencies in Native Hawaiians compared with European Americans (Supplementary Table 1) and an inverse association of European ancestry and T2D risk in this population. Similarly, in Hispanics the first principal component, which is also a measure of European admixture (and ancestry) in this population, was significantly associated with lower T2D risk (P = 2.1 × 10−12 in the MEC) (Supplementary Fig. 2). Adjustment for the first principal component in Hispanics increased the OR and degree of statistical significance for three SNPs that were all less common, although marginally, in Hispanics than in European Americans (rs2237897, KCNQ1; rs4402960, IGF2BP2; and rs7903146, TCF7L2) and diminished significance for rs864745 (JAZF1), which is more common in Hispanics than in European Americans.
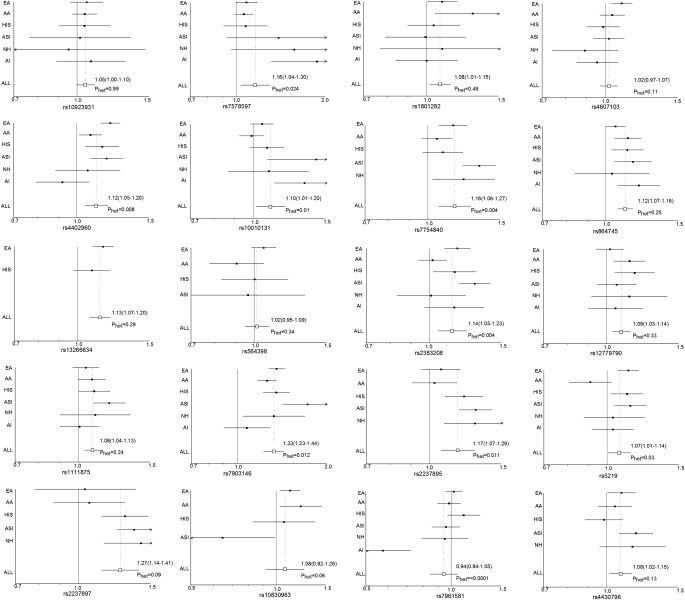
For the most part, the risk allele frequencies of each population tracked with the risk allele frequency of European Americans (Supplementary Fig. 3). Effect estimates were >1 for 69–100% of the SNPs across populations (average: 84%) (Fig. 1). Three variants were significantly associated (P < 0.05) with risk in at least four groups (rs4402960, IGF2BP2; rs864745, JAZF1; and rs7903146, TCF7L2), and of the 17 SNPs evaluated in five or more populations, positive associations were observed with 13 SNPs (OR >1) in at least five groups (Fig. 1). Of the 108 estimated effects (total number of tests: SNP × population), 91 had ORs >1 (84%). Removing European Americans, the population in which most of the original signals were reported, only reduced this percentage to 80%. We observed significant heterogeneity of effect by racial/ethnic group for nine SNPs (Pheterogeneity < 0.05). However, aside from rs7961581 at TSPAN8, eight of these variants (at THADA, IGF2BP2, WFS1, CDKAL1, CDKN2A/CDKN2B [rs2383208], TCF7L2, KCNQ1 [rs2237895], and KCNJ11) were positively associated with risk (OR >1) in at least five populations (Fig. 1). Thus, even for variants that displayed evidence of significant heterogeneity across population, the direction of effect was generally consistent in the majority of the populations.
FIG. 1.
Forest plots for each risk variant. Shown are the effect estimates (squares) and 95% CIs (bars) for each variant by population, as well as overall (hollow square). AA, African American; HIS, Hispanic; AI, American Indian; ALL, random-effects meta-analysis of all populations;ASI, East Asian; EA, European American; NH, Native Hawaiian; Phet, test for heterogeneity across populations.
DISCUSSION
We examined 20 validated risk variants for T2D, representing 18 risk regions, in as many as 16,235 diabetes case and 46,122 control subjects from six major population groups. The vast majority of the variants were positively associated with risk in the five non-European populations. These findings are highly consistent with a previous multiethnic study in the MEC, which contributed a large fraction of the case subjects to this meta-analysis (American Indians 0%, European Americans 11%, African Americans 31%, Hispanics 66%, East Asians 84%, and Native Hawaiians 100%) (37), and suggest that the majority of these variants are likely to be generalized markers of T2D risk across populations.
We did not find evidence of substantial confounding by population stratification in European Americans or African Americans. However, adjustment for population structure using principal components did affect the association with several variants for Native Hawaiians and Hispanics. Native Hawaiians are highly admixed with the three main groups being Polynesian, Asian, and European. The first few principal components capture European admixture, with European ancestry lower in Hawaiian case subjects than in control subjects (41). Therefore, adjustment for European admixture reduced the strength of association for some of the variants that were more common in Polynesians and increased the strength of some of the variants more common in Europeans. Similar differences were noted for some SNPs after principal-components adjustment in Hispanics. Unfortunately, ancestry-informative markers were not available to address the issue of population stratification in the admixed American Indian populations.
The marked directional consistency of association for most genetic variants across populations implies a shared functional common variant in each region. This general pattern of consistency provides little support for the “synthetic association” model (42), which suggests that GWAS signals with common alleles are due to rare alleles, many of which are likely to be ethnically distinct. The inability to replicate associations with variants in populations where statistical power is sufficient may highlight loci for which fine-mapping may be helpful. For example, in African Americans, power was high (≥94%) to detect significant associations, with the index variants at five loci (WFS1, HHEX, CDNK2A/B, THADA, and KCNQ1) that were found to be significantly associated with risk in at least one of the other non-European populations. The lack of a statistically significant association in African Americans at these loci could be because the risk allele is relatively invariant in populations of African ancestry or low linkage disequilibrium between the index signal and the functional allele. Fine-mapping of these loci, and others such as TCF7L2 in American Indians, where we observed no evidence of a significant association (OR 1.08 [95% CI 0.90–1.29]) despite >99% power and despite the suggestion that rs7903146 is the biologically functional variant in African Americans (43) and in genomic studies of open chromatin (44), should be of high priority to extract information about any genetic risk conferred at that locus that may be important for these populations.
This study has a number of limitations. In the design, we allowed for both incident and prevalent diabetes cases as well as different case/control criteria depending on study; however, our sensitivity analysis of the different case groups (Supplementary Data) did not suggest systematic differences in effect sizes based on study design, case definition, or analytic approach. We also had no information about type 1 diabetes in some studies, although case subjects known to be diagnosed before age 30 years were excluded and most participants in these studies were middle-aged or older adults.
This is the largest effort to date to investigate the generalizability of T2D susceptibility variants in the major racial/ethnic groups of the U.S. The consistent patterns of association for these variants provide additional support for the importance of these loci in contributing to T2D risk in multiple populations. Identification of the underlying biological functional allele(s) in each region, through fine-mapping, will be required to determine the extent to which these regions contribute to racial and ethnic disparities in T2D risk.
ACKNOWLEDGMENTS
The PAGE program is funded by the National Human Genome Research Institute, supported by U01HG004803 (CALiCo [Causal Variants Across the Life Course]), U01HG004798 (EAGLE [Epidemiologic Architecture of Genes Linked to Environment]), U01HG004802 (MEC [Multiethnic Cohort]), U01HG004790 (WHI [Women's Health Initiative]), and U01HG004801 (Coordinating Center).
No potential conflicts of interest relevant to this article were reported.
C.A.H. performed experiments, analyzed data, and wrote the manuscript. M.D.F., K.L.S., P.B., V.S.V., P.W., J.H., and N.F. performed experiments, analyzed data, and contributed to writing the manuscript. K.R.M., B.V.H., R.D.J., J.C.F., L.N.K., S.B., R.J.G., S.L., J.E.M., J.B.M., K.W., K.J.M., S.A.P., P.S., L.R.W., L.A.H., J.L.A., K.E.N., U.P., D.C.C., and L.L.M. contributed materials and to the study design, analysis tools, and interpretation of results and contributed to writing the manuscript. J.S.P. performed the experiments, analyzed data, and wrote the manuscript. C.A.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study-specific acknowledgments are listed in the Supplementary Data.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1296/-/DC1.
A complete list of PAGE members can be found at http://www.pagestudy.org.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 2000;26:76–80 [DOI] [PubMed] [Google Scholar]
- 2.Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 2003;52:568–572 [DOI] [PubMed] [Google Scholar]
- 3.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet 2007;39:977–983 [DOI] [PubMed] [Google Scholar]
- 5.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet 2009;41:1110–1115 [DOI] [PubMed] [Google Scholar]
- 6.Saxena R, Voight BF, Lyssenko V, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 7.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 9.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 2007;39:770–775 [DOI] [PubMed] [Google Scholar]
- 10.Tsai FJ, Yang CF, Chen CC, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 2010;6:e1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008;40:1098–1102 [DOI] [PubMed] [Google Scholar]
- 12.Voight BF, Scott LJ, Steinthorsdottir V, et al. MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 14.Zeggini E, Scott LJ, Saxena R, et al. Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeggini E, Weedon MN, Lindgren CM, et al. Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan G, Spurgeon CJ, Tabassum R, et al. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 Indians. Diabetes 2010;59:2068–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Luo Y, Ren Q, et al. Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet 2010;11:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman DM, Hunt KJ, Leach RJ, et al. Haplotypes of transcription factor 7-like 2 (TCF7L2) gene and its upstream region are associated with type 2 diabetes and age of onset in Mexican Americans. Diabetes 2007;56:389–393 [DOI] [PubMed] [Google Scholar]
- 19.Lewis JP, Palmer ND, Hicks PJ, et al. Association analysis in african americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes 2008;57:2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rong R, Hanson RL, Ortiz D, et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes 2009;58:478–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabara Y, Osawa H, Kawamoto R, et al. Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes 2009;58:493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi F, Serizawa M, Yamamoto K, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 2009;58:1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan JT, Ng DP, Nurbaya S, et al. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab 2010;95:390–397 [DOI] [PubMed] [Google Scholar]
- 24.Yan Y, North KE, Ballantyne CM, et al. Transcription factor 7-like 2 (TCF7L2) polymorphism and context-specific risk of type 2 diabetes in African American and Caucasian adults: the Atherosclerosis Risk in Communities study. Diabetes 2009;58:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matise TC, Ambite JL, Buyske S, et al. PAGE Study The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am J Epidemiol 2011;174:849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The ARIC investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol 1989;129:687–702 [PubMed] [Google Scholar]
- 27.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1991;1:263–276 [DOI] [PubMed] [Google Scholar]
- 28.Lee ET, Welty TK, Fabsitz R, et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol 1990;132:1141–1155 [DOI] [PubMed] [Google Scholar]
- 29.North KE, Howard BV, Welty TK, et al. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the strong heart family study. Am J Epidemiol 2003;157:303–314 [DOI] [PubMed] [Google Scholar]
- 30.Chang MH, Lindegren ML, Butler MA, et al. CDC/NCI NHANES III Genomics Working Group Prevalence in the United States of selected candidate gene variants: Third National Health and Nutrition Examination Survey, 1991-1994. Am J Epidemiol 2009;169:54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention. Plan and Operation of the Third National Health and Nutrition Examination Survey, 1988–94. Bethesda, MD, 2004
- 32.Centers for Disease Control and Prevention (CDC) NCfHSN. U.S. Department of Health and Human Services, Hyattsville, MD, 2002
- 33.Steinberg KK, Sanderlin KC, Ou CY, Hannon WH, McQuillan GM, Sampson EJ. DNA banking in epidemiologic studies. Epidemiol Rev 1997;19:156–162 [DOI] [PubMed] [Google Scholar]
- 34.Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol 2000;151:346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Women’s Health Initiative Study Group Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials 1998;19:61–109 [DOI] [PubMed] [Google Scholar]
- 36.Anderson GL, Manson J, Wallace R, et al. Implementation of the Women’s Health Initiative study design. Ann Epidemiol 2003;13(Suppl):S5–S17 [DOI] [PubMed] [Google Scholar]
- 37.Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in europeans in diverse racial and ethnic groups. PLoS Genet 2010;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 2009;30:69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 40.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol 2002;155:478–484 [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Haiman CA, Kolonel LN, et al. Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet 2010;128:165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer ND, Hester JM, An SS, et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes 2011;60:662–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet 2010;42:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]