Abstract
We report the development of the multiplexed nano-flare, a nanoparticle agent that is capable of simultaneously detecting two distinct messenger RNA (mRNA) targets inside a living cell. These probes consist of polyvalent DNA-functionalized gold nanoparticles with multiple DNA sequences, each hybridized to a reporter with a distinct fluorophore label, and each complementary to its corresponding mRNA target. When multiplexed nano-flares are exposed to their targets, they provide a sequence specific signal in both extra- and intracellular environments. Importantly, one of the targets can be used as an internal control, improving detection by accounting for cell-to-cell variations in nanoparticle uptake and background. Compared to single-component nano-flares, these structures allow one to determine more precisely relative mRNA levels in individual cells, improving cell sorting and quantification.
Introduction
The study of messenger RNA (mRNA) is critical for understanding basic biology and identifying therapeutic and diagnostic targets.1 Recently, researchers have determined that many important biological processes rely not just on bulk mRNA expression, but are also highly dependent on cell-to-cell variations in mRNA.2 This is a major challenge in the treatment of heterogeneous diseases like cancer, where a small percentage of cells in a tumor mass can cause major health consequences, such as drug resistance.2 Unfortunately, the most commonly used methods of mRNA detection, such as real-time polymerase chain reaction (RT-PCR)3; 4 and microarray analysis,5; 6; 7 while accurate in detecting relative mRNA expression in bulk samples, are incapable of detecting cell-to-cell variations because they require pooling mRNA from lysates acquired from groups of cells. Therefore, a major challenge exists to accurately detect relative mRNA levels in individual live cells. Alternative approaches for endogenous mRNA analysis, such as in situ hybridization8; 9 and molecular beacons,10; 11; 12; 13; 14; 15 have been used for mRNA measurements in both live and fixed cells. However, these approaches are limited by insufficient cell delivery,16 instability in biological environments,17; 18 high toxicity,19 and immunogenicity.20; 21
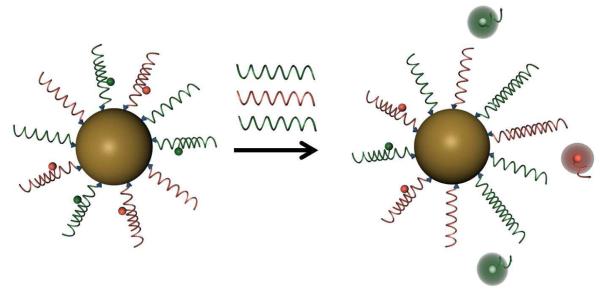
One approach to meeting these challenges utilizes a new class of probe, termed the nano-flare, consisting of a gold nanoparticle (Au NP) core, densely functionalized with duplex oligonucleotides (Figure 1).22; 23 The oligonucleotide capture sequence attached to the nanoparticle hybridizes with short, fluorophore-labeled DNA molecules, termed “flares”. In this conformation, the close proximity of the fluorophore to the Au NP surface leads to quenching of the fluorescence.24 However, when a target mRNA binds to the capture sequence, the concomitant displacement of the flare can be detected as a corresponding increase in fluorescence.25
Figure 1.
Schematic representation of target detection by multiplexed nano-flares. The multiplexed nano-flares bind different target nucleic acids (shown in red and green), displacing the corresponding flare. Once the flare is released, the fluorophore is no longer quenched by the gold surface, and an increase in fluorescence can be measured. By using two different fluorophores, the ratio of each target can be determined in cells.
Nano-flares have several important properties that make them excellent intracellular mRNA detection agents. First, their dense monolayer of oligonucleotides interacts with cell membrane receptors in a unique manner, eliciting high uptake into diverse cell types without the use of transfection agents.26; 27; 28 Second, a high local salt concentration around the nanoparticle inhibits the enzymatic activity of some nucleases, resulting in high stability in biological environments.29; 30 Third, polyvalent oligonucleotide functionalized Au NP conjugates have no known off-target effects, no observable toxicity, and low immunogenicity.20; 31 While nano-flares have previously been shown to be useful for detecting genetic targets, quantifying relative concentrations of these targets has been limited by utilizing only one fluorescent output. With such a single signal method, any mRNA-independent variation in fluorescence between cells is irresolvable. For instance, heterogeneous rates of nanoparticle uptake may result in a varied cell-to-cell fluorescence response. However, the polyvalency of the oligonucleotides on the Au NP scaffold provides a potential solution, since multiple sequences can be included on a single nanoparticle (Figure 1). By developing a nano-flare that targets a second gene along with the mRNA of interest (as is commonly done in RT-PCR), an internal control is included on each particle. Additionally, this work shows that we can simultaneously target multiple sequences via a single structure. Since many diseases and biological processes involve changes in the expression of multiple genes, such a multiplexed assay can provide important information on genetic interactions in these complex systems. Herein, we describe the synthesis of multiplexed nano-flares capable of simultaneously detecting two mRNA targets, thus demonstrating a method for detecting changes of mRNA levels in living cells.
Materials and Methods
Synthesis of materials
Oligonucleotides were synthesized using standard solid-phase phosphoramidite chemistry (Expedite 8909 Nucleotide Synthesis System (ABI)). All reagents were purchased from Glen Research and the oligonucleotides were purified by reverse-phase high performance liquid chromatography (HPLC). The oligonucleotide sequences used in this study are shown below.
Survivin Target: 5′-CAA GGA GCT GGA AGG CTG GG-3′
Survivin Thiol: 5′-CCC AGC CTT CCA GCT CCT TG-(A)5-propylthiol-3′
Survivin Cy5 flare: 5′-(Cy5)-TCA AGG AGC TGG-3′
Survivin Cy3 flare: 5′-(Cy3)-TCA AGG AGC TGG-3′
Actin Target: 5′-GGT CGC AAT GGA AGA AG-3′
Actin Thiol: 5′-CTT CTT CCA TTG CGA CC-(A)5-propylthiol-3′
Actin Cy5 flare: 5′-(Cy5)-TGG TCG CAA TGG-3′
Actin Cy3 flare: 5′-(Cy3)- TGG TCG CAA TGG -3′
To make DNA-functionalized nanoparticles, the thiolated actin and survivin oligonucleotides (1 μM each) were combined with citrate-capped 13 nm gold particles (10 nM) and incubated for 2 hours at room temperature. Next, sodium dodecylsulfate (SDS), phosphate buffer (pH = 7.4), and sodium chloride were added to a final concentration of 0.01%, 5 mM, and 50 mM, respectively. After a 2 hour incubation period, sodium chloride was added to achieve a final concentration of 150 mM, and the particles were stored for 12 hours. Finally, the conjugates were purified into PBS by centrifugation.
The flares were hybridized on the purified DNA-Au NPs (100 nM) in PBS. Cy3 (0.5 μM) and Cy5 (0.5 μM) flares were added at the same time and the solution was heated to 70 °C and slowly cooled to room temperature over 4 h to allow hybridization. The resulting nano-flares were then sterilized using a 0.2 m acetate syringe filter (GE Healthcare) to prevent cell contamination and stored at 4 °C.
Extracellular characterization
Extracellular fluorescence experiments were performed using a Jobin Yvon Fluorolog FL3-22. Each reaction contained 1 mL of PBS with nano-flares (500 pM) and the specified target (100 nM). Cy5 and Cy3 were excited at 630 and 530 nm, respectively. The oligonucleotide target was incubated with nano-flares for 10 minutes before the fluorescence spectrum was recorded.
Cell culture and siRNA treatment
Three human cancer cell lines (HeLa, Jurkat, and MCF-7) were used to test the multiplexed nano-flare. Cells were cultured according to American Tissue Culture Collection (ATCC) recommendations.
To modulate survivin and actin expression levels, siRNA treatment was used. All siRNA was purchased from Santa Cruz Biotechnology. In a typical experiment, cells were grown in a 96-well plate to 80% confluency (~2×104 cells per well), then transfected with either control nonsense siRNA, actin-targeted siRNA, or survivin-targeted siRNA. The transfection was performed using the commercial transfection agent DharmaFECT 1 according to the manufacturer’s recommendations. 0, 10, 20, 50, and 100 nM siRNA was used, as specified above. The cells were then allowed to incubate with the siRNA for 48 hours in Opti-MEM minimal media, after which they were rinsed with PBS pH 7.4, and transfected with multiplexed nano-flares.
Flow cytometry
For flow cytometry analysis, cells were treated with nano-flares for 24 hours (1 nM for Jurkat cells, 0.33 nM for HeLa and MCF-7 cells). After this, cells were prepared by rinsing with PBS, trypsinization, and resuspension in PBS. Cells were analyzed using a Guava® easyCyte™ 8HT Flow Cytometry System (Millipore). Fluorescence channels “Yellow” and “Red2” were used for Cy3 and Cy5 respectively and gated according to the manufacturer’s recommendations. To determine survivin or actin signal from the Cy3 and Cy5 intensity values, first the background was subtracted for each channel (background was measured using cells treated with control DNA-modified Au NPs that lack fluorophores). Next, the ratio of the two intensity values was calculated for individual cells, or for the median of a cell population. Error bars represent the standard error of the mean (SEM) of the median signal, determined from 3 or more independent experiments.
Fluorescence microscopy
Cells were prepared for microscopy the same way they were prepared for flow cytometry except that the nano-flare concentration was 2 nM. The cells were visualized with a Zeiss 510 LSM using HeNe laser excitation at wavelengths of 543 nm (Cy3) and 633 nm (Cy5).
RT-PCR
Cells were treated with anti-survivin and control siRNA (10, 20, 50, and 100 nM), as described above, for 48 hours. The cells were subsequently washed and incubated for an additional 24 hours before being harvested. The cells were harvested, and total RNA was extracted using phenol, guanidine isothiocynate, and chloroform (TRIzol reagent, Invitrogen) followed by treatment with DNAse according to the manufacturer’s protocol (Invitrogen). This procedure is commonly used in antisense experiments because it disrupts the antisense oligonucleotide—RNA duplex, allowing purification of mRNA and preventing interference by the antisense oligonucleotides at later stages in the RNA quantification. 4 g of RNA was reverse transcribed using the Superscript III kit (Invitrogen). Quantitative real-time PCR was performed on the cDNA following the LightCycler 480 SYBR Green 1 Master Mix protocol (Roche) on the LightCycler Real-Time 480 II system. The relative abundance of each survivin mRNA transcript was normalized to actin expression and the relative abundance of each actin mRNA transcript was normalized to survivin expression. Both of these values were compared to untreated cells to determine the increased/decreased expression. The standard deviation was calculated from at least 3 independent experiments. The primers used for the RT-PCR experiment were:
Actin forward, 5′- GCTTCTAGGCGGACTGTGAC-3′;
Actin reverse, 5′- AAAGCCATGCCAATCTCATC-3′;
Survivin forward, 5′- TGCAACCGCCTAGACTTTCT-3′;
Survivin reverse 5′- AACCCTTCCCAGACTCCACT-3′.
Results and Discussions
Multiplexed nano-flares were synthesized starting with citrate-capped 13 nm Au NPs and alkylthiol-modified DNA capture strands capable of binding target mRNA. Survivin was chosen as a target gene in this study, due to its relevance in cancer biology.32 Actin was used as a second target due to its ubiquitous expression across different cell types and its common use as a control gene in conventional molecular biology assays.33; 34 Equal concentrations of Actin and survivin targeted DNA strands were immobilized as a dense (25pmol/cm2) mixed-monolayer on the Au NP surface.35 The single-stranded DNA-Au NP conjugates were then purified by centrifugation, and flares were hybridized to prepare the nano-flare structure. Different fluorophore labels (survivin-Cy3, actin-Cy5) were used for actin and survivin flares to allow independent detection of each target.
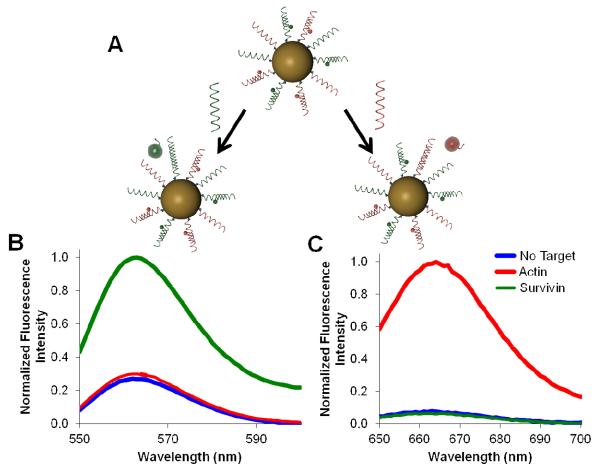
In order to investigate the behavior of the multiplexed nano-flares in a cell free system, the fluorescence signals of the flares were monitored in vitro before and after the addition of synthetic targets (Figure 2). When the actin target was added, fluorescence increased at wavelengths corresponding to the actin flare (Cy5 - 670 nm) but not the survivin flare (Cy3 - 570 nm) (Figure 2b). The reverse of this effect was observed when the survivin target was added (Figure 2c). To confirm that the flare release was sequence specific and detection was fluorophore independent, a new set of nano-flares was synthesized with the fluorophore labels switched (actin-Cy3, survivin-Cy5) and similar results were obtained (Figure S-1). Together, these results demonstrate that multiplexed nano-flares are capable of responding to actin and survivin targets independently and in a sequence specific manner in buffer conditions.
Figure 2.
Sequence specific target detection in extracellular experiments. (A) A scheme of sequence specific flare release in response to two different targets (green shown on the left and red shown on the right). (B, C) Fluorescence spectra of nano-flares in the presence of no target, actin target or survivin target. Actin and survivin flares are labeled with Cy5 and Cy3, respectively. (B) Normalized fluorescence spectra corresponding to Cy3 (survivin flare) signal, with excitation at 530 nm. (C) Normalized fluorescence spectra corresponding to Cy5 (actin flare) signal, with excitation at 630 nm.
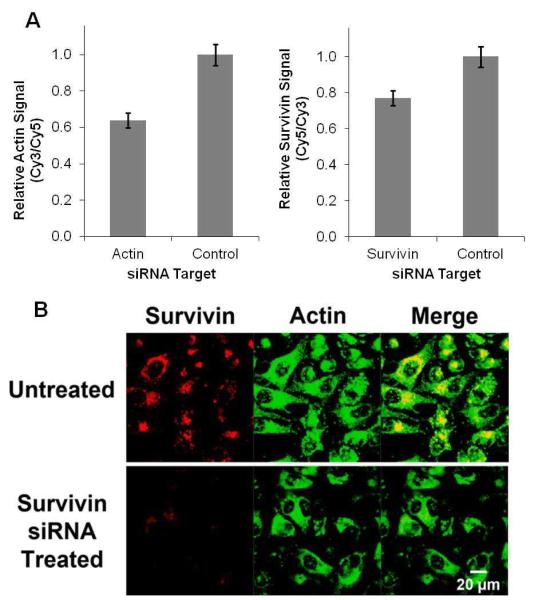
The ability of multiplexed nano-flares to independently detect two targets was then studied in cells. First, the relative levels of survivin and actin expression in HeLa cells were modulated by treatment with siRNA against survivin or actin using DharmaFECT 1, a lipid transfection system. Nonsense siRNA with no target in the human genome was used as a negative control. Next, multiplexed nano-flares were incubated with cells for 24 hours, and intracellular fluorescence was monitored by flow cytometry (Figure 3A). Treatment with survivin siRNA caused a decrease in survivin flare signal. Likewise, cells treated with actin siRNA showed a decrease in actin flare signal, indicating that the multiplexed nano-flare can be used to detect intracellular mRNA levels for two different genes. As in the extracellular experiments, similar intracellular results were obtained when the fluorophore label on each oligonucleotide flare was switched, confirming that the fluorescence signals are fluorophore-independent and flare release is sequence specific (Figure S-2). These experiments were repeated in three different human cell lines (HeLa, Jurkat, and MCF-7) to demonstrate the applicability of the multiplexed nano-flare in a variety of cell types (Figure S-3). When cells are treated with survivin siRNA, the fluorescence intensity from the survivin channel decreases while the fluorescence intensity from the actin channel is largely unaffected. Qualitative analysis was conducted using fluorescence microscopy, and again selective release of the appropriate fluorophore was observed (Figure 3B, S-5). These experiments show the utility of the multiplexed nano-flare for the detection of multiple mRNA targets inside living cells.
Figure 3.
Detection of actin and survivin mRNA expression in cells. Survivin-Cy5 and Actin-Cy3 flares were used. (A) In order to change the relative levels of survivin and actin mRNA the cells were treated with either actin or survivin targeted siRNA. Nonsense siRNA with no target in the cells was used as a negative control. The cell associated fluorescence intensities corresponding to each flare were measured by flow cytometry. (B) In a similar experiment, survivin levels were reduced using siRNA and the change in fluorescence from the multiplexed nano-flares was measured and assessed qualitatively by fluorescence confocal microscopy.
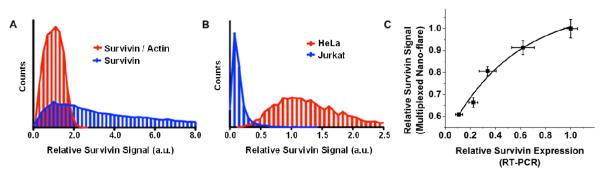
Within a given population of cells, there is observable variation in the rate of nanoparticle uptake and nano-flare degradation, which may change the cell associated fluorescence independent of mRNA levels. This results in a relatively broad distribution of cell-associated fluorescence values, making relative target quantification difficult. We hypothesized that the multiplexed nano-flare could solve this problem by using a second flare as an internal control. To investigate the cell-to-cell variation in survivin mRNA, signal from multiplexed nano-flares was compared to the traditional nano-flare design.22 First, cell-associated fluorescence was measured using traditional mono-functionalized Cy5-survivin nano-flares, and plotted as a histogram with a relatively broad distribution of cell signals (blue trace, Figure 4A). Next, using the multiplexed nano-flare, the survivin mRNA signal was measured as the ratio of Cy5-survivin to Cy3-actin flare associated fluorescence by using actin as a cell-by-cell internal control for target-independent variations in fluorescence readings. This ratiometric approach resulted in a 10-fold reduction in the standard deviation of the survivin fluorescence signal when survivin signal was normalized to the actin fluorescence signal.
Figure 4.
Quantitative and qualitative determination of mRNA expression in living cells. (A) The detection of intracellular survivin mRNA by traditional nano-flares was compared to detection by multiplexed nano-flares, expressed as the ratio of survivin to actin. The cell associated fluorescence intensities were measured in HeLa cells using flow cytometry and expressed as a histogram. (B) In a similar experiment, the multiplexed nano-flares were used to compare the ratio of survivin to actin expression in HeLa and Jurkat cells. (C) The expression of survivin mRNA was measured as a function of survivin siRNA delivered by both the multiplexed nano-flare and RT-PCR. Error bars represent standard error of the mean from at least 3 independent experiments.
Given the narrow signal distribution of the multiplexed nano-flare, we hypothesized that these probes could be used to distinguish two different cell lines, which is a major goal in the field of live cell analysis. Indeed, the differentiation of cancerous cells from non-cancerous cells is crucial for the early diagnosis of cancer. Model cell lines were chosen for this experiment, one with relatively high levels of survivin mRNA (HeLa) and another with relatively low levels (Jurkat) (Figure S-4). Both cell lines were treated with nano-flares at the same concentration and analyzed by flow cytometry (Figure 4B). The multiplexed nano-flares gave 10-fold greater survivin signal in HeLa cells compared to Jurkat cell, allowing for differentiation between these two lines on the basis of gene expression.
In addition to the differentiation of cell types, we hypothesized that the improved detection capability of multiplexed nano-flares could be used to quantify mRNA levels. To test this hypothesis, the survivin mRNA concentration in HeLa cells was modulated by treatment with different concentrations of siRNA and measured using flow cytometry analysis of multiplexed nano-flare fluorescence (Figure 4C). RT-PCR was also used to measure mRNA levels, providing a quantitative standard for the multiplexed nano-flare signal. As the siRNA concentration increased, the survivin signal decreased in both the RT-PCR and the multiplexed nano-flare assays. Based on these results, the relationship between multiplexed nano-flare signal and mRNA levels as measured by RT-PCR could be quantitatively determined (R-squared = .96) for a given cell type (Figure 4C).
Conclusions
The ability to measure relative mRNA levels within live cells is critical for furthering our understanding of fundamental biology and disease. Nano-flares are capable of mRNA detection in living cells, and the development of the multiplexed nano-flare has added the ability to detect two targets at once. Importantly, we have shown that one of the probes can act as an internal control for a cell-by-cell basis by comparing a gene of interest to a second gene. This structure and approach take into account the influence of cell-to-cell variations, such as differences in nanoparticle uptake, allowing the identification of cells based upon mRNA expression. This work significantly extends the scope of the nano-flare platform by achieving three major goals in the field of live cell detection: it allows for the detection of multiple targets in a single experiment, the potential for separation of cells types based upon internal targets, and the relative quantification of mRNA levels within live cells.
Supplementary Material
Acknowledgment
C.A.M. acknowledges a Cancer Center for Nanotechnology Excellence (NCI-CCNE) award for support of this research.
References
- (1).Tyagi S. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- (2).Visvader JE. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- (3).Nolan T, Hands RE, Bustin SA. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- (4).VanGuilder HD, Vrana KE, Freeman WM. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- (5).Brown PO, Botstein D. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- (6).Couzin J. Science. 2006;313:1559–1559. doi: 10.1126/science.313.5793.1559a. [DOI] [PubMed] [Google Scholar]
- (7).Chen JJ. Pharmacogenomics. 2007;8:473–482. doi: 10.2217/14622416.8.5.473. [DOI] [PubMed] [Google Scholar]
- (8).Femino A, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- (9).Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- (11).Liu XJ, Farmerie W, Schuster S, Tan WH. Anal Biochem. 2000;283:56–63. doi: 10.1006/abio.2000.4656. [DOI] [PubMed] [Google Scholar]
- (12).Dubertret B, Calame M, Libchaber AJ. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- (13).Maxwell DJ, Taylor JR, Nie SM. J Am Chem Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- (14).Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, Wood WC, Yang L. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- (15).Santangelo P, Nitin N, Bao G. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- (16).Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Nucleic Acids Research. 2004;32:e58–e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Molenaar C, Marras SA, Slats JCM, Truffert JC, Lemaître M, Raap AK, Dirks RW, Tanke HJ. Nucleic Acids Research. 2001;29:e89–e89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wang L, Yang CJ, Medley CD, Benner SA, Tan W. J Am Chem Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- (19).Zhang J-S, Liu F, Huang L. Advanced Drug Delivery Reviews. 2005;57:689–698. doi: 10.1016/j.addr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- (20).Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Mol Pharmaceut. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Scheule RK, St George JA, Bagley RG, Marshall J, Kaplan JM, Akita GY, Wang KX, Lee ER, Harris DJ, Jiang C, Yew NS, Smith AE, Cheng SH. Hum Gene Ther. 1997;8:689–707. doi: 10.1089/hum.1997.8.6-689. [DOI] [PubMed] [Google Scholar]
- (22).Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Acs Nano. 2009;3:2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dubertret B, Calame M, Libchaber AJ. Nat Biotech. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- (25).Prigodich AE, Lee OS, Daniel WL, Seferos DS, Schatz GC, Mirkin CA. J Am Chem Soc. 2010;132:10638–10641. doi: 10.1021/ja104859j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- (27).Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Letters. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Letters. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Prigodich AE, Alhasan AH, Mirkin CA. J Am Chem Soc. 2011;133:2120–2123. doi: 10.1021/ja110833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Acs Nano. 2010;4:5641–5646. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ambrosini G, Adida C, Altieri DC. Nature Medicine. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- (33).Torres VA, Tapia JC, Rodriguez DA, Parraga M, Lisboa P, Montoya M, Leyton L, Quest AF. J Cell Sci. 2006;119:1812–1823. doi: 10.1242/jcs.02894. [DOI] [PubMed] [Google Scholar]
- (34).Romagnoli M, Trichet V, David C, Clement M, Moreau P, Bataille R, Barille-Nion S. Leukemia. 2007;21:1070–1078. doi: 10.1038/sj.leu.2404602. [DOI] [PubMed] [Google Scholar]
- (35).Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.