Abstract
Rationale
Cardiac progenitor cells (CPCs) in the adult heart are used for cell-based treatment of myocardial damage but factors determining stemness, self-renewal and lineage commitment are poorly understood. Immortal DNA strands inherited through asymmetric chromatid segregation correlate with self-renewal of adult stem cells, but CPCs capacity for asymmetric segregation to retain immortal strands is unknown. Cardioprotective kinase Pim-1 increases asymmetric cell division in vivo but the ability of Pim-1 to enhance asymmetric chromatid segregation is unknown.
Objective
Demonstrate immortal strand segregation in CPCs and the enhancement of asymmetric chromatid distribution by Pim-1 kinase.
Methods and Results
Asymmetric segregation is tracked by incorporation of bromodeoxyuridine (BrdU). CPC DNA was labeled for several generations and then blocked in second cytokinesis during chase to determine distribution of immortal versus newly synthesized strands. Binucleated CPCs with BrdU intensity ratio of 70:30 or more between daughter nuclei indicative of asymmetric chromatid segregation occur with a frequency of 4.57% and asymmetric chromatid segregation is demonstrated at late mitotic phases. Asymmetric chromatid segregation is significantly enhanced by Pim-1 overexpression in CPCs (9.19% vs 4.79% in eGFP expressing cells, p=0.006).
Conclusions
Asymmetric segregation of chromatids in CPCs is increased nearly twofold Pim-1 kinase overexpression, indicating Pim-1 promotes self renewal of stem cells.
Keywords: immortal DNA strand hypothesis, asymmetric chromatid segregation, asymmetric cell division, BrdU, cytochalasin B
Introduction
The immortal strand hypothesis states that adult stem cells selectively retain chromatids with old DNA strands (the mother strand) whereas all differentiating progeny acquire newly synthesized daughter to protect stem cells from DNA replication errors1 and aging2. The silent sister hypothesis presumes template strand segregation of all or some chromatids to regulate gene expression to determine cell fate3, 4. The immortal strand and silent sister hypotheses are supported by observations of absolute to partial asymmetric chromatid segregation (ACS) in adult stem cells like mouse intestinal and colon epithelial stem cells5, 6, neural stem cells7, satellite cells8, 9 and many cancer stem cells10, 11. Stem cells may also undergo symmetric segregation on an intermittent basis and switch back to asymmetric segregation7, 8, 10, 11. The silent sister hypothesis is also supported by the correlation of ACS with asymmetric cell division (ACD), self-renewal and fate determination7–10. Discovery of resident adult stem cells specific to the heart termed cardiac progenitor cells (CPCs)12 has opened new possibilities for cell-based treatment of heart disease which could be mitigated by improved myocardial regeneration13. CPCs found in cardiac-niches are c-kit positive, clonogenic, multipotent, self-renewing, and capable of asymmetric division12, 14. Results of a phase I clinical trial with CPCs are promising15, but clinical potential of CPCs is limited by donor age and disease state16, 17. CPCs in aged myocardium are senescent with limited replication potential17. Therefore, a subset of CPCs capable of ACS would be desirable to preserve youthful self-renewal of the stem cell phenotype. ACS in CPCs is enhanced two fold in our study by overexpression of Pim-1, a kinase that enhances cardiac repair18, increases asymmetric cell division19, and promotes CPC self-renewal.
Methods
Cell culture, treatments, BrdU immuno-staining and image quantification are explained in online supplementary methods. Experimental design for label release and retention assay are explained below and depicted in Online Figure I.
Label Release Assay
‘Immortal’ strands are never labeled as per Carins initial postulate1. Short time BrdU labeling for a single S phase results in CPCs with one labeled and one unlabeled DNA strand (Online Figure IA). After completion of mitosis when cells are chased in BrdU free media, chromatids with unlabeled stands segregate from chromatids with newly made BrdU-labeled strands. Therefore stem cells release the label due to retention of the oldest immortal strands not being labeled throughout the assay7, 11.
Label Retention Assay
Long term BrdU exposure of CPCs allows for labeling of both DNA strands (Online Figure IB) assuming switching occurs between symmetric and asymmetric segregation as previously published6–8, 10, 11. Following completion of a single mitotic division, chasing cells in BrdU free medium results in CPCs with one labeled and one unlabeled strand in a chromatid. Following a second round of mitosis, labeled immortal strands will segregate from the unlabeled new strands, therefore label retention will occur for CPCs continuing to asymmetrically segregate their chromatids6, 7, 10.
Results
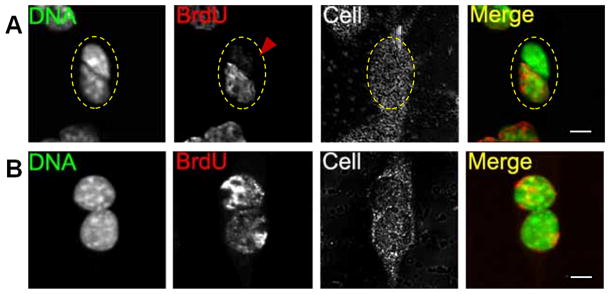
Cell cycle distribution of CPCs after drug-induced mitotic synchronization of the cell population is shown in Online Figure II. Use of a label release assay (Online Figure IA) was devised to demonstrate presence of unlabeled ‘immortal stands’. Asymmetric segregation of unlabeled immortal strands from BrdU labeled newly synthesized strands is revealed by rare binucleated CPCs with one daughter nucleus having little or no BrdU staining while the other nucleus is positive for BrdU (Fig. 1A). CPCs exhibiting daughter nuclei with equal BrdU intensity indicate symmetric segregation (Fig. 1B).
Figure 1. CPCs retain unlabeled immortal strands in label release assay.
A, Binucleated cell with one daughter nuclei exhibiting low BrdU immuno-staining (red arrowhead) and another showing intense BrdU signal. B, Binucleated cell with daughter nuclei exhibiting symmetric BrdU immunostaining. Binucleated CPCs are confirmed by reflection scanning (Cell). Scale bar = 10 μm.
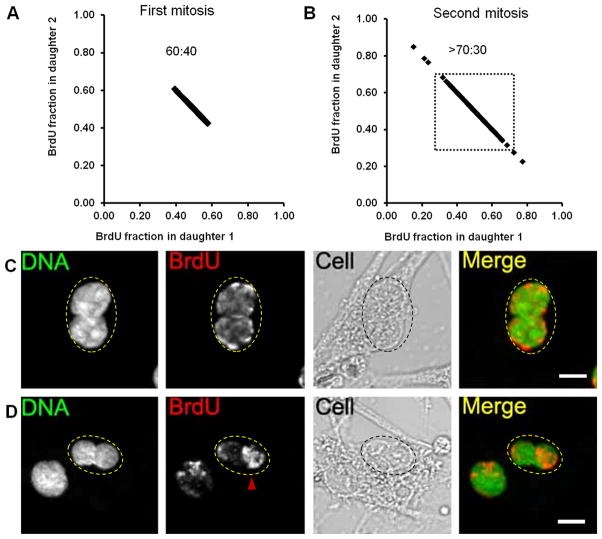
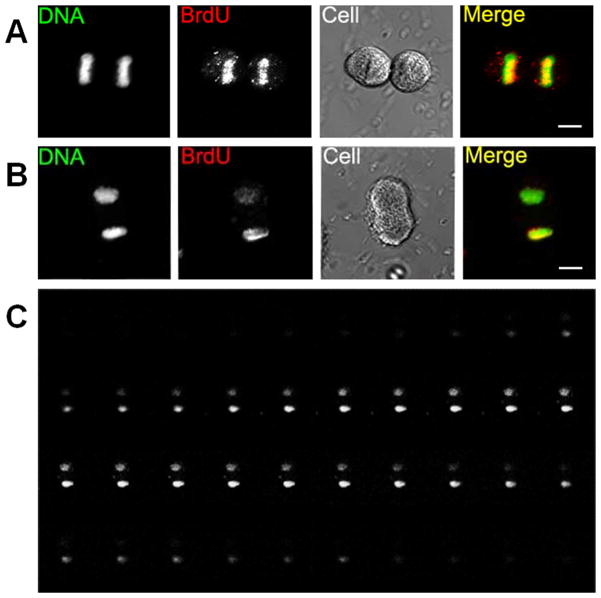
Label retention assay (Online Figure IB) validates results of the release assay that is limited to CPCs undergoing DNA synthesis at the time of labeling. Both strands are BrdU labeled in the label retention assay, in which CPCs are continuously labeled and passaged at least four times to dilute slowly cycling and exclusively asymmetrically segregating CPCs6–8, 10. Daughter nuclei with approximately 50:50 BrdU intensity ratio (only symmetric segregation) are observed in CPCs blocked at first mitosis during chase (Fig. 2A), consistent with model predictions (Online Figure IB). Both symmetric and asymmetric BrdU staining is observed at second mitosis (Fig. 2B-D). Asymmetric segregation of BrdU labeled chromatids is demonstrated by binucleated cells having daughter nuclei with disproportionate BrdU intensity (Fig. 2B and 2D). CPCs never exhibited a completely negative paired with positive BrdU daughter nuclei pair, consistent with previous reports of punctate BrdU staining10, 20. Therefore ACS is defined as daughter nuclei pair having BrdU ratio of 70:30 or higher similar to chromosome in-situ hybridization experiment5 instead of absolute ratio (100:0) confirming that not all chromatids are asymmetrically segregated4, 21, 22. CPCs with asymmetric segregation of chromatids with BrdU intensity ratio of 70:30 or higher between the daughter nuclei are found in 4.59% of the population (n=4, 53–194 binucleated cells per experiment). CPCs at late stage mitosis collected through mitotic shake off exhibit asymmetric (Fig. 3A) as well as symmetric BrdU intensity (Fig. 3B) demonstrating that ACS occurs without requiring drug-induced cell cycle arrest. Intensity differences between daughter nuclei are not due to differences in focal plane as confirmed by confocal optical sectioning (Fig. 3C), validating asymmetric segregation of BrdU labeled chromatids.
Figure 2. CPCs asymmetrically segregate BrdU labeled chromatids.
A and B, BrdU intensity distribution among daughter nuclei in binucleated cells at first (A) and second (B) mitosis during chase in label retention assay. 70:30 distribution threshold for BrdU intensity ratio is shown as dotted box in panel B and events above this threshold are considered asymmetrically segregating cells. C and D, Representative binucleated cells with symmetric (C) and asymmetric (D) BrdU immuno-stain.
Figure 3. Asymmetric BrdU segregation at late mitotic stages.
A, A telophase cell with symmetric BrdU staining. B, An anaphase cell with asymmetric BrdU segregation. C, Z stacks of BrdU immuno-staining of B. Mitotic phases are confirmed by differential interference contrast (DIC) images, labeled as Cell.
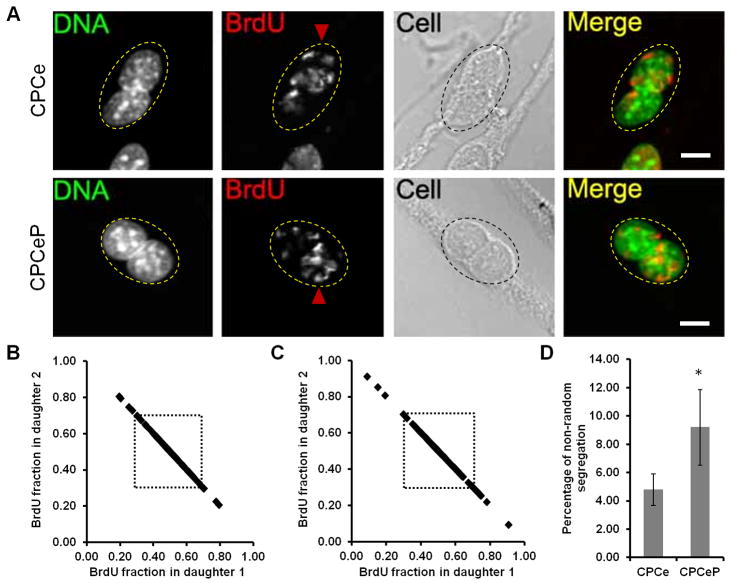
CPCs over expressing Pim-1 and eGFP (CPCeP) or eGFP alone (CPCe) were used to test the capacity of Pim-1 to enhance ACS by label retention assay. Morphological differences were not observed between CPCs, CPCe, or CPCeP indicating that the genetic modification does not significantly affect cell function. Binucleated CPCs with daughter nuclei exhibiting marked differential BrdU intensity are observed in both CPCe and CPCeP (Fig. 4A). Asymmetric segregation of chromatids is increased nearly two-fold in CPCeP (9.19%, n=5, 72–135 binucleated cells per experiment) compared to CPCe (4.79%, p=0.006, Fig. 4B–D) with no significant difference between non-transduced control versus CPCe (4.59% vs 4.79%, p=0.375).
Figure 4. Pim-1 kinase increases asymmetric DNA segregation.
A, Representative binucleated cell at second mitosis for CPCe (upper) and CPCeP (lower) with asymmetric BrdU staining indicated by red arrow head. B and C, BrdU distribution at second mitosis between daughter nuclei of CPCe and CPCeP, respectively. Dotted box represents 70:30 threshold of BrdU intensity ratio. D, Percentage of asymmetrically segregating cells from five independent experiments. Asterisk indicates p=0.006 compared to CPCe.
Discussion
Autologous cell therapy with CPC is an efficacious treatment for heart failure15. Beneficial effects of naïve cells are improved by modification and selection to promote reparative potential in the harsh milieu of a post-infarct heart.16 Modification with Pim-1 facilitates CPC survival and proliferation after infarction injury18, 23 in addition to numerous other signaling molecules and pathways that facilitate stem cell mediated repair (reviewed in16). However, CPC-mediated regeneration could be further augmented by identification of a cell subset that possesses enhanced potential for self-renewal as evidenced by ACS. Therefore, there is a need to identify signal transduction cascades that enhance ACS to confer superior self-renewal properties.
The presence of immortal/template strands and ACS in adult stem cells remains controversial2–4. The immortal strand hypothesis assumes ability of stem cells to retain old strands to minimize transformational risk associated with DNA replication errors1. Our findings of predominant rather than total ACS are consistent with direct observation of chromatid segregation5 and punctate BrdU retention in non-myocardial stem cell studies10, 20 indicating partial ACS might be stem cell-type specific. The silent sister hypothesis presumes ACS to regulate differential gene expression profile to determine cell fate and self-renewal3, 4, 24, which is strengthened by the observation of co-segregation of template strands with self-renewal markers in stem cells7–10 and of conformational changes in histone variant H2A.Z on template strands20. Functional significance of ACS through study of live cells is technically challenging due to protocols of fixation and immunolabeling required for detection by fluorescent cell sorting to isolate asymmetrically segregating cells, although utilization of directly labeled thymidine analogues has recently been used in cancer stem cells to circumvent fixation requirements25, 26. Future studies will capitalize upon the functional characterization of asymmetrically segregating CPCs to determine their regenerative potential relative to comparable CPCs undergoing symmetric segregation, with the expectation that asymmetric segregation correlates with enhanced myocardial repair.
Previous research in our lab established multiple beneficial roles for Pim-1 including enhancement of proliferation, survival and commitment of CPCs without causing tumor formation after transplantation18, 19. Pim-1 mediated increase in ACS can be explained by the multifaceted participation of Pim-1 in mitosis and transcriptional regulation. Pim-1 co-localizes at spindle poles during mitosis to phosphorylate NuMA and promote complex formation with HP1β, dynein and dynactin during chromosome segregation27. The left-right dynein motor is necessary for selective segregation of mouse chromosome 722. Pim-1 also binds and phosphorylates HP1γ on serine rich clusters, modulating its transcriptional repression activity28. Pim-1 mediated phosphorylation of HP1 proteins, which are involved in chromatin structure, indicates Pim-1 likely modulates chromatin dynamics. Chromatin dynamic regulation by Pim-1 could explain the functional significance of ACS in conjunction with the silent sister hypothesis and stem cell self-renewal. Pim-1 increases asymmetrically dividing CPCs in the heart after pathological injury to balance self-renewing versus differentiated cell populations19. Pim-1 decreases spontaneous differentiation of ES cells29 and is highly expressed in long term populating HSC30, supporting the premise that Pim-1 regulates stemness. Future studies are warranted to address the direct role of Pim-1 in ACS and stem cell self-renewal to further enhance the regenerative potential of CPCs and other stem cell types.
Supplementary Material
Novelty and Significance.
What is known?
Preferential partitioning of old versus newly synthesized DNA strands known as asymmetric segregation is hypothesized to regulate self-renewal and aging in several cell types.
The cardioprotective kinase Pim-1 increases proliferation, asymmetric cell division, and regenerative potential of cardiac progenitor cells (CPCs)
What new information does this article contribute?
Asymmetric partitioning of old versus newly synthesized DNA formed during mitosis occurs in a small subpopulation of CPCs.
Asymmetric segregation of chromatids into daughter CPCs is enhanced by Pim-1 kinase overexpression.
CPCs are effective as agents of cell-based regenerative treatment for myocardial damage, but factors determining self-renewal and lineage commitment of CPCs are poorly understood. So too, the regenerative potential of aged CPCs may be compromised in eldery patients suffering from chronic heart disease. Subsets of CPCs exhibiting preservation of proliferative potential and enhanced self-renewal will be valuable for use in the clinical setting to augment stem cell-based repair. Superior self-renewal properties and protection from phenotypic characteristics of aging are associated with asymmetric chromatid segregation wherein old DNA strands preferentially distribute to a single daughter cell during mitosis. This study confirms that asymmetric DNA segregation occurs in CPCs at a low frequency consistent with rates observed for other adult stem cell types. Moreover, the frequency of asymmetric DNA segregation in CPCs can be significantly increased by genetic modification to overexpress Pim-1 kinase. The observation of increased asymmetric segregation with Pim-1 overexpression may help explain the enhanced regenerative properties of CPCs engineered to overexpress Pim-1 and provide another mechanistic basis for increasing self-renewal of CPCs that is essential for effective myocardial repair.
Acknowledgments
We thank the Sussman lab members for their critical review of the manuscript and Brett Hilton of SDSU FACS Core facility for his help in flow cytometry analysis.
Sources of Funding:
M.A.S. was supported by National Heart, Lung, and Blood Institute Grants 1R21HL102714-01, 2 R01 HL067245, IR37 HL091102-01, 5 P01HL085577-05, RC1HL100891-02, 1 R21 HL102613-02, 1 R21 HL102714-02, 1 R21 HL104544-02 and R01 HL105759-01. C.T.C. is supported by the Rees-Stealy Research Foundation, the Achievement Rewards for Colleges Scientists Foundation, American Heart Association Predoctoral Fellowship 10PRE3060046, and an Inamori Foundation Fellowship. M.H.K is supported by Deutsche Forschungsgemeinschaft (DFG) Grant KO 3900/1-1.
Non standard Abbreviations and Acronyms
- ACD
asymmetric cell division
- ACS
asymmetric chromatid segregation
- BrdU
5-bromo-2′-deoxyuridine
- CPC
Cardiac Progenitor Cell
- CPCe
CPC expressing eGFP
- CPCeP
CPC expressing eGFP and Pim-1
Footnotes
Disclosures: None.
References
- 1.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 2.Charville GW, Rando TA. Stem cell ageing and non-random chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 2011;366:85–93. doi: 10.1098/rstb.2010.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Tajbakhsh S. Stem cell identity and template DNA strand segregation. Curr Opin Cell Biol. 2008;20:716–722. doi: 10.1016/j.ceb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Falconer E, Chavez EA, Henderson A, Poon SS, McKinney S, Brown L, Huntsman DG, Lansdorp PM. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463:93–97. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 7.Karpowicz P, Morshead C, Kam A, Jervis E, Ramunas J, Cheng V, van der Kooy D. Support for the immortal strand hypothesis: Neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 9.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc Natl Acad Sci U S A. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62:6791–6795. [PubMed] [Google Scholar]
- 12.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 13.Anversa P, Kajstura J, Leri A, Bolli R. Life and death of cardiac stem cells: A paradigm shift in cardiac biology. Circulation. 2006;113:1451–1463. doi: 10.1161/CIRCULATIONAHA.105.595181. [DOI] [PubMed] [Google Scholar]
- 14.Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, Bearzi C, Boni A, Bolli R, Kajstura J, Anversa P, Leri A. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci U S A. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (scipio): Initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Mohsin S, Siddiqi S, Collins B, Sussman MA. Empowering adult stem cells for myocardial regeneration. Circ Res. 2011;109:1415–1428. doi: 10.1161/CIRCRESAHA.111.243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesselli D, Beltrami AP, D’Aurizio F, Marcon P, Bergamin N, Toffoletto B, Pandolfi M, Puppato E, Marino L, Signore S, Livi U, Verardo R, Piazza S, Marchionni L, Fiorini C, Schneider C, Hosoda T, Rota M, Kajstura J, Anversa P, Beltrami CA, Leri A. Effects of age and heart failure on human cardiac stem cell function. Am J Pathol. 2011;179:349–366. doi: 10.1016/j.ajpath.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer KM, Cottage CT, Wu W, Din S, Gude NA, Avitabile D, Quijada P, Collins BL, Fransioli J, Sussman MA. Enhancement of myocardial regeneration through genetic engineering of cardiac progenitor cells expressing pim-1 kinase. Circulation. 2009;120:2077–2087. doi: 10.1161/CIRCULATIONAHA.109.884403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cottage CT, Bailey B, Fischer KM, Avitable D, Collins B, Tuck S, Quijada P, Gude N, Alvarez R, Muraski J, Sussman MA. Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ Res. 2010;106:891–901. doi: 10.1161/CIRCRESAHA.109.208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huh YH, Sherley JL. Molecular cloaking of h2a.Z on mortal DNA chromosomes during nonrandom segregation. Stem Cells. 2011;29:1620–1627. doi: 10.1002/stem.707. [DOI] [PubMed] [Google Scholar]
- 21.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311:1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 22.Armakolas A, Klar AJ. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science. 2007;315:100–101. doi: 10.1126/science.1129429. [DOI] [PubMed] [Google Scholar]
- 23.Mohsin S, Khan M, Toko H, Bailey B, Cottage CT, Wallach K, Nag D, Lee A, Siddiqi S, Lan F, Fischer KM, Gude N, Quijada Q, Avitabile D, Truffa S, Dembitsky W, Wu JC, Sussman MA. Human cardiac progenitor cells engineered with pim-1 kinase enhance myocardial repair. J Am Coll Cardiol. 2012 doi: 10.1016/j.jacc.2012.04.047. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajbakhsh S, Gonzalez C. Biased segregation of DNA and centrosomes: Moving together or drifting apart? Nat Rev Mol Cell Biol. 2009;10:804–810. doi: 10.1038/nrm2784. [DOI] [PubMed] [Google Scholar]
- 25.Hari D, Xin HW, Jaiswal K, Wiegand G, Kim BK, Ambe C, Burka D, Koizumi T, Ray S, Garfield S, Thorgeirsson S, Avital I. Isolation of live label-retaining cells and cells undergoing asymmetric cell division via nonrandom chromosomal cosegregation from human cancers. Stem Cells Dev. 2011;20:1649–1658. doi: 10.1089/scd.2010.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xin HW, Hari DM, Mullinax JE, Ambe CM, Koizumi T, Ray S, Anderson AJ, Wiegand GW, Garfield SH, Thorgiersson SS, Avital I. Tumor initiating label-retaining-cancer-cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. 2012 doi: 10.1002/stem.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya N, Wang Z, Davitt C, McKenzie IF, Xing PX, Magnuson NS. Pim-1 associates with protein complexes necessary for mitosis. Chromosoma. 2002;111:80–95. doi: 10.1007/s00412-002-0192-6. [DOI] [PubMed] [Google Scholar]
- 28.Koike N, Maita H, Taira T, Ariga H, Iguchi-Ariga SM. Identification of heterochromatin protein 1 (hp1) as a phosphorylation target by pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of hp1(1) FEBS Lett. 2000;467:17–21. doi: 10.1016/s0014-5793(00)01105-4. [DOI] [PubMed] [Google Scholar]
- 29.Aksoy I, Sakabedoyan C, Bourillot PY, Malashicheva AB, Mancip J, Knoblauch K, Afanassieff M, Savatier P. Self-renewal of murine embryonic stem cells is supported by the serine/threonine kinases pim-1 and pim-3. Stem Cells. 2007;25:2996–3004. doi: 10.1634/stemcells.2007-0066. [DOI] [PubMed] [Google Scholar]
- 30.Hu YL, Passegue E, Fong S, Largman C, Lawrence HJ. Evidence that the pim1 kinase gene is a direct target of hoxa9. Blood. 2007;109:4732–4738. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.