Abstract
The Charophycean green algae (CGA) occupy a key phylogenetic position as the evolutionary grade that includes the sister group of the land plants (embryophytes), and so provide potentially valuable experimental systems to study the development and evolution of traits that were necessary for terrestrial colonization. The nature and molecular bases of such traits are still being determined, but one critical adaptation is thought to have been the evolution of a complex cell wall. Very little is known about the identity, origins and diversity of the biosynthetic machinery producing the major suites of structural polymers (i. e., cell wall polysaccharides and associated molecules) that must have been in place for land colonization. However, it has been suggested that the success of the earliest land plants was partly based on the frequency of gene duplication, and possibly whole genome duplications, during times of radical habitat changes. Orders of the CGA span early diverging taxa retaining more ancestral characters, through complex multicellular organisms with morphological characteristics resembling those of land plants. Examination of gene diversity and evolution within the CGA could help reveal when and how the molecular pathways required for synthesis of key structural polymers in land plants arose.
Keywords: charophycean green algae, model organisms, next generation sequencing, phylogeny, whole genome duplication
Approximately 470–500 million years ago, members of the Charophycean green algae (CGA) developed the capacity to colonize progressively drier habitats, ultimately giving rise to the first land plants (embryophytes).1 This remarkable transition from an aquatic to a terrestrial environment would have necessitated a wide range of structural and physiological adaptations in order to tolerate the stresses associated with life on land, such as desiccation and UV radiation, and also to succeed in the absence of water to support the plant body and to distribute progeny.1,2 However, while various features that are critical for life on land can be inferred, the molecular bases of these adaptations are still not well understood. Moreover, it is clear that the identities of many biochemical and regulatory pathways that were critical in this remarkable phase of plant evolution are still to be revealed.
One approach to address these questions is to study members of the extant CGA, comprising diverse taxonomic lineages of a phylogenetic grade (Figure 1) that range morphologically from single‑celled algae to structurally complex multicellular organisms that morphologically resemble embryophytes. The question of the precise phylogenetic relationships of various clades within the CGA and the placement of the immediate ancestor of land plants is still controversial; however, recent evidence supports either the Zygnematales or a Zygnematales/Coleochaetales clade as sister to the land plants3 and not the Charales, as previously speculated.4 Current determinations of the evolutionary relationships between CGA taxa have been based largely on sequencing only a few genes across representative species from the major CGA groups, which provides limited resolution. This will undoubtedly change in the very near future as new DNA sequencing technologies allow rapid, high-throughput and cheap access to genomic and transcriptomic information.5,6 Indeed, while there is not yet a reference genome available for any CGA species, the first larger data sets derived from several CGA transcriptomes have recently been published.3,7
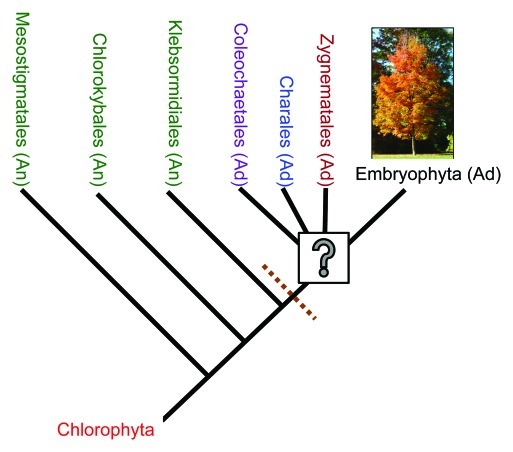
Figure 1.
Phylogenetic relationships of green plant taxa. The question mark refers to the currently unresolved question of which group represents the immediate sister group to land plants. The dashed line marks the split between clades with “ancestral” (An) cell walls and those with features indicative of “advanced” (Ad), or land plant-like cell walls. It should be noted that Embryophyta has diverged substantially to occupy diverse habitats and consequently a wide range of morphologies are recognized. The image here shows a single species from one of the approximately 60 orders within the APG (Angiosperm Phylogeny Group) III system.19
In addition to DNA sequencing, studies of specific aspects of plant biology are helping to resolve some of the adaptive traits that were necessary in the first land plants. One of the most important was likely to have been the development of a complex cell wall, which would have been needed to provide both structural support and defense against biotic stresses. Indeed, a recent study of the cell wall architecture and composition8 of a variety of CGA species suggested an evolutionary pattern that is largely consistent with established phylogenies based on ultra-structural and molecular data (Figure 2). Specifically, the CGA can be divided into a ‘ancestral’ group relative to land plants and an advanced group that have more complex cell wall compositions and morphologies (Figure 1). The results further indicate that the capacity to synthesize key cell wall polysaccharide architectural networks, as well as the major wall component lignin, arose at some point during divergence and radiation of the CGA. Accordingly, a substantial expansion in cell wall polysaccharide diversity8 and genetic evidence of biosynthetic capacity (unpublished data) is seen specifically in the Zygnematales when compared to earlier diverging clades such as Mesostigmatales and Chlorokybales. This trend is evidenced by the relative representation and abundance of genes that can be associated with the synthesis and modification of cellulose, hemicelluloses and pectins.7,8
Figure 2.
Cladogram showing the relationships amongst 10 CGA species based on cell wall polymer structure, interpreted from glycan array and linkage analysis data (www.bioinf.ebc.ee/EP/EP/EPCLUST; default settings with UPGMA clustering based on Euclidean distance). Colors indicate clade of species analyzed: Coleochaetales (purple), Charales (blue), Zygnematales (brown), Klebsormidiales (orange), Chlorokybales (green).
The evolutionary origins of other structures that are crucial for the success of land plants, such as a hydrophobic cuticle, and other mechanisms for reproduction and survival are likely to be revealed through similar comparative studies of the CGA. We propose that one important driving force underlying the development of many essential adaptations could have been whole genome duplication (WGD‑polyploidy9). This evolutionary process has not only been suggested to be responsible for the remarkable radiation of angiosperms, but is also likely to be responsible for up to 15% of angiosperm speciation events.10 WGD allows for gene families and even whole pathways to expand and develop through processes such as gene retention, sub-functionalization and neo‑functionalization.11,12 There has been considerable speculation about this issue for some time, but while there is some evidence that polyploidy exists in specific CGA lineages, such as Charales, Coleochaetales and Zygnematales,13 strong experimental evidence for nuclear genome condition within the CGA has not been reported.
In addition to the evolutionary questions, CGA species offer potentially valuable experimental model systems to study many aspects of plant biology. Specifically, it has proven to be difficult to elucidate which gene products are responsible for wall biosynthesis, modification and degradation of cell wall components, apparently in part because of the redundancy of the members of large gene‑families.14-16 Initial surveys suggest that while CGA that share more recent ancestry with land plants have walls that are remarkably similar to those of land plants, their families of cell wall biosynthetic genes are generally smaller. Moreover, CGA species provide an opportunity to work with unicellular organisms that are readily cultured and manipulated17 and a recent report of successful transformation of Closterium18 adds to the value of the CGA as model organisms. We conclude that CGA species are emerging as valuable systems to study not just cell walls, but many other aspects of plant growth, development and environmental adaptations.
Acknowledgments
Support for this research was provided to IS by an individual postdoc stipend from the VKR foundation, Denmark, and to JKCR by a grant from the National Science Foundation (Plant Genome Program, DBI-0606595). Support for DSD and JJD was provided by grants from the National Science Foundation NSF MCB-0919925 and NSF IOS-0744306, respectively. We thank R. Sørensen for photographic assistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18574
References
- 1.Niklas KJ, Kutschera U. The evolution of the land plant life cycle. New Phytol. 2010;185:27–41. doi: 10.1111/j.1469-8137.2009.03054.x. [DOI] [PubMed] [Google Scholar]
- 2.Niklas KJ. The cell walls that bind the tree of life. Bioscience. 2004;54:831–41. doi: 10.1641/0006-3568(2004)054[0831:TCWTBT]2.0.CO;2. [DOI] [Google Scholar]
- 3.Wodniok S, Brinkmann H, Glöckner G, Heidel AJ, Philippe H, Melkonian M, et al. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–3. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- 5.Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, Liang H, et al. Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics. 2009;10:347. doi: 10.1186/1471-2164-10-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timme RE, Delwiche CF. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010;10:96. doi: 10.1186/1471-2229-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sørensen I, Pettolino FA, Bacic A, Ralph J, Lu F, O'Neill MA, et al. The Charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011;68:201–11. doi: 10.1111/j.1365-313X.2011.04686.x. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Shiu S-H, Thoma S, Li W-H, Patterson SE. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006;7:R87. doi: 10.1186/gb-2006-7-9-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–9. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–61. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 12.Coate JE, Schlueter JA, Whaley AM, Doyle JJ. Comparative evolution of photosynthetic genes in response to polyploid and nonpolyploid duplication. Plant Physiol. 2011;155:2081–95. doi: 10.1104/pp.110.169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapraun DF. Nuclear DNA content estimates in green algal lineages: Chlorophyta and Streptophyta. Ann Bot. 2007;99:677–701. doi: 10.1093/aob/mcl294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010;10:282. doi: 10.1186/1471-2229-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen JK, Kim H, Cocuron JC, Orler R, Ralph J. Wilkerson CG. The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 2011;66:387–400. doi: 10.1111/j.1365-313X.2010.04475.x. [DOI] [PubMed] [Google Scholar]
- 16.Kong Y, Zhou G, Yin Y, Xu Y, Pattathil Z, Hahn MG. Molecular Analysis of a Family of Arabidopsis Genes Related to Galacturonosyltransferases. Plant Physiol. 2011;155:1791–805. doi: 10.1104/pp.110.163220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domozych DS, Brechka H, Britton A, Toso M. Cell Wall Growth and Modulation Dynamics in a Model Unicellular Green Alga—Penium margaritaceum: Live Cell Labeling with Monoclonal Antibodies. J Bot. 2011;2011 [Google Scholar]
- 18.Abe J, Hori S, Tsuchikane Y, Kitao N, Kato M, Sekimoto H. Stable Nuclear Transformation of the Closteriumperacerosum-strigosum-littorale Complex. Plant Cell Physiol. 2011;52:1676–85. doi: 10.1093/pcp/pcr103. [DOI] [PubMed] [Google Scholar]
- 19.The angiosperm phylogeny group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–21. doi: 10.1111/j.1095-8339.2009.00996.x. [DOI] [Google Scholar]