Abstract
Cell separation processes, such as abscission, are critical for plant development and play key roles from sculpting the form of the plant to scattering seeds. It is however essential that such processes are under tight temporal and spatial regulation. Floral organ abscission in Arabidopsis thaliana is regulated by a ligand-receptor module consisting of the signaling peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and the two receptor-like kinases HAESA (HAE) and HAESA-LIKE 2 (HSL2), and it is the restricted expression pattern of IDA that hinders cell separation from occurring in the abscission zones (AZs) of other organs where HAE and HSL2 are present. In the July issue of The Plant Cell we report on the identification of additional components acting downstream in the IDA signaling pathway. Through a screen for mutations that restore floral organ abscission in ida mutants, we identified two new alleles of the KNOTTED-LIKE HOMEOBOX gene BREVIPEDICELLUS (BP)/KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1) and show that BP/KNAT1 is important in regulating the timing of floral abscission by controlling AZ cell size and by regulating KNAT2 and KNAT6.
Keywords: cell elongation, cell separation, cell signaling, floral organ abscission, HAE, HSL2, IDA, KNOX proteins
Intercellular signaling mediated by secretory peptide ligands are important for regulating various aspects of plant growth and development.1 Abscission, a physiological process that involves programmed changes in cellular adhesion of specialized abscission zone (AZ) cells, allows the plant to discard non-functional or infected organs. The correct temporal and spatial regulation of abscission is crucial during plant development. Premature abscission of reproductive organs or immature seeds can comprise reproduction, and unrestricted cell separation can interfere with tissue integrity. INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) encodes a protein with features compatible with it being a peptide ligand2 and is essential for controlling floral organ abscission in Arabidopsis thaliana.3 IDA, like the peptide ligand CLAVATA 3 (CLV3), which is required to hinder the overproduction of flowers and floral organs in conjunction with the receptors CLV1, CLV2, CORYNE (CRN) and RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2),4-7 has a conserved C-terminal domain (the EPIP domain) that contains the active peptide2,8,9 which is likely to undergo posttranslational modifications such as hydroxyprolination of key proline residues.
Floral Abscission in Arabidopsis
The floral organ abscission process in Arabidopsis is divided into four distinct developmental stages.10 The first stage, the differentiation of several layers of specialized AZ cells at the base of the developing organs, is required for the later separation from the plant. If the AZ cells are not differentiated, abscission will not take place, as is the case in the double mutant of BLADE-ON-PETIOLE 1and 2 (BOP1 BOP2) which does not portray anatomically distinct AZ cells.11 The second phase starts as the organs prepare to separate from the plant and the AZ cells become competent to respond to abscission signals. Subsequently, the pectin in the cell walls between a subset of cells within the AZ start to degrade followed by an expansion in the size of the cells at the proximal side of the AZ. Ultimately, the separation of the organ supervenes and a lignified protective layer develops on the distal side of the AZ.
The relationship between AZ cell enlargement and organ separation is unclear.10 It has been proposed that the functional role of AZ cell expansion might be to create the tension needed for the final mechanical rupture of the AZ.12 In support of this, the receptor-like kinase (RLK) EVERSHED (EVR) has recently been implicated in regulating the proper timing of floral organ abscission in part by restricting AZ cell size.13 Furthermore, when Arabidopsis plants overexpress (OX) IDA under the control of the cauliflower mosaic virus 35S promoter (35S), precocious floral organ abscission is observed together with an enlargement of the AZ cells.14 There is a significant increase in the number of cells undergoing separation in response to the IDA signal. 35S:IDA plants also display ectopic abscission of pedicels and leaves, organs not normally shed in Arabidopsis, but even if IDA is abundantly expressed in all organs OX of IDA does not induce middle lamella breakdown between all cells.14 This indicates that for cells to respond to IDA signaling some indispensable components, such as downstream activators of a signaling pathway, must be present for IDA-induced abscission to occur.
The IDA Signaling Pathway
A number of mutants have been identified with changes in floral organ abscission.15 Recently, it was discovered that HAESA (HAE) and HAESA-LIKE2 (HSL2), a pair of leucine-rich repeat (LRR)-RLKs, are redundantly required for regulating cell separation during floral organ abscission.2,16 hae hsl2 double mutants are phenotypically similar to ida plants and both mutants have morphologically normal AZ cells compared with wild type (WT) plants and are largely unaffected during the initial steps of organ abscission.3,16 However, expansion of the AZ cells prior to organ separation is not observed and both ida and hae hsl2 mutants fail to undergo the final cell separation step.3,16 Genetic interaction studies have shown that HAE and HSL2 are essential for relaying an IDA signal2,16 by activating a MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascade including MAPK KINASE 4 (MKK4), MKK5, MPK3 and MPK6.16
Class I KNOTTED-Like Homeobox Proteins Control Floral Abscission
In our recent publication17 additional components of the IDA signaling pathway were identified by screening the progeny of mutagenized ida seeds for suppressor mutants showing normal floral organ abscission. Of the 13 revertant lines identified, two of them had, in addition to normal organ shedding, the characteristic downward pointing silique phenotype conferred by mutations in the KNOTTED-LIKE HOMEOBOX (KNOX) gene BREVIPEDICELLUS (BP)/KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1).18,19 Sequencing of the BP/KNAT1 gene identified point mutations in each of the two revertant lines, leading to a premature stop codon and a change in the splice acceptor of intron one, respectively.17 Morphological analysis by scanning electron microscopy (SEM) of the petal AZ of the revertant lines together with measurements quantifying the force needed to remove the petals from the plant (petal breakstrength, pBS)20 confirmed the complete rescue of the abscission defect of ida. To substantiate the ability of a mutation in BP/KNAT1 to rescue the abscission phenotype of ida and to investigate the genetic relationship with the hae hsl2 mutant, a null-allele deletion mutant of BP/KNAT1, bp-3,21 was crossed to both ida and hae hsl2. As expected for a downstream component of the IDA signaling pathway, the bp-3 mutant was capable of reverting the floral organ abscission defect of both ida and hae hsl2.17 Interestingly, HAE and BP/KNAT1 are both expressed at the base of the pedicel,18,22,23 where a vestigial AZ is found14 and where OX of IDA induces ectopic abscission.
pBS measurements and SEM analysis of AZ cells showed that bp-3 had a precocious dissolution of the middle lamella and was morphologically similar to plants OX IDA with enlarged AZ cells, indicating that BP/KNAT1 functions as an inhibitor of abscission and is important for regulating the proper timing of cell separation by controlling cell expansion.17 Interestingly, EVR is downregulated in the bp mutant.24 However, unlike bp-3, evr is not capable of rescuing the abscission defect of ida or hae hsl2.13 Thus, EVR inhibits abscission independently of IDA, but BP/KNAT1 may mediate crosstalk between the IDA-HAE/HSL2 and the EVR pathways.
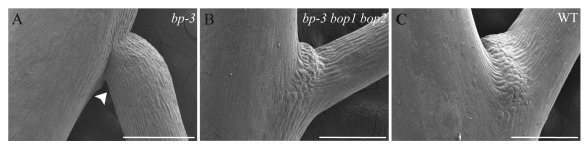
It has previously been suggested that BP/KNAT1, or other class I KNOX genes, could be involved in regulating AZ cell formation or differentiation.11,24 However, floral organ abscission was not reestablished in a bop1 bop2 bp-3 triple mutant showing that mutations in BP/KNAT1 are not sufficient to initiate AZ formation, suggesting that the observed abscission phenotype in the bp-3 mutant is dependent on the presence of morphologically distinct AZ cells. Surprisingly, the downward-pointing silique phenotype of the bp-3 mutant was absent in the bp-3 bop1 bop2 mutant and the uneven cell division and elongation of the epidermal cells on the abaxial and adaxial side of the bp-3 pedicel was reverted to WT (Fig. 1). This suggests that in the absence of pedicel AZ cells;11 there is no effect of a mutation in BP/KNAT1 on pedicel development.
Figure 1. Pedicel phenotype of bp-3 and bp-3 bop1 bop2 mutants. (A) bp-3 mutants have down-ward pointing pedicels due to unsymmetrical cell differentiation, elongation and growth, which is more affected on the abaxial (arrow head) than the adaxial side. (B) The downward-pointing silique phenotype of the bp-3 mutant was absent in the bp-3 bop1 bop2 mutant and the cell morphology of the pedicel was reverted to WT (C). Bars = 400 μM (A) 300 μM (B and C).
During inflorescence development, BP/KNAT1 is known to restrict the expression of the two KNOX domain genes KNAT2 and KNAT6 to promote correct pedicel growth. Expression studies from AZ cells using promoter constructs of KNAT2 (KNAT2pro:GUS) and KNAT6 (KNAT6pro:GUS) showed that BP/KNAT1 also restricts the expression of these genes in AZ cells. Consistent with what is observed in the pedicel,23 a double mutant of knat2 knat6 rescues the premature abscission and enlargement of AZ cells observed in bp-3 mutants.17 This indicates that KNAT2 and KNAT6 act as positive regulators of cell separation during floral abscission downstream of BP/KNAT1. When IDA was OX in the knat2 knat6 mutant background the early abscission phenotype was lost, conversely when KNAT2 or KNAT6 were OX in the ida mutant background, a rescue of the ida abscission defect was observed. This is consistent with the hypothesis that IDA signaling positively regulates KNAT2 and KNAT6 and indeed KNAT2pro:GUS and KNAT6pro:GUS expression was absent from the AZ of both the ida and hae hsl2 mutants.17
Signaling Model
Results from our recent publication,17 in addition to experiments performed by several scientists in the recent years, allow us to propose the following signaling model for the IDA peptide (Fig. 2). A posttranslationally modified IDA peptide binds the extracellular LRR of either a homo or heterodimer including HAE and/or HSL2 causing autophosphorylation of the kinase receptor domains and the transduction of a phosphorelay signal through a MAPK cascade that acts to regulate BP/KNAT1. Since the transcriptional level of BP/KNAT1 is unaltered in ida and hae hsl2,17 the regulation is likely to be at the protein level and may include dimerization with another homeodomain protein, such as BEL1-like (BELL) proteins, or posttranscriptional modifications of BP/KNAT1. Upon activation of the IDA signaling pathway, the BP/KNAT1 restriction of KNAT2 and KNAT6 is elevated, and these transcription factors may in turn positively regulate the transcription of genes encoding proteins involved in cell separation, such as cell wall remodeling (CWR) enzymes.
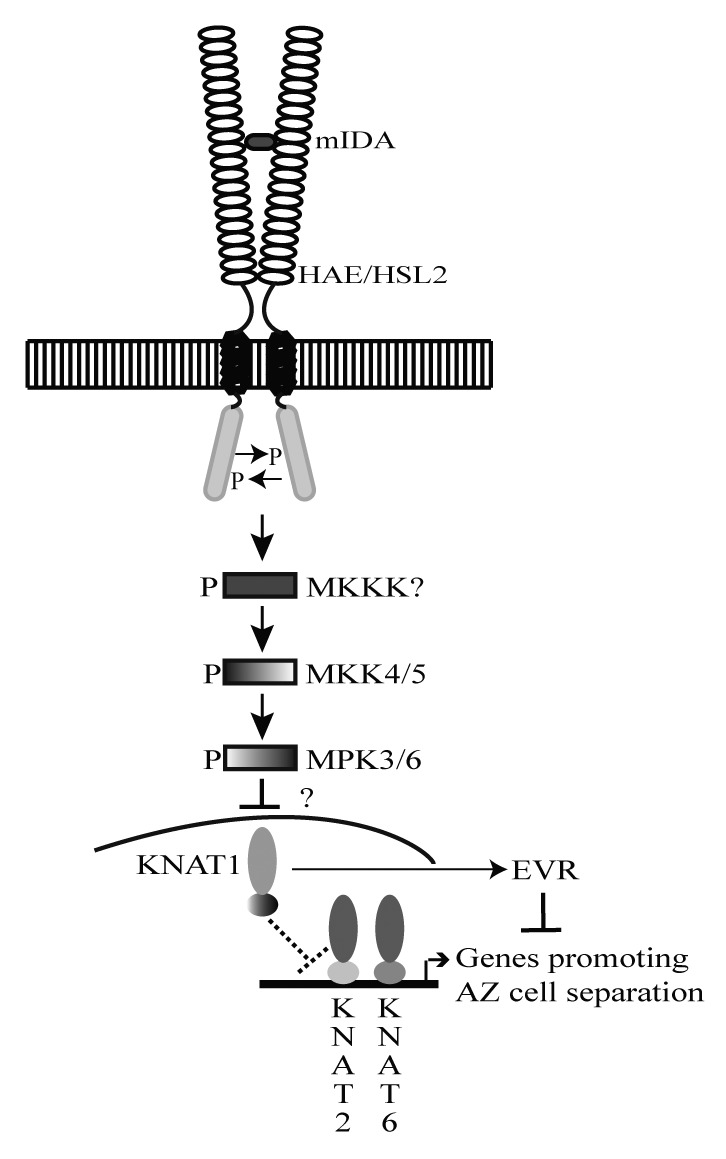
Figure 2. Model of IDA signaling. A mature posttranslational modified IDA peptide consisting of the EPIP, or part of the EPIP, domain may bind the extracellular LRRs of a HAE homodimer, a HSL2 homodimer, or a HAE-HSL2 heterodimer and cause autophosphorylation (P) of the kinase receptor domains. The signal from IDA-HAE/HSL2 is suggested to be transduced via a phosphorelay involving MKK4, MKK5, MPK3 and MPK6. Genetic evidence suggests that BP/KNAT1 acts as a negative downstream component of this signaling pathway by inhibiting cell separation and the expression of KNAT2 and KNAT6. Upon activation of the IDA signaling pathway, the BP/KNAT1 restriction of KNAT2 and KNAT6 is elevated, and these transcription factors can act as positive regulators of floral organ separation by positively regulating genes promoting AZ cell separation. In addition, BP/KNAT1 is a positive regulator of EVR, which influences cell separation by a separate parallel pathway.
Future experiments investigating the cellular and subcellular localization of BP/KNAT1 during WT abscission compared with ida will hopefully give us an insight as to how the IDA signaling pathway is regulating the action of BP/KNAT1. Furthermore, the genetic identification of the remaining revertant lines will surely provide more detailed information on the IDA signaling system.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18379
References
- 1.Butenko MA, Aalen RB. Receptor ligands in development. In: Receptor-like kinases in plants: from development to defense: Springer Verlag Series Signaling and Communication in Plants. Kemmerling B, Tax F (eds), Vol. 13; 2012. [Google Scholar]
- 2.Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–17. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butenko MA, Patterson SE, Grini PE, Stenvik G-E, Amundsen SS, Mandal A, et al. INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15:2296–307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–4. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–20. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- 6.Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–46. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–406. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–8. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 9.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–80. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 10.Patterson SE. Cutting loose. Abscission and dehisence in Arabidopsis. Plant Physiol. 2001;126:494–500. doi: 10.1104/pp.126.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKim SM, Stenvik GE, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, et al. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development. 2008;135:1537–46. doi: 10.1242/dev.012807. [DOI] [PubMed] [Google Scholar]
- 12.Sexton R, Redshaw AJ. The role of cell expansion in the abcission of Impatiens leaves. Ann Bot (Lond) 1981;48:745–57. [Google Scholar]
- 13.Leslie ME, Lewis MW, Youn JY, Daniels MJ, Liljegren SJ. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development. 2010;137:467–76. doi: 10.1242/dev.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell. 2006;18:1467–76. doi: 10.1105/tpc.106.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis MW, Leslie ME, Liljegren SJ. Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol. 2006;9:59–65. doi: 10.1016/j.pbi.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:15629–34. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi C-L, Stenvik G-E, Vie AK, Bones AM, Pautot V, Proveniers M, et al. Arabidopsis Class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell. 2011;23:2553–67. doi: 10.1105/tpc.111.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell. 2002;14:547–58. doi: 10.1105/tpc.010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, et al. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:4730–5. doi: 10.1073/pnas.072626099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lease KA, Cho SK, Walker JC. A petal breakstrength meter for Arabidopsis abscission studies. Plant Methods. 2006;2:2. doi: 10.1186/1746-4811-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rim Y, Jung J-H, Chu H, Cho WK, Kim S-W, Hong JC, et al. A non-cell-autonomous mechanism for the control of plant architecture and epidermal differentiation involves intercellular trafficking of BREVIPEDICELLUS protein. Plant Biol. 2009;36:280–9. doi: 10.1071/FP08243. [DOI] [PubMed] [Google Scholar]
- 22.Jinn T-L, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14:108–17. [PMC free article] [PubMed] [Google Scholar]
- 23.Ragni L, Belles-Boix E, Gunl M, Pautot V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell. 2008;20:888–900. doi: 10.1105/tpc.108.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X-Q, Xu W-H, Ma L-G, Fu Z-M, Deng X-W, Li J-Y, et al. Requirement of KNAT1/BP for the Development of Abscission Zones in Arabidopsis thaliana. J Integr Plant Biol. 2006;48:15–26. doi: 10.1111/j.1744-7909.2005.00085.x-i1. [DOI] [Google Scholar]