Abstract
The voltage-dependent anion channels (VDACs), known as a major group of outer mitochondrial membrane proteins, are present in all eukaryotic species. In mammalian cells, they have been established as a key player in mitochondrial metabolism and apoptosis regulation. By contrast, little is known about the function of plant VDACs. Recently, we performed functional analysis of all VDAC gene members in Arabidopsis thaliana, and revealed that each AtVDAC member has a specialized function. Especially, in spite of similar subcellular localization and expression profiling of AtVDAC2 and AtVDAC4, both the T-DNA insertion knockout mutants of them, vdac2–2 and vdac4–2, showed severe growth retardation. These results suggest that AtVDAC2 and AtVDAC4 proteins clearly have distinct functions. Here, we introduced the AtVDAC2 gene into the vdac2–2 mutant, and demonstrated that the miniature phenotype of vdac2–2 plant is abolished by AtVDAC2 expression.
Keywords: Arabidopsis thaliana, AtVDAC2, AtVDAC4, mitochondrial porin signature, plant growth, voltage-dependent anion channel
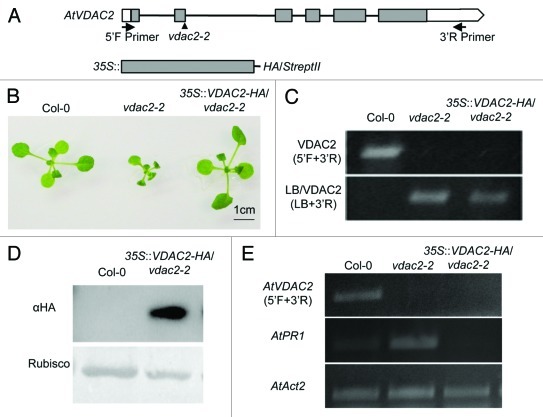
The voltage-dependent anion channels (VDACs), one of the most abundant proteins in outer mitochondrial membrane (OMM), are thought to mediate the transport of metabolites between mitochondria and the cytoplasm.1 The functional analysis of VDACs has been extensively studied in animals, and now it is recognized that VDAC is involved in many physiological and pathological processes such as male reproduction, the central nervous system, glucose homeostasis, and mitochondrial-mediated apoptosis.2,3 In contrast, physiological significance of plant VDACs is not well known.1 Recently, we have performed functional analysis of the plant VDACs and revealed that some of Nicotiana benthamiana VDAC members and Arabidopsis thaliana AtVDAC1 are involved in plant innate immunity through regulation of hydrogen peroxide production.4,5 Moreover, we demonstrated, through characterization of all Arabidopsis VDAC members (AtVDAC1 to AtVDAC4), that plant VDACs were also important for vegetative and reproductive growth.5 According to the phylogenetic analysis, dicotyledonous plant VDACs are clearly classified to two groups: One group is formed by AtVDAC1 and AtVDAC3, the other by AtVDAC2 and AtVDAC4. The former retains the conserved eukaryotic mitochondrial porin signature (MPS) and localizes only in the mitochondrion, while the latter has divergent MPS motif and localizes not only in mitochondria but also in other organelle. In addition, the latter VDACs’ expression profiles were almost similar.5 We, therefore, thought that AtVDAC2 and AtVDAC4 have a redundant function in planta. However, T-DNA knockout mutant of AtVDAC2, vdac2–2, showed retarded growth and abnormal pollen development even if AtVDAC4 gene was highly expressed. Moreover, some of the stress marker genes including PR (pathogenesis-related) genes are constitutively expressed in normal growth condition. The AtVDAC4 knockout plant, vdac4–2, showed a similar phenotype with vdac2–2.5 To confirm that the absence of AtVDAC2 is the real causal factor for the severe retarded growth observed in vdac2–2, we have generated the complementation plant using a heterozygous T-DNA insertion mutant (VDAC2/vdac2–2) because the homozygous knockout mutant (vdac2–2/vdac2–2) did not set fertile seed. And yet, we selected the vdac2–2/vdac2–2 plant expressing HA-tagged VDAC2 (described as a 35S::VDAC2-HA/vdac2–2) via self-pollination of the VDAC2/vdac2–2 heterozygous plant expressing VDAC2-HA. As shown in Figure 1, T-DNA insertion was confirmed in vdac2–2 as well as in 35S::VDAC2-HA/vdac2–2 plant (C), and also AtVDAC2 RNA expression were at null levels (E). In 35S::VDAC2-HA/vdac2–2 plant expressing VDAC2-HA (D), the growth retardation was recovered and the abnormal expression of AtPR1 gene was totally attenuated (B, E).
Figure 1. Complementation analysis of AtVDAC2 knockout plant, vdac2–2. (A) Schematic structures of AtVDAC2 genomic DNA (top) and HA-tag fused AtVDAC2 construct cloned into pPZP2H3a (+) vector used for making the complementation plant (bottom). Arrows show the positions and orientations of the primers used in PCR (5′F; 5′- GTCGTAATTCTTCTATCTTGATATCTT -3′ and 3′R; 5′- GCGGAACTATTTATTGATTCCA -3′). Arrowhead shows the T-DNA insertion site in vdac2–2. Filled boxes, exons; open regions, untranslated regions; black lines, introns. (B) Phenotypes of Col-0, vdac2–2 and 35S::VDAC2-HA/vdac2–2 plants. All plants were grown at 22°C on 1/2 MS plate for 20 d with 16h light /8h dark condition. Transgenic seeds were selected by 1/2 MS plate with Hygromycin and BASTA, and the other method was followed as described previously.5 (C) Confirmation of the T-DNA insertion by genomic PCR analysis in Col-0, vdac2–2 and 35S::VDAC2-HA/vdac2–2 plants. The homozygous line was identified using the left border primer of the T-DNA (LB) and the gene-specific primer (3′R). (D) VDAC2-HA protein detected in 35S::VDAC2-HA/vdac2–2 plants. Total proteins were extracted from 16 d-old plants according to the method by Chinchilla et al.6 VDAC2-HA protein was detected by anti-HA antibody (Roche, Switzerland). (E) RT-PCR analysis of AtVDAC2 and AtPR1 genes in Col-0, vdac2–2 and 35S::VDAC2-HA/vdac2–2 leaves. AtAct2 is used as a loading control. The primers information was described previously.5
Some of the plant VDACs could compromise VDAC1-disrupted yeast phenotype.7,8 Overexpression of rice OsVDAC4 induces apoptosis in the Jurkat T-cell line, which can be blocked by Bcl-2 protein.9 These findings strongly suggest that VDACs share some common function in eukaryotes. Therefore, we were surprised that AtVDAC2 and AtVDAC4 have distinct function in A. thaliana. It is likely that they form a heterocomplex and act as multifunctional channels depend on the site of organelle localization because they show the loose mitochondrial localization. Mammalian VDACs, once thought to be exclusively located in the OMM, are localized to non-mitochondrial cell compartments such as plasma membrane and endoplasmic reticulum.10,11 In plant VDACs also seem to localize in mitochondria, however, some of the Lotus japonicus VDACs are localized in small vesicles at the cell periphery.7 Maize VDAC identified as a sucrose synthase (SUS)-interacting protein is localized not only in mitochondria but also in nuclei along with SUS.12 These evidences imply that VDACs have more novel functions. Further studies are needed.
Acknowledgments
We thank Masahiro Yano for providing vectors and Thomas Berberich for critically reading the manuscript. This work was supported in part by Grant-in-Aid from Japan Society for the Promotion of Science to CT (21–6801).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18394
References
- 1.Kusano T, Tateda C, Berberich T, Takahashi Y. Voltage-dependent anion channels: their roles in plant defense and cell death. Plant Cell Rep. 2009;28:1301–8. doi: 10.1007/s00299-009-0741-z. [DOI] [PubMed] [Google Scholar]
- 2.Craigen WJ, Graham BH. Genetic strategies for dissecting mammalian and Drosophila voltage-dependent anion channel functions. J Bioenerg Biomembr. 2008;40:207–12. doi: 10.1007/s10863-008-9146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med. 2010;31:227–85. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Tateda C, Yamashita K, Takahashi F, Kusano T, Takahashi Y. Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Rep. 2009;28:41–51. doi: 10.1007/s00299-008-0630-x. [DOI] [PubMed] [Google Scholar]
- 5.Tateda C, Watanabe K, Kusano T, Takahashi Y. Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. J Exp Bot. 2011;62:4773–85. doi: 10.1093/jxb/err113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 7.Wandrey M, Trevaskis B, Brewin N, Udvardi MK. Molecular and cell biology of a family of voltage-dependent anion channel porins in Lotus japonicus. Plant Physiol. 2004;134:182–93. doi: 10.1104/pp.103.031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkeles A, Breiman A, Zizi M. Functional differences among wheat voltage-dependent anion channel (VDAC) isoforms expressed in yeast. J Biol Chem. 1997;272:6252–60. doi: 10.1074/jbc.272.10.6252. [DOI] [PubMed] [Google Scholar]
- 9.Godbole A, Varghese J, Sarin A, Mathew MK. VDAC is a conserved element of death pathways in plant and animal systems. Biochim Biophys Acta. 2003;1642:87–96. doi: 10.1016/S0167-4889(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 10.Shoshan-Barmatz V, Zalk R, Gincel D, Vardi N. Subcellular localization of VDAC in mitochondria and ER in the cerebellum. Biochim Biophys Acta. 2004;1657:105–14. doi: 10.1016/j.bbabio.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 11.De Pinto V, Messina A, Lane DJR, Lawen A. Voltage-dependent anion-selective channel (VDAC) in plasma membrane. FEBS Lett. 2010;584:1793–9. doi: 10.1016/j.febslet.2010.02.049. [DOI] [PubMed] [Google Scholar]
- 12.Subbaiah CC, Huber SC, Sachs MM, Rhoads D. Sucose synthase: expanding protein function. Plant Signal Behav. 2007;2:28–9. doi: 10.4161/psb.2.1.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]