Abstract
Encoding a conserved protein of unknown function, the Medicago truncatula RDN1 gene is involved in autoregulation of nodulation through signaling in the root. In contrast, the SUNN kinase in M. truncatula has been shown by grafting of mutant scions to control nodule number in the root by communication of a signal from the shoot to the root. GUS staining patterns resulting from expression of the SUNN promoter fused to uidA showed expression of SUNN in most parts of plant including the root, but confined to the vascular tissue, a pattern that overlaps with that published for RDN1. Real Time qRT-PCR analysis showed levels of both SUNN RNA and RDN1 RNA did not change significantly during early nodulation signaling (0–72 h after inoculation). The similarity in expression in cell types strongly suggests vascular signaling for nodule number regulation, while the lack of changes over early nodule development suggest post transcriptional mechanisms such as protein association or phosphorylation transmit the signal.
Keywords: autoregulation of nodulation, gene expression, nodulation
The symbiosis between legumes and rhizobia involves signaling, response and regulation by both the bacterial and plant partners. The regulation of nodule number by the plant, termed “autoregulation,” includes a long distance signal transduction pathway involving both the shoot and the root. Because supporting active nodules has an energy cost to the plant in the range of 12–17 g of carbon per gram of nitrogen obtained,1 regulation of nodule number by the plant presumably balances the need for fixed nitrogen with the cost of supporting the bacteria. Isolation of plant mutants unable to regulate the number of nodules that form on the roots identified multiple genes encoding proteins that control nodule number from different parts of the plant (for review, see refs. 2 and 3). Grafting experiments in which mutant shoots were grafted onto wild-type roots and the reciprocal experiments of wild type shoots grafted onto mutant roots revealed that for some genes, the genotype of the shoot determined the number of nodules that formed on the root, while for other genes, it was the genotype of the root which determined the number of nodules that formed.2,3 We recently identified a gene, RDN1 (ROOT DETERMINED NODULATION 1), that controls nodule number from the root but is expressed in both shoots and roots.4 A fusion of the RDN1 promoter to the uidA (GUS) gene gave staining in the vasculature of transgenic roots.4
The SUNN gene in M. truncatula encodes a leucine-rich repeat receptor kinase that controls nodule number from the shoot but like RDN1 is expressed in both shoots and roots.5 To assess tissue-level expression of SUNN, we constructed a transgenic M. truncatula plant containing 1360 bp of sequence upstream of SUNN driving expression of an mGFP-GUS gene fusion. This construct includes the entire region of 5′ DNA between the start of the SUNN coding sequence up to and including the next genetic landmark, a genomic MITE insertion. The construct was transformed into wild type (A17) M. truncatula using the protocol of Zhou, et al.6 and detection of GUS was via the protocol of Jefferson et al.7
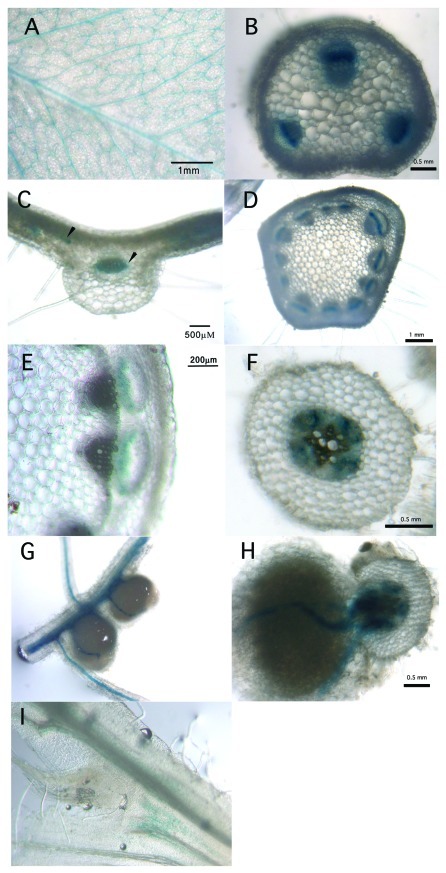
The expression of this construct in T3 plants gave a very similar pattern of expression to that reported for the RDN1 reporter construct. The GUS staining pattern indicated expression in the vasculature of many tissues including leaves, petioles, stems and roots (Fig. 1). The staining was often in cell layers adjacent to phloem cells in tissues examined under higher magnification. For example, staining was seen in the procambium in petioles (Fig. 1B) and stems (Fig. 1D and E), and the cells surrounding the primary phloem in roots (Fig. 1F). Despite the fact that SUNN regulates nodule formation, nodule expression was limited to the vasculature (Fig. 1G and H). No staining was observed in shoot meristematic tissue with faint staining in the vasculature leading to the meristem (Fig. 1I). These results were consistent with those of Nontachaiyapoom et al. who examined the orthologous soybean NARK and Lotus HAR1 promoters in a L. japonicus background.8 They reported sequence conservation between the promoters of the Lotus, Medicago and Glycine orthologs including a sequence element driving vascular specific expression and our results confirm this experimentally in the Medicago background.
Figure 1. Expression of a SUNNpro:mGFP-GUS fusion in wild-type plants. A Nikon E600 microscope with a Retiga EXi-FAST monochrome CCD 12-bit camera was used for visualization and imaging of some stained samples and a Zeiss Lumar.V12 stereoscope equipped with AxioCAM MRC and AxioVision software (Zeiss) for others. Leaf, stem and root transverse sections were made by hand using a razor blade. The sections were routinely ~0.2mm in thickness. (A) Staining in the veins in leaflets; (B) cross section of petioles; (C) cross section of leaflet with arrows indicating staining in vasculature; (D) cross section of stem; (E) close up of stem showing staining in phloem; (F) cross section of root; (G) mature nodules; (H) cross section of root and nodule and (I) faint staining in the vasculature leading to the meristem from the same plant used for (E), but no meristematic staining.
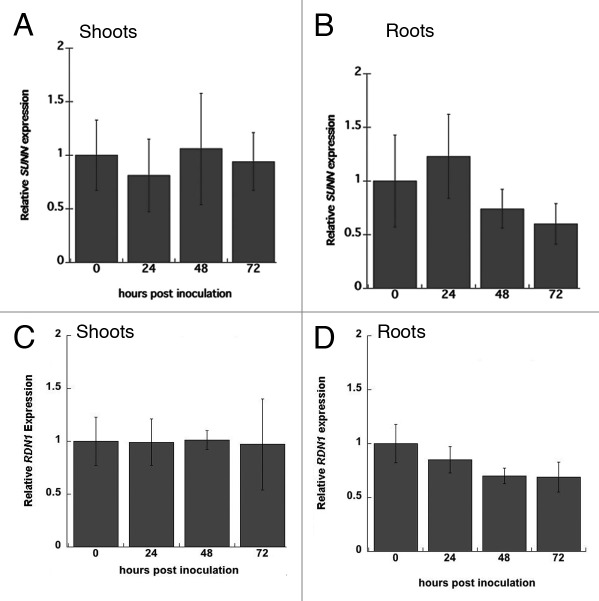
Because both the RDN1 and SUNN genes were expressed in the vasculature and regulate nodule number, we asked if SUNN or RDN1 transcription had similar expression level patterns in response to rhizobia. We examined SUNN and RDN1 expression in wild-type shoots and roots over a time course of early nodulation with Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR), using protocols from our previous work.9 We saw little change in expression levels in either gene in root and shoot tissue at 24, 48, or 72 h after addition of rhizobia (Fig. 2). Values show no statistical difference with p > 0.05 for all comparisons by Student’s t test.
Figure 2. Quantification of gene expression. Results of reverse transcription quantitative PCR of SUNN and RDN1 gene expression in shoots and roots of wild type plants during early nodulation signaling. Expression is shown relative to expression at time 0 (no rhizobia) for SUNN in shoots (A) and roots (B) as well as for RDN1 in shoots (C) and roots (D). Mean of three independent biological replicates of 3–5 plants per time point and three technical replicates. Error bars are standard error of the mean.
The presence of SUNN and RDN1 in vascular tissue has mechanistic implications. We speculate that the function of the SUNN kinase and/or RDN1 may affect phloem loading/unloading of auxin or other molecules. Outside of legumes, the SUNN kinase is most closely related by sequence to the Arabidopsis CLV1 kinase and related proteins in monocots involved in meristem maintenance,5 and AtCLV1, in addition to its well characterized expression in apical meristems, is expressed in the phloem companion cells.10 Combined with the observation that the RDN1 protein is completely unknown in function but highly conserved across all green plants including moss,4 the functions of RDN1 and SUNN cannot be limited to signal transduction in the autoregulation of nodulation. The fact that they are expressed in the vasculature with little change in expression in response to rhizobia could suggest the two proteins are involved in the same signal transduction event and we are investigating this further. The observation that SUNN regulates from the shoot while RDN1 regulates from root, yet both are expressed in both shoot and root tissues, most likely indicates there are other as yet undiscovered genes involved in long distance nodulation signaling events.
Acknowledgments
Support for this work is from the National Science Foundation (grant # IOB-0641848) and undergraduate research support through the Clemson Calhoun Honors College and the Howard Hughes Medical Institute to J.F. We would like to thank past undergraduate researcher Richard Gillette for contributions to the work and Rachael Keith for delivering the M. truncatula calli. We would also like to thank Harry Kurtz, Jr. for the use of his microscope. The preparation of the manuscript was supported in part by the Genomic Science Program, US. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area (pmi.ornl.gov). Technical Contribution # 5182 of the Clemson Experiment Station.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18491
References
- 1.Crawford N, Kahn M, Leustek T, Long S. Nitrogen and sulfur in Buchanon, BB, Gruissem, W. and Jones, RL, eds. Biochemistry and Molecular Biology of Plants, Rockville, MD: American Association of Plant Biologists, 2000, p. 786-849. [Google Scholar]
- 2.Reid DE, Ferguson BJ, Hayashi S. Lin, Y. and Gresshoff, PM. Molecular mechanisms controlling legume autoregulation of nodulation. Ann Bot (Lond) 2011;108:789–95. doi: 10.1093/aob/mcr205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortier V, Holsters M, Goormachtig S. Never too many? How legumes control nodule numbers. Plant Cell Environ. 2011 doi: 10.1111/j.1365-3040.2011.02406.x. In Press. [DOI] [PubMed] [Google Scholar]
- 4.Schnabel EL, Kassaw TK, Smith LS, Marsh JF, Oldroyd GE, Long SR, et al. ROOT DETERMINED NODULATION 1 regulates nodule number in M. truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 2011;157:328–40. doi: 10.1104/pp.111.178756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnabel E, Journet EP, Carvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN gene encoding a CLV1-like leucine-rich repeat receptor kinase regulates both nodule number and root length. Plant Mol Biol. 2005;58:809–22. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, Chandrasekharan MB, Hall TC. High rooting frequency and functional analysis of GUS and GFP expression in transgenic Medicago truncatula A17. New Phytol. 2004;162:813–22. doi: 10.1111/j.1469-8137.2004.01065.x. [DOI] [PubMed] [Google Scholar]
- 7.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–7. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nontachaiyapoom S, Scott PT, Men AE, Kinkema M, Schenk PM, Gresshoff PM. Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants. Mol Plant Microbe Interact. 2007;20:769–80. doi: 10.1094/MPMI-20-7-0769. [DOI] [PubMed] [Google Scholar]
- 9.Schnabel E, Mukherjee A, Smith L, Kassaw T, Long S, Frugoli J. The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol. 2010;154:1390–1402. doi: 10.1104/pp.110.164889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011;21:345–52. doi: 10.1016/j.cub.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]