Abstract
Bacteriophages offer interesting alternatives to antibodies for the specific capture and detection of pathogenic bacteria onto biosensing surfaces. Procedures for the optimal chemical immobilization of lytic bacteriophages onto surfaces are presented. More specifically, the removal of lysate contaminants from bacteriophage suspensions by size exclusion chromatography significantly increases the resultant planar surface density of immobilized bacteriophages. E. coli T4 and Salmonella enterica serovar Typhimurium P22 phage systems seem to undergo highly heterogeneous adsorption to the surface, possibly explaining the observed phage clustering at higher surface densities. The T4 phage and its E. coli host were initially employed as a model system where we discovered an optimal planar surface density of phages for best bacterial capture: 18.9 ± 0.8 phages/μm2 capturing 18.0 ± 0.3 bacteria/100 μm2. Phage surface clustering ultimately limits the T4 phage-immobilized surface’s ability to specifically capture its host bacteria. Nevertheless, this is to our knowledge the largest surface capture density of E. coli reported using intact T4 bacteriophages. Two additional purified bacteriophage systems (P22 and Campylobacter jejuni phage NCTC 12673) were then similarly studied for their ability to capture their corresponding host bacteria (Salmonella enterica serovar Typhimurium and Campylobacter jejuni respectively) on a surface.
Keywords: bacteriophage, biosensor, food contamination, pathogenic bacteria, purification, surface adsorption, virus immobilization
Introduction
Food-borne infectious diseases are a major global health concern. Enteric diseases are the second leading cause of child death worldwide killing nearly 1.7 million children every year.1 According to the World Health Organization (WHO), the bacteria: Campylobacter, Salmonella and E. coli O157:H7 are the three most prominent disease-causing food-borne contaminants.2 The development of a quick, low-cost, easy to use, portable food-testing device would be transformative in the establishment of adequate food safety programs throughout the developing world—diminishing the reliance on costly laboratory infrastructure.
Bacteria are routinely detected and identified by microscopy, colony-forming assay, PCR3 and ELISA.4 More recently bacteriophages have been used in a phage amplification assay5 and in fluorescence microscopy with labeled phages.6 These methods however are time-consuming, labor-intensive, and require specialized laboratory skills. There are rapid biosensor platforms being developed for microcantilever, surface plasmon resonance, quartz crystal microbalance and impedometric-based detection.7 However, these systems are dependent on the capture of the analyte on an interface.
Bacteriophages have several advantages over antibodies that are conventionally used as probes for bacterial detection. Bacteriophages are stable macromolecular assemblies that are relatively insensitive to temperature, pH, and ionic strength compared with antibodies. In fact, many phages can maintain their ability to infect for decades.8 They are also easy to produce by simple infection of their host bacteria whereas antibody production (monoclonal and polyclonal) is expensive and complicated.9
Bacteriophages initiate infection of their hosts by adsorption and then molecular recognition of the bacterial cell surface. The phage tails that bind to host cell surface polysaccharides or proteins mediate the recognition.10,11 Phage recognition of its host is commonly specific enough to differentiate between strains of the same species and this unique recognition makes bacteriophages an excellent choice as probes for selective detection of their host pathogen. Furthermore, bacteriophages are considered the most widely distributed biological entity in the biosphere, with an estimated population density of ~10 million/cm3 in any environmental niche where bacteria reside.12 We believe that this incredible biodiversity is a major strength of the intact phage approach.
Reporter bacteriophages are unique systems that have been developed for detection of bacteria exploiting the specific recognition of these viruses. A reporter bacteriophage carries a reporter gene that is delivered into the host bacteria upon infection and is expressed by the bacterial molecular machinery enabling their identification. Bacteriophages by themselves are incapable of expressing the gene and do not show signal until the gene is delivered into the host and thus a positive expression of the gene is a direct indicative of the presence of the host bacterium. Several reporter phages such as luciferase reporter phages (lux13 and luc14), ice nucleation reporter phages,15 fluorescent dye labeled phages,16 lacZ reporter phages17 etc have been used for target organisms including Salmonella,13 E. coli,16 Listeria18 and Mycobacterium.14 Hagens et al. give a detailed account of use of reporter phages for the detection of food born pathogens19 while Smartt et al. describe the general application of this technology in a recent review.20 However, use of biosensors for bacterial detection has gained tremendous popularity for improved detection limits and possibility of developing point of care devices for fast and accurate assessment. Improving the strategy for bacteriophage immobilization on a biosensor platform has therefore become a field of active research in the recent years.
All previous literature discussing surface-immobilized bacteriophages for the capture of bacteria use partially-purified phage suspensions.21-24 Propagated phages and the resultant lysate are full of contaminants derived from the bacterial host, such as lipopolysaccharides (endotoxin), peptidoglycan fragments, flagella and proteins. These previous studies do describe some preliminary purification steps from the crude lysate. Bacteriophage purification and concentration by CsCl gradient, PEG precipitation, ultrafiltration, and ultracentrifugation are the most common methods. However, these methods are either not efficient enough to remove most contaminants from the preparation after one purification, are time-consuming or produce a low yield. Recent advances in bacteriophage purification are chromatographic methods. Ion exchange chromatofocusing is possible, but would requires determination of the phage pI and stability of the phage through a pH range for each phage system under study.25 Sephacryl S-500 size exclusion chromatography (SEC) has also been demonstrated,26 but most phages would be larger than its exclusion limit and would elute in the void volume with other large contaminants.
SEC by Sephacryl S-1000 proves to be an excellent, simple, and versatile method for purification of entities such as bacteriophages < 400 nm in diameter27—which constitute a very large set of the known range of phage diversity. Concentrated phage preparations can easily be loaded onto the column and purified phage eluent collected automatically as is typically done by most FPLC systems. The separation is non-destructive and can occur under mild conditions (pH 7, room temperature, PBS eluent).
Previous work with unpurified T4 suspensions has shown that the use of covalent bonding in surface attachment gives a density of 18 ± 0.15 phages/µm2.24 We report here a substantial increase of phage surface density when chromatographically purified suspensions are rather used, resulting in an improved surface coverage for the purpose for capturing the host pathogen. This has in turn resulted in a marked improvement of E. coli capture density. Phage surface clustering ultimately limits the T4 phage-immobilized surface’s ability to specifically capture its host bacteria. Nevertheless, this is to our knowledge the largest surface capture density of E. coli reported using intact T4 bacteriophages. We extended this study to two other phage suspensions (P22 and NCTC 12673), which also show significant improvement in phage surface density.
Most importantly, such improvement of phage binding allowed a rigorous study of the surface attachment isotherm. Our analysis reveals that phage attachment to the surface does not obey the idealized Langmuir isotherm, but rather fits closest to the Brouers-Sotolongo isotherm,28 suggesting that a highly heterogeneous surface exists. We assert that phages initially attaching to the surface could be providing lower-energy sites for additional phage attachment, thus explaining the extensive surface aggregation, or clustering of phages, observed at higher phage titers. Finally, we have also applied these improvements to demonstrate the real-time capture of E. coli using surface plasmon resonance (SPR) with a T4-immobilized surface.
Results and Discussion
Purification of bacteriophages by size exclusion chromatography
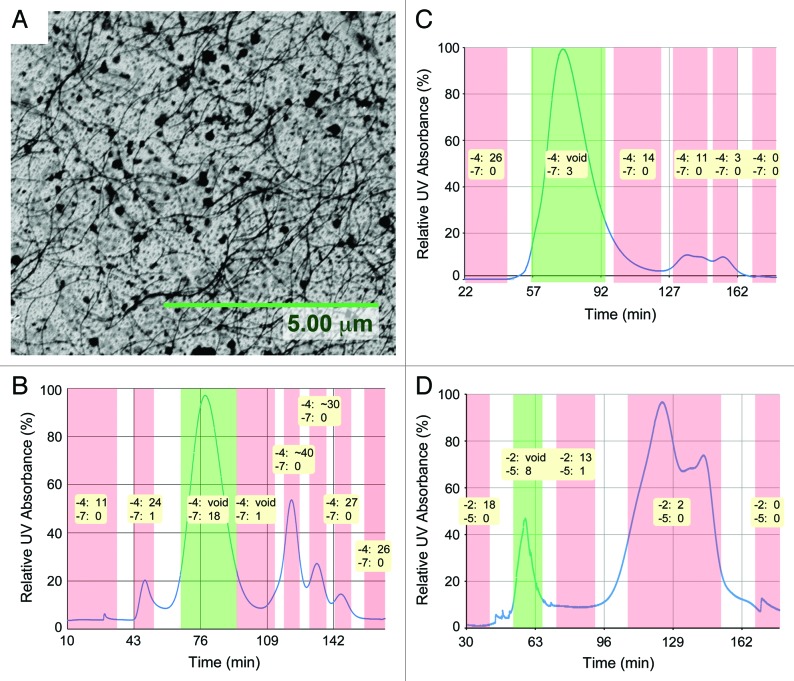
This study was initially prompted by the observation of large quantities of Salmonella flagella fragments in ultracentrifugation-purified P22 phage preparations (Fig. 1A). It was quickly observed that the presence of these fragments on the capture surface severely interfered with the capture of the host Salmonella by the immobilized phage. Similar flagella fragments can also be seen by AFM in other work on P22-immobilized surfaces.22 Thus, there was a need for an alternative purification method to remove these bacterial contaminants after phage propagation. We used size exclusion chromatography to further purify the ultracentrifuged phage preparation. In size exclusion chromatography (SEC), the parameter governing the retention of a solute is its hydrodynamic volume or Stokes radius.29 The flagella fragments are expected to have a much longer Stokes radius and were therefore likely to elute first, as was observed by the first peak in the P22 chromatogram (Fig. 1B).
Figure 1. (A) Hair-like contaminating flagella fragments are commonly seen on unpurified P22-immobilized surfaces. Size exclusion chromatograms of (B) P22, (C) T4 and (D) NCTC 12673 phage preparation run on Sephacryl S-1000 solid support. Plaque assay results (10−4 and 10−7 for T4 and P22; 10−2 and 10−5 for NCTC) are shown for each collected fraction; “void” results are multiple overlapping plaques that are uncountable, creating a large void in the bacteria overlay.
The first run for a new phage sample was done on the XK 16/70 column, loading a 1 mL sample of ultracentrifuged phage preparation. Of the resultant chromatogram, each major fraction was diluted by 10−4 and 10−7 and then plaque assayed to identify the phage peak. The phage peak is the first peak in the T4 (Fig. 1C) and NCTC 12673 (Fig. 1D) chromatograms, while it is the second peak in the P22 chromatogram. The presence of other peaks confirms that other contaminating proteins exist in the samples.
Phage purification was scaled up to a 12 mL sample delivery on the XK 26/70 column; the sample volume to column volume ratio was correspondingly scaled up from 0.83% to 3.77%. With the increase in sample proportion, the column height equivalent of theoretical plate (HETP) reduced for P22 and NCTC 12673, but increased for the T4 samples. The purification of a 12 mL sample of phage lysate could be achieved successfully in 45 min, however the entire experiment including column equilibration, sample loading, elution and column washing could be performed in less than 3 h.
CsCl gradient ultracentrifugation has been the most popular method of phage lysate purification for removal of bacterial protein contaminants. Gradient ultracentrifugation as a stand-alone technique for purification however does not result in complete removal of the contaminant protein despite repeated cycles.30 It has been successfully coupled with PEG precipitation to achieve better purification but is still time consuming (several hours) and laborious since it involves PEG precipitation, several rounds of ultra-high centrifugation followed by ultrafiltration and subsequent removal of CsCl from the purified phage sample.31 Comparatively, our method is much faster than CsCl gradient ultracentrifugation and achieve the purification of 12 mL of phage lysate in less than 3 h considering the column equilibration time, sample loading and elution time and column washing time.
Covalent attachment of bacteriophages to reactive surfaces
The phage surface density improves significantly to 54 ± 7 phages/μm2 using Sephacryl S-1000 purified T4 bacteriophages, a 9-fold improvement over ultracentifugation-purified T4 phages. Similarly with purified P22, phage density improves to 199 ± 2 phages/μm2, a 25-fold improvement over unpurified P22 phages (Fig. 2).
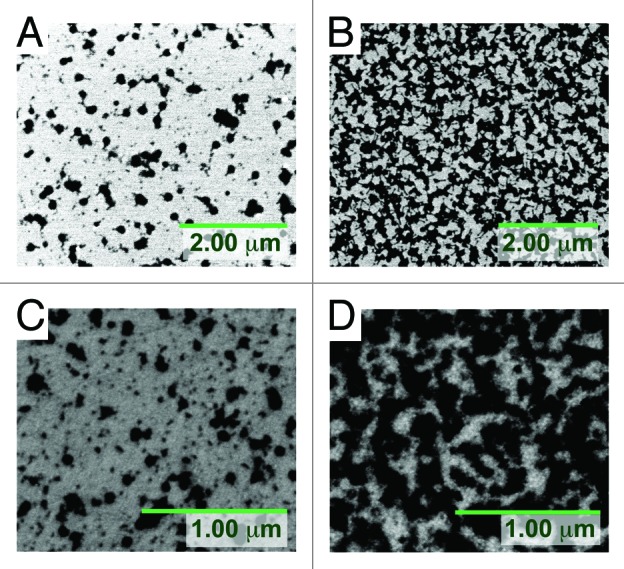
Figure 2. Covalent attachment of unpure and purified bacteriophages on planar Au-DTSP surface: (A) unpure T4: 5.8 ± 0.7 phages/μm2; (B) pure T4: 54 ± 7 phages/μm2; (C) unpure P22: 8.2 ± 0.1 phages/μm2; (D) pure P22: 199 ± 2 phages/μm2.
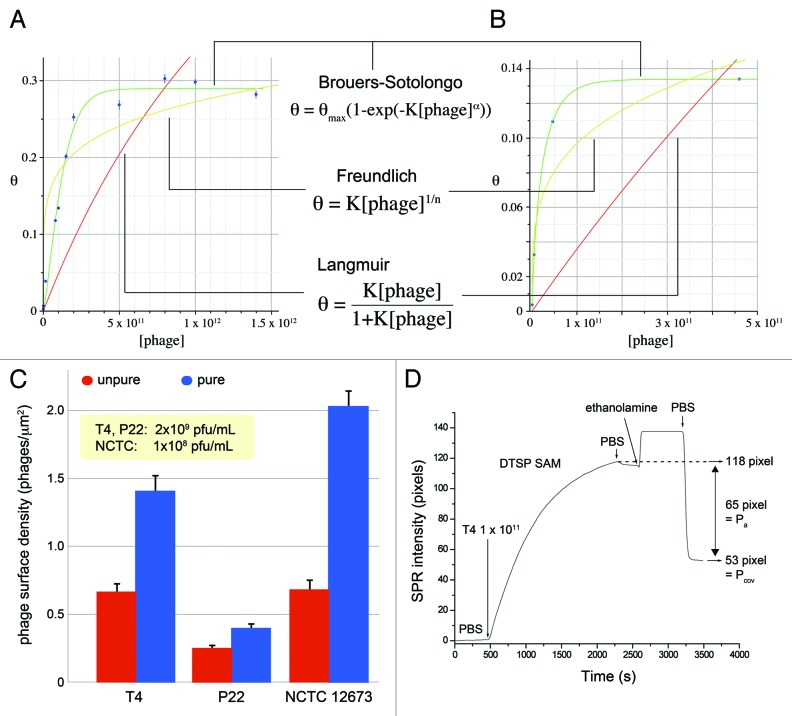
The use of purified phages for covalent attachment is allowing us to approach a jamming (maximum) surface coverage (Fig. 3A and B). The jamming coverage is the steric limit to further adsorption, at a specific surface attachment reaction temperature. To the best of our knowledge, this has never been demonstrated before with tailed bacteriophages. The purified T4 samples show a jamming surface coverage of 0.29, while purified P22 show a jamming coverage of 0.13 at 40°C. We could not observe jamming surface coverage for NCTC 12673 due to its low sample titer.
Figure 3. θ vs. [phage] curves for two phage systems, (A) T4 and (B) P22. θ is the fraction of total area of adsorbed analyte over the total surface area, called surface coverage. Langmuir: T4, R2 = 0.42; P22, R2 = 0.15. Freundlich: T4, R2 = 0.81; P22, R2 = 0.87. Brouers-Sotolongo: T4, R2 = 0.99, θmax = 0.29, K = 4.96 × 10−15, α = 2.68; P22, R2 = 0.99, θmax = 0.13, K = 5.26 × 10−9, α = 0.80. (C) Diffusion-controlled transport of bacteriophages to Au-DTSP planar surface, using low bulk concentration of phage to prevent phage clustering and steric hindrance to attachment. Pure/unpure ratios: T4W = 2.1x; P22 = 1.6x; CP1 = 3.0x. (D) Surface plasmon resonance plot of purified T4 immobilization to planar Au surface treated with reactive DTSP self-assembled monolayer.
Figure 3C shows that the purified phage suspensions exhibit higher surface densities. There should be diffusion-controlled transport to the substrate surface within a thin layer (known as a boundary layer) over the reactive surface.32 Assuming the contaminating proteins and phage are spherical particles, the smaller contaminating proteins should diffuse faster through this layer as governed by the Einstein-Stokes equation. They would thus out-compete the bacteriophages in adsorbing to and reacting on the surface. Following Adamczyk et. al’s studies of diffusion-controlled irreversible adsorption of micron-sized latex particles to mica surfaces,33 we attempted to approach diffusion-controlled transport to our Au-DTSP surfaces, minimizing the role of convection or external force. We also observe differences between the three bacteriophage species in attachment efficiency to the Au-DTSP surface which is probably due to the difference in phage surface amino acid composition (varying number of displayed amino groups).
Our SPR study reveals the online surface attachment kinetics for purified bacteriophages (Fig. 3D). Total phage attachment to the surface approaches equilibrium at around 2200 sec for T4 with a bulk suspension concentration of ~1 × 1011 pfu/mL. After an ethanolamine blocking step and PBS rinses, ~65 SPR pixels of phage washed off the surface, with 53 pixels remaining. It is the remaining phages that are likely to be primarily covalently attached to the surface, while those that washed off were only physically adsorbed.
Phage surface attachment model and clustering of bacteriophages on planar surfaces
Purified bacteriophage suspensions allow an opportunity to study their surface attachment more rigorously. In Figure 3A and B the data are empirically fit to adsorption isotherms by a nonlinear fitting procedure. Covalent attachment of purified phages to the surface does not seem to follow the idealized Langmuir adsorption isotherm, which assumes that all available adsorption sites have equivalent sorption energies. The empirical Freundlich equation is based on sorption onto a heterogeneous surface.32 However, in our data where a saturation regime is clearly observed, the Freundlich isotherm and its surface heterogeneity assumption will no longer be appropriate.34 The Brouers-Sotolongo isotherm (BSI) (a deformed Weibull exponential isotherm) has been employed in previous studies to analyze sorption processes on highly heterogeneous surfaces.28,34 It was observed that the BSI empirically best fits to our data as well.
Nevertheless, a limitation with this adsorption isotherm fitting analysis is that it assumes that only an adsorption process occurs and that all the adsorbed phages remain on the surface prior to characterization. We know that this is not true as indicated by our SPR real-time phage immobilization study (Fig. 3D). The SPR analysis shows that at the end of the phage immobilization step, the substrate surface is populated by covalently attached and adsorbed phages. Therefore we do not account for the covalent attachment process in our understanding of phage surface attachment in Figure 3A and B.
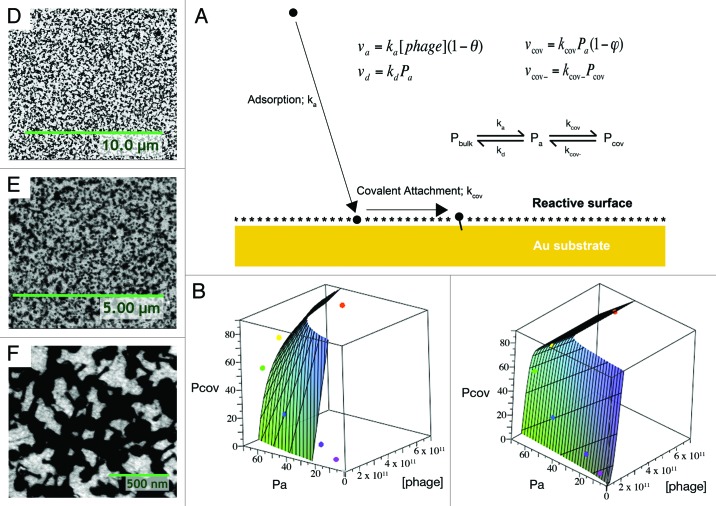
Thus, a two-step phage surface attachment model was derived (Fig. 4A). Based on SPR online binding plots at several T4 phage concentrations, we approach a limit in the phage attachment step where d(Pa + Pcov)/dt ≈ 0. From this assumption, we can derive phage surface attachment models based on Langmuir-type adsorption (Equ. 1):
Figure 4. (A) Schematic of theoretical phage surface attachment mechanism and rate equations (assuming Langmuir-type adsorption). θ is the surface coverage of the phage; φ is the fraction of reactive surface sites hydrolyzed. (B) Fit to Langmuir-derived model; R2 = 0.89, AIC = 69.1, KL = 0.019, ε = 118. (C) Fit to BSI-derived model: R2 = 0.98, AIC = 67.6, θmax = 2.69, KBSI = 1.04 × 10−30, α = 2.75. Observed phage surface clustering: (D) T4 and (E) P22 phage-immobilized surfaces; (F) a T4 surface at 50k magnification exhibiting tails buried within clusters.
| (1) |
where Pcov is SPR pixels corresponding to covalently bonded phages (as illustrated on an SPR phage binding plot, Fig. 3D), Pa is SPR pixels corresponding to adsorbed phages, that are removed by washing (as illustrated on an SPR phage binding plot, Fig. 3D), Atot is the total surface area of the SPR chip accessible for phage binding, Aph is the estimate of the planar surface area of surface attached phages, determined by SEM images, KL is the Langmuir adsorption constant, [phage] is phage titer in suspension, β is Aph error term and ε is overall error term.
The equation (Equ. 2) for BSI-type adsorption similarly can be given as:
| (2) |
where θmax is the maximum surface coverage (can be empirically seen on θ vs. [phage] plots, Fig. 3A and B), KBSI is the Brouers-Sotolongo adsorption constant and α is a constant defined in the derivation of the Brouers-Sotolongo adsorption isotherm28 that is a “measure of the average sorption energy and width of the sorption energy distribution”.
Our SPR data for the surface attachment of T4 phage fits somewhat to the Langmuir-derived model (Fig. 4B), but it fits best to the BSI-derived model (Fig. 4C). This model also has a lower Akaike Information Criterion (AIC) and thus its better fit is not due to excessive free parameters or overfitting.35 The Brouers-Sotolongo isotherm is derived from the notion that there is a Pareto distribution of sorption energies on the surface. Also, α is a measure of the average sorption energy and width of the sorption energy distribution28; both decrease when α increases.
For the T4 system, the α derived from fitting to the surface attachment model (2.75) and that derived from the θ vs. [phage] curve (2.68) are relatively close. Therefore fitting to θ vs. [phage] data may be a reasonable representation of surface attachment behavior for deriving α. In the P22 system the α derived from a θ vs. [phage] curve is much lower (0.80). The significant difference in α between the two phage systems suggest that it is the phages themselves that are perhaps involved in imparting this surface heterogeneity. Therefore, we suggest that initial attachment of phages is perhaps providing lower energy sites for subsequent adsorption.
In fact it follows that SEM micrographs show significant phage surface clustering (Fig. 4D–F), particularly when approaching the jamming coverage. In the case of T4 phages, one can clearly observe phage tails buried within clusters (Fig. 4F), which may render them surface inaccessible to specifically capture their bacterial hosts. T4 starts to noticeably form surface clusters beyond 2 × 1011 pfu/mL bulk concentration (phage surface density of 23.7 ± 0.1 phages/μm2). The same observation was made for P22 phage immobilization beyond bulk concentration of 4.6x1010 pfu/mL (phage surface density of 33.6 ± 0.2 phages/μm2). The smaller α (larger average sorption energy) for P22 may also explain why we observe more favorable clustering with this phage. We did not observe significant phage clustering with NCTC 12673 due to the low sample titers obtained for these phages. Elsewhere, filamentous phage “bundling” has also been reported.36
Further work on mitigating phage surface clustering could involve varying the ionic strength to dampen any possible electrostatic interactions between phages. Surface patterning approaches37 to guide phage attachment should also be explored.
Strain-specific bacterial capture by bacteriophage-coated surfaces
The capture of bacteria by bacteriophage-immobilized surfaces is highly strain-specific as has been reported previously.24 In that work, the control non-host strains of E. coli (6M1N1, NP10 and NP30) do not show significant binding to the T4 phage-immobilized surface. The SEM images in Figure 5A–C shows the successful specific capture of the three host bacterial pathogens (E. coli K12, C. jejuni 11168H and S. Typhimurium (ATCC 19585)) on the corresponding phage-immobilized surfaces (T4, NCTC 12673 and P22 respectively).
Figure 5. Specific capture of (A) E. coli K12, (B) C. jejuni 11168H and (C) Salmonella Typhimurium (ATCC 19585). (D) E. coli K12 capture density vs. T4 phage surface density plot. Bacterial capture density peaks at ~19 phages/μm2. (E) Real-time and rapid surface plasmon resonance detection of 107 and 108 cfu/mL E. coli K12 captured by the T4 phage-immobilized surface.
Intuitively one might expect that maximizing phage surface density (approaching the jamming coverage) should correspond to maximal specific bacterial binding to the surface. However, we show that this is not true as is indicated from the bacterial capture density analysis in Figure 5D. Instead, we determined a near-optimal phage surface density for best bacterial capture for the model T4 system. We observed that 18.9 ± 0.8 phages/μm2 phage surface density gives the best host bacterial capture density (18.0 ± 0.3 bacteria/100 μm2). To the best of our knowledge, this is the largest surface capture density of E. coli reported using intact T4 bacteriophages. The host bacterial capture density drops off above this optimal phage surface density. This is likely due to increasing phage surface clustering that is causing some tails to become inaccessible, thus decreasing the effective molecular recognition probe density on the surface.
Similar behavior was observed for the P22 system, with a near-optimal phage surface density of 10 ± 1 phages/μm2 capturing 4.1 ± 0.1 bacteria/100 μm2; although Salmonella counting was difficult due to their autoagglutination or clumping. For C. jejuni, the NCTC 12673 phage sample titer we could produce was too low (108 pfu/mL), making it exceedingly difficult to generate a relevant range of phage surface densities.
But, at similar phage surface densities, the three different phage-immobilized surfaces were compared for their bacterial capture efficiency (Table 1). We determine that both P22 and NCTC 12673 surfaces have larger capture efficiencies than T4, at this relatively low phage surface density where phage clustering is not observed.
Table 1. Comparative bacterial capture efficiencies.
| Phage | Phage Surface Density (phage/μm2) | Bacterial Capture Efficiency (number of surface-immobilized phages per captured bacteria) | x more efficient than T4 |
|---|---|---|---|
| T4 |
3.65 |
1887 |
1 |
| P22 |
1.15 |
41 |
46 |
| NCTC 12673 | 1.37 | 3 | 612 |
Finally, SPR was employed to detect real-time E. coli bacterial binding to a T4 phage-immobilized surface (Fig. 5E). 107 and 108 cfu/ml concentration of the host E. coli K12 strain was employed to illustrate the bacterial capture while 109 cfu/ml non-host 6M1N1, NP10 and NP30 were used as controls that show negligible bacterial capture on the surface (data not shown). The host bacterial capture shows a distinct concentration dependent SPR intensity change that has been demonstrated by us previously.38
Building a generalized scheme to uncover other bacteriophages suitable for specific capture of any bacteria of interest
In this work, we have evaluated the ability of three bacteriophages to specifically capture their corresponding bacterial pathogens onto a surface. From our experience, we realize that a basic screening process can be elucidated (Fig. 6) that can be employed to evaluate a set of candidate bacteriophages against a bacterial pathogen of interest. First, each candidate phage should be amplified or concentrated to a titer of ~10^12 pfu/mL. The phage suspension should then be purified by size exclusion chromatography to remove contaminating proteins. For each phage system, the experiment should be designed to test the specific bacterial capture density across a range of phage surface densities (Fig. 5D). It is possible that one candidate phage may demonstrate a superior absolute host bacterial capture density on the surface, and a significantly better bacterial capture efficiency. This phage, selected through this screening process, can then be exploited for applications such as developing surface-based bacterial biosensor.
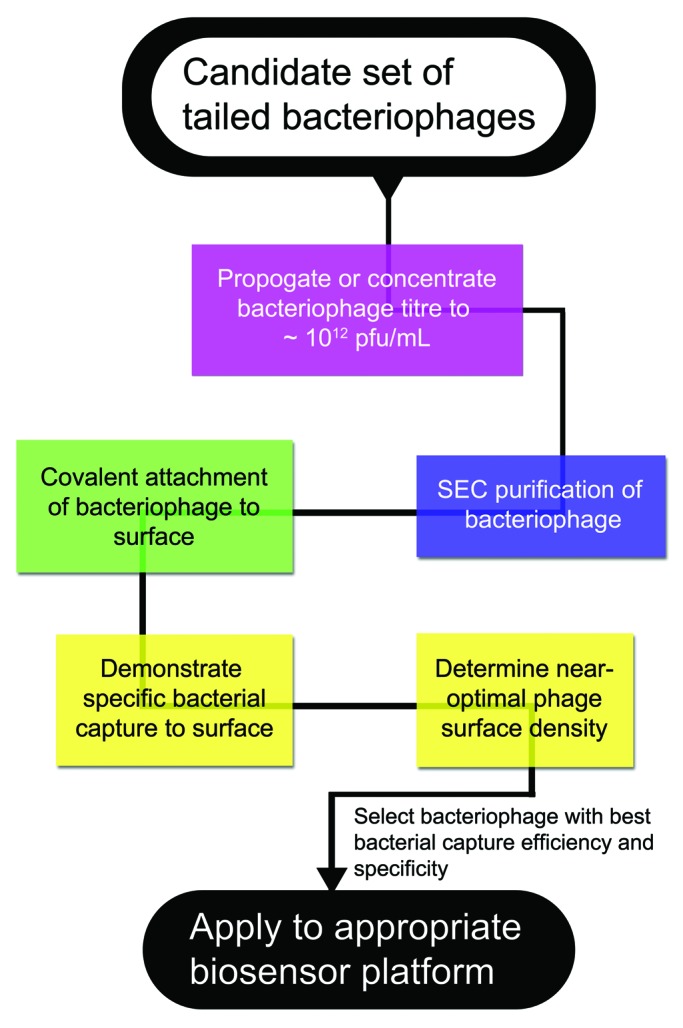
Figure 6. A generalized scheme for evaluating bacterial capture efficiency for a set of candidate phages.
Materials and Methods
Materials
Dithiobis(succinimidyl propionate) (DTSP), glutaraldehyde, cysteamine and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (D3669, G7651, M9768 and A7906 respectively). Tween-20 was obtained from MP Biomedicals, Inc. (11TWEEN201).
SPR gold chips were obtained from GWC Technologies and were washed successively in acetone, isopropanol, ethanol and water for 5 min each before use. The gold substrates were fabricated using piranha cleaned 3′′ silicon (100) substrate by sputtering a 5 nm thick chrome adhesion layer followed by 25 nm thick gold layer. The sputtered substrates were diced into 5 mm × 7 mm rectangular chips using a dicing saw machine.
E. coli K12 and T4 bacteriophages were kindly provided by Dr. Mansel Griffiths (University of Guelph).
Bacteriophage preparation
The amplification of wild-type, intact T424 and P22 phages39 was done using an established protocol described elsewhere. Briefly, T4 and P22 phages were amplified in E. coli K12 and Salmonella enterica serovar Typhimurium hosts respectively. T4 phages were amplified by incubating 100 μL of 102 pfu/mL phages in 4 mL of fresh log-phase E. coli K12 bacteria for 15 min. This mixture was then added to 250 mL of LB media at 37°C for 6 h. The LB media that turns turbid after the incubation was centrifuged at 4000 rpm for 10 min to remove bacteria followed by filtration of supernatant through sterile 0.22 μm filter. The filtered supernatant was ultracentrifuged and the pellet was resuspended in 1 mL SM buffer for titration. Similarly, 900 μL of 107 pfu/ml P22 phages were mixed with 3.6 ml of Salmonella culture and incubated at room temperature for 10 min. This mixture was added to 630 ml of LB and was incubated at 37°C, while shaking at 150 rpm for 15 h. The amplified phages were then centrifuged at 2500 rpm for 20 min to remove bacterial cells. The supernatant was filtered, ultracentrifuged and pellet was resuspended in 1ml SM buffer, filtered and titrated.
For NCTC 12673 phages, C. jejuni strain 12661 was grown on Mueller Hinton (MH) agar under microaerobic conditions for 18 h at 37°C. MH broth containing salts (filter sterilized 10 mM MgSO4 and 1 mM CaCl) was added to the plates and the cells were scraped off into tubes and placed in microaerobic conditions without shaking for 4 h. C. jejuni bacteriophage NCTC 12673 was serially diluted to 106 phage/mL in 1× SSC (150 mM NaCl, 15 mM sodium citrate, pH 7.0). Then, 100 μL of C. jejuni 12661 was added to 100 μL of the 106 phage/mL suspension and incubated together for 10 min at room temperature. The mixture was then added to MH broth with salts at 37°C under microaerobic conditions with shaking (125 rpm) for 24 h. The bacteriophage lysed culture was centrifuged at 4,300 g for 10 min at 4°C. The supernatant containing the phage was filtered through a 0.22 μm filter into tubes and stored at 4°C. The phage concentration was determined using a plaque assay (see below).
For all three phages, the filtered phage preparations were then ultracentrifuged at 310,000 g (at max. radius of fixed angle rotor) for 1 h at 20°C. The phage pellets were re-suspended in SM buffer overnight at 4°C.
A GE Life Sciences XK 16/70 column was packed with Illustra Sephacryl S-1000 Superfine beads to a bed height of ~60 cm. For the first run of a new phage, 1 mL of SM suspended unpurified phage sample was loaded onto the column. A Bio-Rad Biologic LP FPLC system was used to pump PBS buffer through the column at a 30 cm/h linear flow rate. Then, 1 mL fractions were collected and combined according to peak or chromatographic feature; then each fraction group was plaque assayed. From this the phage peak was identified.
For preparation, only the phage peak eluent was collected and concentrated by ultrafiltration using a Millipore Amicon Ultra-15 centrifugal filter device. The resultant purified phage filtrant was re-suspended in SM buffer, ready for latter surface attachment steps.
Plaque and colony assays
Bacterial enumeration was done by the plate count method and was expressed in cfu/mL, while the phage suspension concentration was determined by the soft agar overlay technique and expressed in plaque-forming units (pfu/mL).40 Specifically for NCTC 12673 phage quantification, phage samples were serially diluted in 1× SSC buffer. Each dilution was spotted (10 μL) onto MH agar plates overlaid with 0.6% MH agar containing 10 μL of C. jejuni cells and grown under microaerobic conditions for 24 h.
Reactive thiol monolayer formation on Au surface
First, 5 × 7 mm Au substrates were sonicated for 5 min in acetone followed by cleaning in isopropanol and ethanol for 5 min each. The surfaces were irradiated with UV light in ozone for 10 min and finally rinsed in Milli-Q water for 5 min prior to their surface functionalization. These cleaned Au substrates were immersed into a pre-reduced 2 mg/mL solution of dithiobis(succinimidyl propionate) (DTSP) in acetone for 30 min at room temperature on a shaker for self-assembled monolayer (SAM) formation. After this, the surfaces were rinsed with acetone to remove any unbound DTSP followed by rinsing in water. The DTSP modified surfaces were utilized immediately for immobilization of bacteriophages.
Immobilization of bacteriophages on surface
The DTSP SAM immobilized gold substrates were further washed in isopropyl alcohol, ethanol and PBS for 5 min each on an orbital shaker. The washed substrates were incubated in desired phage solution for 1 h to facilitate immobilization of the phages. The phage immobilized substrates were washed in PBS followed by 10% solution of ethanolamine in PBS to remove any physically adsorbed phages as well as neutralize any uncoordinated DTSP molecule. The surfaces were finally rinsed in PBS to remove excess of ethanolamine and were incubated for 30 min in 1 mg/ml solution of bovine serum albumin (BSA) in PBS. The BSA blocked surfaces were finally washed twice in PBS for 5 min on an orbital shaker and were used for bacterial capture studies.
Bacterial capture on phage immobilized surfaces
Fresh cultures of host bacteria were grown to obtain a concentration of 109 cfu/mL. The culture (1 mL) was then centrifuged and re-suspended in 1 mL of phosphate buffered saline (PBS). The phage-immobilized substrates were immersed in the bacterial culture for 20 min at room temperature. The substrates were then washed in TSB to remove excess stain and further thoroughly washed in 0.05% Tween-20 in PBS to remove loosely bound bacteria. For SEM imaging of bacterial capture, the samples were fixed in a 2% aqueous solution of glutaraldehyde for 1 h followed by washing twice with deionized water for 5 min on an orbital shaker. The samples were then dried under nitrogen gas flow prior to SEM characterization.
SPR studies
Surface plasmon resonance measurements (GWC Technologies) have been performed to reveal the online binding kinetics of purified T4 phages and their interaction with E. coli K12 as model system. The SF-10 glass substrate coated with 9 isolated circular spots of 45 nm gold was used for each SPR measurements. All the SPR measurements were acquired selecting one region of interest (ROI) from each spot and the data was plotted as the average from all the ROIs. The baseline for the SAM modified electrode is established using PBS (0.01 M, pH 7.2) for 400 sec, followed by the injection of purified phage suspensions into the SPR flow cell at a flow rate of 100 μL/min. The flow of T4 suspensions was performed for 30 min and then PBS was injected to wash the surface. Then, 10% v/v solution of ethanolamine in PBS was introduced into the flow cell and kept flowing for 10 min to block free succinimidyl groups and to remove physically adsorbed T4 phages from the surface. Flowing PBS at the flow rate of 200 μL/min further washed the surface and the change in the SPR pixels before and after T4 binding is recorded. The bacterial capture analysis was performed by flowing 107 and 108 cfu/ml concentration of host bacteria while 109 cfu/ml E. coli 6M1N1, NP10 and NP30 were used as non-host control strains which show negligible binding (data not shown). The bacterial suspension in PBS were flown on bacteriophage immobilized surface at a flow rate of 100 μL/min for 20 min followed by flowing PBS at the flow rate of 200 μL/min to facilitate the removal of loosely bound bacteria. The reduction in SPR intensity during the washing step confirms the removal of loosely/non-specifically bound bacteria from the surface.
To quantify the amount of binding, the SPR instrument is calibrated with different concentrations of ethanol in water, which revealed that a change of 9 SPR pixels corresponds to a change of 0.006 refractive index of surface. The change in refractive index of 0.001 in SPR corresponds to 1 ng/mm2 binding of protein.
Summary and Conclusion
There exists an incredible biodiversity of bacteriophages, with an accompanying body of basic scientific research that has accumulated over more than 50 years. Intact bacteriophages are therefore very attractive to be exploited as a new class of specific molecular recognition probes against bacteria. This is not the first publication demonstrating their usefulness, however we do present significant improvements. We reveal a current upper-limit on the covalently-attached tailed bacteriophage ability for specific bacterial capture to a planar surface. Further methods, such as patterned placement of phages to mitigate phage surface clustering, would need to be explored to raise this limit.
Alternative methods involve employing M13 filamentous phage display technology to display protein fragments for specific recognition of bacteria. Biotinylation of phage capsid proteins—to bias phage orientation more favorably—has been done, but requires genetic modification.41 Finally, one may derive proteins responsible for bacterial host recognition directly from phage tails, and then apply them onto a surface as a monolayer.39 While promising, there is currently no systematic way of identifying these proteins genomically—owing to the complex evolutionary history and biodiversity of bacteriophages. Also, tail-spike protein monolayers will be unable to initiate the lytic pathway; conversely this has been demonstrated by intact bacteriophage-immobilized surfaces.39 Bacterial pathogen lysis (killing) on the surface could be a useful additional functionality of intact phage-immobilized surfaces. For instance, it could be applicable to the development of antibacterial tubing and devices for use in hospitals to prevent biofilm formation.
Our method is simple and provides a platform to quickly evaluate many bacteriophages and their specificities. Although we demonstrate the application of bacteriophages for biosensor development, it is conceivable that the methodology will be useful for any other bioengineering applications involving the attachment of viruses to surfaces, and to basic and applied microbiological research.
Acknowledgments
Financial support for these studies was from the National Research Council of Canada and the Alberta Glycomics Centre. C.M.S. is an Alberta Innovates Scholar.
Glossary
Abbreviations:
- BSI
Brouers-Sotolongo isotherm
- cfu
colony forming unit
- pfu
plaque forming unit
- SAM
self-assembled monolayer
- SEC
size exclusion chromatography
- SEM
scanning electron microscope
- SPR
surface plasmon resonance
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/19079
References
- 1.Global burden of disease. World Health Organization, 2004. [PMC free article] [PubMed]
- 2.Food safety and foodborne illness. World Health Organization, 2007.
- 3.Oyofo BA, Thornton S, Burr D, Trust T, Pavlovskis O, Guerry P. Specific detection of Campylobacter jejuni and Campylobacter coli by using polymerase chain reaction. J Clin Microbiol. 1992;30:2613–9. doi: 10.1128/jcm.30.10.2613-2619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janyapoon K, Korbsrisate S, Thamapa H, Thongmin S, Kanjanahareutai S, Wongpredee N, et al. Rapid detection of Salmonella enterica serovar choleraesuis in blood cultures by a dot blot enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2000;7:977–9. doi: 10.1128/cdli.7.6.977-979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart GS, Jassim S, Denyer S, Newby P, Linley K, Dhir V. The specific and sensitive detection of bacterial pathogens within 4 h using bacteriophage amplification. J Appl Microbiol. 1998;84:777–83. doi: 10.1046/j.1365-2672.1998.00408.x. [DOI] [PubMed] [Google Scholar]
- 6.Mosier-Boss PA, Lieberman S, Andrews J, Rohwer F, Wegley L, Breitbart M. Use of fluorescently labeled phage in the detection and identification of bacterial species. Appl Spectrosc. 2003;57:1138–44. doi: 10.1366/00037020360696008. [DOI] [PubMed] [Google Scholar]
- 7.Zourob M, ed. Recognition Receptors in Biosensors. 1. Springer, 2010. [Google Scholar]
- 8.Kutter E, Sulakvelidze A, eds. Bacteriophages: Biology & Applications. Boca Raton, FL: CRC Press, 2005. [Google Scholar]
- 9.Pancrazio JJ, Whelan J, Borkholder D, Ma W, Stenger D. Development and application of cell-based biosensors. Ann Biomed Eng. 1999;27:697–711. doi: 10.1114/1.225. [DOI] [PubMed] [Google Scholar]
- 10.Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys J. 1996;71:2040–8. doi: 10.1016/S0006-3495(96)79402-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinbacher S, Baxa U, Miller S, Weintraub A, Seckler R, Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. 0-antigen receptors. Proc Natl Acad Sci USA. 1996;93:10584–8. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karam JD. Bacteriophages: the viruses for all seasons of molecular biology. Virol J. 2005;2:19. doi: 10.1186/1743-422X-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Griffiths MW. Salmonella detection in eggs using lux + bacteriophages. J Food Prot. 1996;59:908–14. doi: 10.4315/0362-028X-59.9.908. [DOI] [PubMed] [Google Scholar]
- 14.Bardarov S, Dou H, Eisenach K, Banaiee N, Ya S, Chan J, Jacobs WR, Riska PF. Detection and drug-susceptibility testing of M.tuberculosis from sputum samples using luciferase reporter phage: comparison with the Mycobacteria Growth Indicator Tube (MGIT) system. Diagn Microbiol Infect Dis. 2003;45:53–61. doi: 10.1016/S0732-8893(02)00478-9. [DOI] [PubMed] [Google Scholar]
- 15.Wolber PK, Green RL. Detection of bacteria by transduction of ice nucleation genes. Trends Biotechnol. 1990;8:276–9. doi: 10.1016/0167-7799(90)90195-4. [DOI] [PubMed] [Google Scholar]
- 16.Goodridge L, Chen J, Griffiths M. The use of a fluorescent bacteriophage assay for detection of Escherichia coli O157:H7 in inoculated ground beef and raw milk. Int J Food Microbiol. 1999;47:43–50. doi: 10.1016/S0168-1605(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 17.Goodridge L, Griffiths MW. Reporter bacteriophage assays as a means to detect foodborne pathogenic bacteria. Food Res Int. 2002;35:863–70. doi: 10.1016/S0963-9969(02)00094-7. [DOI] [Google Scholar]
- 18.Loessner MJ, Rudolf M, Scherer S. Evaluation of luciferase reporter bacteriophage A511:luxAB for detection of Listeria monocytogenes in contaminated foods. Appl Environ Microbiol. 1997;63:2961–5. doi: 10.1128/aem.63.8.2961-2965.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagens S, Loessner MJ. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol. 2007;76:513–9. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- 20.Smartt AE, Ripp S. Bacteriophage reporter technology for sensing and detecting microbial targets. Anal Bioanal Chem. 2011;400:991–1007. doi: 10.1007/s00216-010-4561-3. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian S, Sorokuvlova I, Vodyanoy V, Simonian A. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus - a surface plasmon resonance spectroscopic study. Biosens Bioelectron. 2007;22:948–55. doi: 10.1016/j.bios.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Handa H, Gurczynski S, Jackson M, Auner G, Walker J, Mao G. Recognition of Salmonella typhimurium by immobilized phage P22 monolayers. Surf Sci. 2008;602:1392–400. doi: 10.1016/j.susc.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shabani A, Zourob M, Allain B, Marquette C, Lawrence M, Mandeville R. Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal Chem. 2008;80:9475–82. doi: 10.1021/ac801607w. [DOI] [PubMed] [Google Scholar]
- 24.Singh A, Glass N, Tolba M, Brovko L, Griffiths M, Evoy S. Immobilization of bacteriophages on gold surfaces for the specific capture of pathogens. Biosens Bioelectron. 2009;24:3645–51. doi: 10.1016/j.bios.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Brorson K, Shen H, Lute S, Perez J, Frey D. Characterization and purification of bacteriophages using chromatofocusing. J Chromatogr A. 2008;1207:110–21. doi: 10.1016/j.chroma.2008.08.037. [DOI] [PubMed] [Google Scholar]
- 26.Zakharova MY, Kozyr A, Ignatova A, Vinnikov I, Shemyakin I, Kolesnikov A. Purification of filamentous bacteriophage for phage display using size-exclusion chromatography. Biotechniques. 2005;38:194–8. doi: 10.2144/05382BM04. [DOI] [PubMed] [Google Scholar]
- 27.GE Healthcare UK. Illustra Sephacryl S-1000 Superfine Product Booklet. Buckinghamshire, UK: GE Healthcare UK Ltd, 2006:1-23. [Google Scholar]
- 28.Brouers F, Sotolongo O, Marquez F, Pirard J. Microporous and heterogeneous surface adsorption isotherms arising from Levy distributions. Physica A. 2005;349:271–82. doi: 10.1016/j.physa.2004.10.032. [DOI] [Google Scholar]
- 29.Mikkelsen SR, Corton E, eds. Bioanalytical Chemistry. Hoboken, NJ: John Wiley & Sons, 2004. [Google Scholar]
- 30.Osborn M, Weiner A, Weber K. Large Scale Purification of A-Protein from Bacteriophage R 17. Eur J Biochem. 1970;17:63–7. doi: 10.1111/j.1432-1033.1970.tb01134.x. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey SB, Stanton B, Jensen S, Zuerner R. Purification and Characterization of VSH-1, a Generalized Transducing Bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 1997;179:323–9. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrade J, Hlady V. Protein adsorption and materials biocompatibility: a tutorial review and suggested hypotheses. In: Advances in Polymer Science: Biopolymers/Non-Exclusion HPLC. Berlin: Springer, 1986:1-63; DOI: 10.1007/3-540-16422-7_6. [Google Scholar]
- 33.Adamczyk Z, Szyk L. Kinetics of irreversible adsorption of latex particles under diffusion-controlled transport. Langmuir. 2000;16:5730–7. doi: 10.1021/la991433m. [DOI] [Google Scholar]
- 34.Ncibi M, Altenor S, Seffen M, Brouers F, Gaspard S. Modeling single compound adsorption onto porous and non-porous sorbents using a deformed Weibull exponential isotherm. Chem Eng J. 2008;145:196–202. doi: 10.1016/j.cej.2008.04.001. [DOI] [Google Scholar]
- 35.Sakamoto Y, Ishiguro M, Kitagawa G, eds. Akaike Information Criterion Statistics. Springer, 1986. [Google Scholar]
- 36.Huang S, Yang H, Lakshmanan R, Johnson M, Chen I, Wan J, et al. The effect of salt and phage concentrations on the binding sensitivity of magnetoelastic biosensors for Bacillus anthracis detection. Biotechnol Bioeng. 2008;101:1014–21. doi: 10.1002/bit.21995. [DOI] [PubMed] [Google Scholar]
- 37.Vega R, Maspoch D, Salaita K, Mirkin C. Nanoarrays of Single Virus Particles. Angew Chem. 2005;117:6167–9. doi: 10.1002/ange.200501978. [DOI] [PubMed] [Google Scholar]
- 38.Arya S, Singh A, Naidoo R, Wu P, McDermott M, Evoy S. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst. 2011;136:486–92. doi: 10.1039/c0an00697a. [DOI] [PubMed] [Google Scholar]
- 39.Singh A, Arya S, Glass N, Hanifi-Moghaddam P, Naidoo R, Szymanski C, et al. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens Bioelectron. 2010;26:131–8. doi: 10.1016/j.bios.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Russell DW, eds. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]
- 41.Gervais L, Gel M, Allain B, Tolba M, Brovko L, Zourob M, et al. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens Actuators B Chem. 2007;125:615–21. doi: 10.1016/j.snb.2007.03.007. [DOI] [Google Scholar]