Abstract
We investigate genes of lytic, Bacillus thuringiensis bacteriophage 0305ϕ8-36 that are non-essential for laboratory propagation, but might have a function in the wild. We isolate deletion mutants to identify these genes. The non-permutation of the genome (218.948 Kb, with a 6.479 Kb terminal repeat and 247 identified orfs) simplifies isolation of deletion mutants. We find two islands of non-essential genes. The first island (3.01% of the genomic DNA) has an informatically identified DNA translocation operon. Deletion causes no detectable growth defect during propagation in a dilute agarose overlay. Identification of the DNA translocation operon begins with a DNA relaxase and continues with a translocase and membrane-binding anchor proteins. The relaxase is in a family, first identified here, with homologs in other bacteriophages. The second deleted island (3.71% of the genome) has genes for two metallo-protein chaperonins and two tRNAs. Deletion causes a significant growth defect. In addition, (1) we find by “in situ” (in-plaque) single-particle fluorescence microscopy that adsorption to the host occurs at the tip of the 486 nm long tail, (2) we develop a procedure of 0305ϕ8-36 purification that does not cause tail contraction, and (3) we then find by electron microscopy that 0305ϕ8-36 undergoes tail tip-tail tip dimerization that potentially blocks adsorption to host cells, presumably with effectiveness that increases as the bacteriophage particle concentration increases. These observations provide an explanation of the previous observation that 0305ϕ8-36 does not lyse liquid cultures, even though 0305ϕ8-36 is genomically lytic.
Keywords: bacteriophage, deletion mutant, DNA sequencing, electron microscopy, fluorescence microscopy, informatics, long-genome, microbial biofilm
Introduction
Bacteriophages isolated in the past were selected for characteristics useful either for studies of molecular biology or for applications in the management of infectious disease-causing bacteria.1,2 The methods of bacteriophage isolation included both liquid culture in various broths and single-plaque propagation in broth-containing agar gels. A plaque-supporting agar gel typically had an agar concentration between 0.4 and 1.0%, as described in references recently reviewed.1 However, we have found that some bacteriophages do not propagate sufficiently to clear hosts during use of either of these methods. These latter bacteriophages include two aggregating RNA bacteriophages3 and an aggregating double-stranded DNA bacteriophage, 0305φ8-36 of Bacillus thuringiensis.4 The aggregation characteristic of these bacteriophages included the propagation-associated formation of aggregates that varied in size, and often had hundreds to thousands of bacteriophage particles. Subsequently, eukaryotic RNA virus, HTLV-1, was found to form similar aggregates during propagation.5,6 The aggregating bacteriophages were propagated in agarose gels with unusually low concentrations of 0.05–0.15% (reviewed in ref. 7).
Aggregating bacteriophages are potentially useful either for studies of ecology or for management of bacteria that have not been successfully managed with either antibiotics or the lytic bacteriophages traditionally used. Bacteriophage 0305φ8-36 is genomically lytic8 and is also phenotypically lytic when propagated in dilute agarose gels. The dilute agarose apparently contributes to the degree of host cell lysis because liquid cultures do not visibly clear during infection in the same broth, although the bacteriophage does propagate in liquid culture to a limited extent.9 These observations suggest a new paradigm in which difficult-to-detect, aggregating bacteriophages constitute a significant percentage of environmental bacteriophages. As previously reviewed, bacteriophages are present in biofilms10-12 and aggregating bacteriophages are potentially also a component of environmental biofilms.
Indeed, electron microscopy of thin sections provides compelling evidence that bacteriophage aggregates are at least sometimes a major structural component of human dental plaque, a biofilm.13 The biofilm-forming, aggregating bacteriophage of reference 13 has a nucleic acid (presumably DNA) that is contained in a shell that is indistinguishable in radius from that of 0305φ8-36. The tail is also similar. Whatever the details, aggregating bacteriophages are likely to have genes that were selected for advantages provided in the environment only. These genes will be non-essential in the laboratory. In the current communication, we test for such genes by isolating deletion mutants of 0305φ8-36. We continue by exploring extracellular events that might explain the finding that 0305φ8-36 is more virulent when forming plaques than it is in liquid culture.
Bacteriophage 0305φ8-36 is an appropriate model virus for selecting deletion mutants, because the 0305φ8-36 genome has been found8 to be non-permuted (all DNA molecules have the same ends) and terminally repetitious (the sequence at one end is repeated at the other). Based on work with other bacteriophages,14,15 a non-permuted, terminally repetitious genome is produced by cleavage from a concatemer of mature genomes by a sequence-specific nuclease. If a bacteriophage genome is non-permuted, the genome becomes shorter when genes are deleted, while the DNA-encapsulating shell remains the same size. The reasons are that (1) shell size is determined before packaging and (2) no mechanism exists to add redundant genomic DNA to fill the shell.14,15 The shortened genome of a deletion mutant exerts reduced pressure on the capsid, thereby making deletion mutants physically selectable because they are physically more stable (more resistant to elevated temperature) than the wild type bacteriophage.16,17
In contrast, elongation of a terminal repeat occurs when genes are deleted from bacteriophages with permuted genomes, which are also cleaved and packaged from a concatemer. The reason is that the volume of the internal cavity of the capsid’s shell determines the mature DNA length.18,19 Thus, in this case, deletion mutants are not more physically stable than the wild type bacteriophage. Unfortunately for deletion analysis, the genomes of all sufficiently characterized long-genome (> 200 Kb) bacteriophages, other than 0305φ8-36, are permuted to our knowledge. Thus, the work described here appears to be the first deletion analysis for any long-genome bacteriophage.
Results
The first deletion mutant, Δ(118,397–124,982)
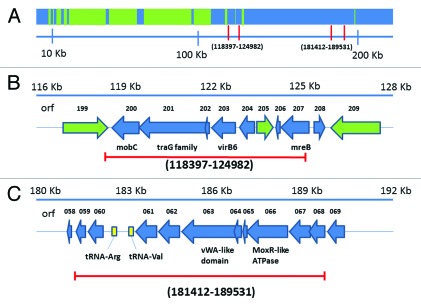
After selection for mutants stable to elevated temperature, we isolated three independent mutants, total, two of which were deletion mutants, based on the restriction endonuclease analysis described in the Materials and Methods Section. In Figure 1A, a pair of vertical red lines indicates each of the regions deleted, within a low-resolution map of the entire 218.948 Kb genome (6.479 Kb terminal repeat; 247 identified orfs).
Figure 1.
Maps of the bacteriophage 0305φ8-36 genome and its deletions. (A) A low-resolution map of the entire genome. (B) A high-resolution map of the Δ(118,397–124,982) deletion with distance from the left end (Kb) at the top, orf number in the next line and homolog just below the corresponding arrow. (C) A high-resolution map of the Δ(181,412–189,531) deletion, with the same annotations. Orfs indicated by a green arrow were found8 by protein mass spectrometry to encode a protein of the mature bacteriophage capsid. Orfs indicated by a blue arrow were not found to encode a protein of the mature bacteriophage capsid. The direction of an arrow indicates the direction of transcription and translation. Red lines [vertical in (A); horizontal in (B and C)] indicate the span of a deletion. The numbers in parentheses are the numbers of the nucleotides deleted. The protein encoded by gene 205 is a minor capsid protein with location not yet identified in electron micrographs.
The first 0305φ8-36 deletion mutant was missing nucleotides 118,397–124,982 (3.01% of the genome) and will be called Δ(118,397–124,982). Orfs 200–207 were deleted, as shown at higher resolution in Figure 1B. We found a single deletion and its approximate position by restriction endonuclease analysis that detected a single deletion in the 110,000–130,000 area (not shown). We obtained confirmation and a more precise position by PCR amplification across the site of the deletion. Finally, we located the deletion precisely by Sanger sequencing of the PCR fragment. Details are in the Materials and Methods Section. As described below, the genes deleted included a DNA translocation operon.
The propagation phenotype of Δ(118,397–124,982)
Next, we determined whether the Δ(118,397–124,982) mutant had a propagation defect in relation to the wild type phage. The following characteristics were compared: average plaque diameter (D) and average infectious particle number per ml of a plaque (I). The value of I was assayable because the plaque-supporting, 0.1% agarose gel was weak enough so that a 0.2 ml portion of a plaque (about 25% of the total cleared region) was pipeted for titering. No significant difference between wild type and Δ(118,397–124,982) deletion mutant was observed for either D or I (Table 1). That is to say, no growth defect was detected for the Δ(118,397–124,982) mutant.
Table 1. Propagation phenotypes of deletion mutants.
| Bacteriophagea | (D)b cm | (I)b pfu/ml |
|---|---|---|
|
Wild Type |
1.43 ± 0.05 |
5.5 ± 1.9 x 108 |
|
Δ(118397–124982) |
1.37 ± 0.04 |
4.4 ± 1.3 x 108 |
| Δ(181412–189531) | 0.85 ± 0.04 | 7.7 ± 1.5 x 108 |
The Results section describes the bacteriophages.
The Materials and Methods section describes procedures for determining D and I.
Deletion of a DNA relaxase gene in the Δ(118,397–124,982) mutant
The genes deleted in Δ(118,397–124,982) included orf200, which was found via multi-iteration PsiBlast and reverse PsiBlast searches to be related to the DNA relaxase from the DNA translocation operon of Enterobacter cloacae plasmid CloDF13 (mobC).20 The weakest link in this association was challenged using HHpred as described in the Materials and Methods Section. Alignments of proteins clearly related to either orf200 or CloDF13 mobC were developed using SAM with a stringent inclusion threshold of E = 1.0 × 10−9. Each was picked out of the library with the other as queried by HHpred, with an E value of 4.4 × 10−22 (Table 2, row 1). However, both families also matched a variety of helix-turn-helix domains in their N-terminal domains (Table 2, row 2).
Table 2. Homologs of the 0305φ8-36 relaxase, coupling ATPase and membrane binding protein.
| Querya | Family Membersb | Target of interest | Target Family Membersc | E valued | |
|---|---|---|---|---|---|
| |
Relaxase Similarities |
||||
|
1 |
0305φ8-36 orf200 |
165 |
CloDF13mobC |
66 |
4.4e-22 |
|
2 |
0305φ8-36 orf200 N-ter. domain |
165 |
Various HTH families |
|
3.4e-5 to 1e-3 |
|
3 |
0305φ8-36 C-ter. domain orf200 |
166 |
CloDF13mobC |
53 |
3.9e-7 |
|
4 |
0305φ8-36 orf200 |
1 |
SPO1 gp27.3 |
1 |
3.0e-10 |
|
5 |
0305φ8-36 C-ter. domain orf200 |
1 |
SPO1 gp27.3 |
1 |
0.009 |
|
6 |
0305φ8-36 C-ter. domain orf200 |
1 |
GBSV1 |
43 |
4.2e-7 |
|
7 |
Gamma-77020183 C-ter. domain |
1 |
Bacillus Virus 1 gp40 |
1 |
2e-9 |
|
8 |
Gamma-77020183 C-ter. domain |
72 |
CloDF13 mobC |
85 |
0.005 |
| |
Coupling ATPase Similarities |
||||
|
9 |
0305φ8-36 orf201 |
1 |
SPO1 gp27.4 |
1 |
4e-22 |
|
10 |
0305φ8-36 orf201 |
1 |
ING plasmid traG NP_858410 |
15 |
4.5e-5 |
|
11 |
0305φ8-36 orf201 N-ter. domain |
151 |
ING plasmid traG |
15 |
0.006 |
| |
Membrane Binding Protein Similarities |
||||
| 12 | 0305φ8-36 orf207 | 1 | SPO1 gp27.9 | 1 | 2e-5 |
Source sequence for a protein query family. bNumber of query family members for each source sequence. cNumber of target family members. dValues for HHpred matches, without secondary structure prediction, between the families of the proteins indicated. The context is an 80,000 family library, except for 1 sequence to 1 sequence matches, which were blastP results in nr plus env nr.
To test that orf200 and CloDF13 mobC were related beyond both simply having helix-turn-helix domains, the entire operation was repeated with their C-terminal domains only. The result was finding of significant homology (Table 2, row 3). Thus, it is clear that 0305φ8-36 orf200 contains a DNA relaxase domain, with homologs that include CloDF13 mobC, the latter described in reference 21. Conjugation relaxases are usually encoded adjacent to a DNA translocase that acts as a coupling protein for transfer of the nicked DNA into the conjugation system.21,22
Deletion of a coupling protein/translocase gene in the Δ(118,397–124,982) mutant
The candidate for coupling protein in 0305φ8-36 is the adjacent orf201, originally annotated8 as a VirD4-like protein, based on the closest annotated sequences found by PsiBlast. More detailed analysis revealed that the similarity is confined to a C-terminal ATPase domain. Query with this domain hits Pfam family AAA_10, an ATPase domain frequently found in conjugation-associated ATPases (annotator's comments; PF12846).
However, 0305φ8-36 orf201 differed from both VirD4 and mobB in not having a N-terminal domain composed primarily of transmembrane helix. The N-terminal domain of 0305φ8-36 orf201 had no detectable transmembrane helical segment. To determine whether this absence of a transmembrane helical segment occurs in other, similar genes, we searched for genes similar to 0305φ8-36 orf201 and found them among genes hit by queries from family models of both 0305φ8-36 orf201 and a homolog, ING1 (Table 2, rows 10 and 11), the latter also a homolog of the traG translocase. Although traG has a N-terminal transmembrane helix domain,23,24 about 75% of hits from queries with both ING1 and 0305φ8-36 orf201 models had no transmembrane helices (not shown). Thus, (1) the absence of a transmembrane domain is not unusual among translocases and (2) orf200 and 201 appear to have a relatively standard organization to be the core of a conjugation relaxase/translocase operon.
Completion of the operon: Membrane attachment sites
Mobilization complexes have to make an association with the membrane-bound conjugation system in order to function. Hence, the absence of a transmembrane domain in orf201 might mandate that a separate protein(s) provides for membrane association. If so, the genes in the 0305φ8-36 version of this operon should include at least one orf that encodes a protein with a membrane attachment site. For the following reasons, we conclude that orf203 and orf207 are orfs of this type. Orf203 is homologous to the virB6 locus, which encodes a membrane-associated protein that interacts with the DNA mobilization complex and the sex pilus during Ti plasmid-mediated conjugation.25 Orf207 is homologous to mreB, which encodes a bacterial protein that is structurally homologous to actin and that associates with the cell membrane where it influences cell shape and is an anchor for another arm of the DNA translocase family involved in chromosome segregation.26,27 Thus, the translocation operon includes orfs 200, 201, 203 and 207, no two of which have homologs from a single known bacterial operon.
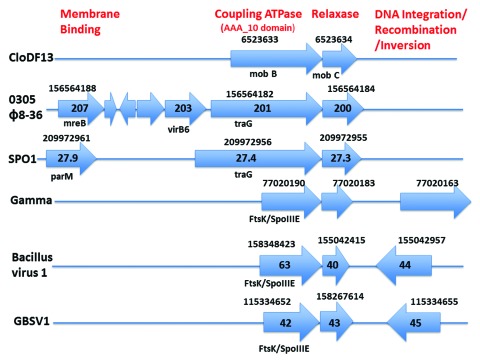
However, we found that a partial version of a similar 0305φ8-36 DNA translocation operon does exist in other bacteriophages, based initially on hits from both entire relaxase genes and their C-terminal segments. The data for the following bacteriophages are in Figure 2 (bacteriophage, followed in succession by gi number, orf number and the row(s) in Table 2 that have data for either the entire gene or the C-terminal domain): (1) Bacillus subtilis bacteriophage SP01 (209972955; 27.3; rows 4–5), (2) Geobacillus bacteriophage GBSV1 (158267614; 43; row 6), (3) Bacillus virus 1 (155042415; 40; row 7), (4) Bacillus anthracis bacteriophage Gamma (77020183; orf not numbered; row 8) and (5) close relatives of Gamma, including Cherry, Fah22 and WBeta (data not shown).
Figure 2.
Homologs of genes from the proposed DNA translocation operon of bacteriophage 0305φ8-36. The plasmid or bacteriophage name is at the left. The NCBI accession number is above an arrow. The orf number is within an arrow, when given at the NCBI site. The family of some orfs is below the arrow and orf number. The functions of homologs in a given column are at the top in red. All distances are proportional to physical distances.
The similarity with the entire 0305φ8-36 operon is highest for lytic bacteriophage SPO1, which has a homolog for orfs 201 (coupling ATPase; Table 2, row 9) and orf207 (Table 2, row 12), not present in the other genomes (Fig. 2). The other bacteriophages, all lysogenic, also have neighboring genes for integration/site specific recombination that 0305φ8-36 does not have (Fig. 2, rightmost genes in the bottom three rows).
Yet, we did not find a single bacteriophage-borne member of any of the other mobilization/relaxase families (reviewed in ref. 28). Hence, there may be something about this particular mobilization system that adapts it for function within a phage genome, but the reason for its peculiar association with phages is not known.
Remainder of the genes missing in the Δ(118,397–124,982) mutant
The genes deleted in Δ(118,397–124,982) also included orf205 (Fig. 1B), an orf that had no detected character of a gene with a function in translocation. Orf205 encodes a protein previously identified as a capsid protein.8 In addition, orf205 is inverted relative to the other deleted orfs, suggesting that it is a comparatively recent addition to the genome and is in the class of genes sometimes called morons.29 We assume, therefore, that the function of orf 205 is different from the function of the orfs in which it is embedded.
The remaining orfs in the deleted region have no known function and presumably represent at least one more function, making a total of at least three functions in an island deletable without any detected effect on propagation of 0305φ8-36 in the laboratory.
The second deletion mutant, Δ(181,412–189,531)
The second deletion mutant, Δ(181,412–189,531), was missing a second island (8.119 Kb; 3.71% of the genome) that also had genes of at least three different functions. This deletion mutant was mapped by the procedures used for the first deletion mutant. The deleted genes include genes for (1) two metallo-protein chaperonins (orf063 and orf066), (2) two tRNAs and (3) six unknown functions (Fig. 1C). Bacteriophages T430 and T531 also have deletable tRNA genes. Deletion of tRNA genes lowers both burst size and rate of protein synthesis in the case of T4.32 Orf66 encodes one of the Mox-R ATPase-like metal chelatase proteins,33 which provide energy for inserting metals into other proteins and are in a diverse family of macromolecule remodeling, P-loop ATPases called AAA+ proteins.34 Orf63 encodes a vWA-like protein, which are metal binding and usually found to work with AAA+ proteins.33,34
Both the Δ(118,397–124,982) and the Δ(181,412–189,531) deletions are in the right half of the 0305φ8-36 genome (Fig. 1A). The Δ(181,412–189,531) deletion did cause a significant growth defect, as seen via significant reduction of D, but not I (Table 1).
Bacteriophage dimerization
To begin analysis of both the aggregation pathway of 0305φ8-36 and the possible role of aggregation in the phenotype of 0305φ8-36, we further characterized 0305φ8-36 bacteriophage particles. We initially developed an ultracentrifugal purification procedure that did not inactivate 0305φ8-36. We had to do this because centrifugation in a cesium chloride density gradient, the procedure previously used, causes inactivation accompanied by contraction of the tail sheath. The contraction initiates at the tail tip.8,9 The procedure developed is based on centrifugation through a sucrose gradient (Materials and Methods Section).
After the revised ultracentrifugation, we removed the sucrose by dialysis and observed the purified bacteriophages by electron microscopy of a negatively stained specimen. The result was that the tail sheath contraction was not observed in electron micrographs of wild type bacteriophage particles. However, most of the wild type bacteriophage particles were in large aggregates, making problematic their further characterization (not shown).
Thus, we repeated the revised purification with a spontaneously generated, reduced-aggregation 0305φ8-36 mutant that had been selected by serial propagation (beyond the propagation used for the original cloning) in the laboratory. As judged by the in situ (in-plaque) fluorescence microscopy of reference 4, the mutant had less than 1% of the aggregation previously observed for the wild type bacteriophage in reference 4 (not shown).
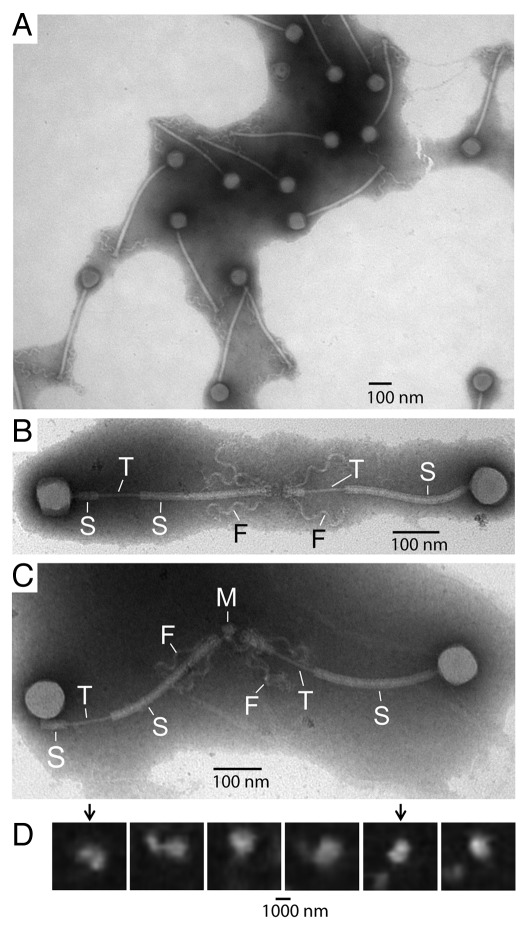
In confirmation of the results obtained with the wild type bacteriophage, the revised purification yielded mutant bacteriophage particles without tail tip-initiated sheath contraction. A field of several particles is in Figure 3A. Nonetheless, sheath contraction was observed for the mutant after purification in a cesium chloride density gradient (not shown), as previously seen for wild type bacteriophage. The bacteriophage particles are monomeric in Figure 3A, even though the sample is concentrated enough so that bacteriophages are close to overlapping.
Figure 3.
Electron microscopy of bacteriophage 0305φ8-36 after improved fractionation. After fractionation of reduced aggregation bacteriophage particles in a sucrose step gradient, fluorescence microscopy was performed with particles from the phage band-region of the gradient (undialyzed). The sucrose was then removed by dialysis and the bacteriophage particles were prepared for electron microscopy (EM) by negative staining, by use of procedures described in the Materials and Methods Section. (A) Low magnification EM image of a field of several bacteriophages. (B and C) Higher magnification EM images of bacteriophage dimers. (D) Captured video frames from fluorescence microscopy of DAPI-stained particles; the frames are separated by 0.5 sec. Legend for (B and C): S, segment of tail sheath; T, exposed segment of tail tube; M, probable fragment of the host; F, tail fiber.”
However, some bacteriophage particles were not monomeric, as most conclusively seen in more dilute areas of a specimen; these particles were less than 10% of the total. The multimeric bacteriophage particles were always joined tail tip-to-tail tip and over 90% of them were dimeric; a few trimers were seen. Dimers are shown in the higher magnification images of Figure 3B and C. The tail tip-to-tail tip, multimer-forming interaction was confirmed by the following transformation, seen only in multimers. The tail sheaths underwent splitting into two segments. “S” in Figure 3B and C indicates each of the two sheath segments. The splitting left a section of the tail tube exposed, as indicated by “T” in Figure 3B and C. The remainder of the tube is presumably encased within the sheath. The sheath splitting is different from the previously observed8,9 sheath contraction because the point of sheath breakage varies throughout the tail in sheath splitting, whereas the point of breakage is only atthe tail tip in sheath contraction. Tail-to-tail dimerization apparently places the entire tail sheath under stress. We do not know whether the sheath splitting occurs before or during preparation for electron microscopy.
Although some dimers appeared to consist only of bacteriophage particles, as in Figure 3B, others appeared to have an additional component, indicated by the arrow labeled “M” in Figure 3C. Although the composition of this M component is not known from direct characterization, the M component varies in size and shape among different dimers. Thus, we think likely that this component is part the host, probably part of the cell wall or cell membrane.
Tail-to-tail dimerization occurred before electron microscopy, as judged by single-particle fluorescence microscopy of pre-dialyzed versions of the same specimens. The procedures are described in the Materials and Methods Section. A presumed dimer was revealed via two separate, resolution-limited bright regions that, during motion, were tethered together and separated by about two tail lengths (about 1,000 nm or about 4× the resolution limit of the fluorescence microscope) in some projections. These particles were undergoing tumbling and translational motion in the specimen. A series of video images, framed to include the same particle (0.5 sec between frames), is in Figure 3D. The two bright regions are seen separately from each other in frames indicated with an arrow; rotation brings these regions into coincidence in the other frames. The dimer was translating throughout this video series. In the second frame from the left, the leftmost bright region is from a second particle that drifted into apparent contact with the dimer; the framed particle is distinct in the other images.
By this criterion, ~10% of the particles observed were dimers. If, in analogy with the behavior of other myoviruses,35 0305φ8-36 attaches to a host cell via its tail, then dimerization will inhibit the infection of host cells.
Bacteriophage adsorption to a host cell
The tail of bacteriophage 0305φ8-36 is long enough so that we determined the orientation of host-attached bacteriophage particles by in situ (in-plaque) fluorescence microscopy of DAPI-stained (Materials and Methods Section) samples of 0305φ8-36 plaques. If the bacteriophage adsorbs tail tip-to-cell surface, then this orientation will be revealed by a DAPI-stained, resolution-limited spot (packaged DNA) at a distance from the cell surface that is maintained during translational and tumbling motion. Given the 486 nm rigid tail and the potentially flexible, wavy tail fibers of 0305φ8-36, this distance depends on the extent to which the tail fibers are extended, but would be between 486 and ~700 nm in any case.
Figure 4 shows a single host cell with at least eight bacteriophages attached, the latter as revealed by resolution-limited spots derived from DNA-bound dye fluorescence. One spot is at the upper right and five distinct spots are at the lower left of the bacterial cell. Two of these latter spots are produced by fluorescence from two bacteriophages. The additional bacteriophages (1) were obscured by superposition in the frame in Figure 4, (2) were seen in other frames as distinct, after tumbling generated a change in orientation (not shown) and (3) were revealed for the view in Figure 4 via asymmetry of the lower two spots; asymmetry can also be generated by limited time resolution during motion. Seven of the eight spots (packaged DNAs) are at a distance from the cell surface consistent with attachment of a bacteriophage particle via the tail tip or tail fibers.
Figure 4.
In situ fluorescence microscopy of host-bacteriophage attachment. A portion of the clear region of a 0.1% agarose-supported plaque was pipeted, mixed with dye and examined by fluorescence microscopy, as described in the Materials and Methods Section. The bar is 1,000 nm (1 μm) long.
The uppermost spot at the lower left, on the other hand, appeared to be attached to the cell surface at a distance too small to be tail tip-to-cell surface attachment. This condition was maintained during tumbling, although this resolution-limited spot was slightly separated from the surface in one orientation.
We conclude that bacteriophage 0305φ8-36 is selected to adsorb to its host by the tip of its tail, but that alternative patterns of adsorption can occur. We further conclude that the observed tail tip-tail tip dimerization (Fig. 3B and C) inhibits initiation of infection by 0305φ8-36. The significance of this latter conclusion is discussed in the next section.
Parenthetically, we note that seven of the bacteriophages of Figure 4 had adsorbed to the host cell before the microscopy began. But, the bacteriophage at the upper right appeared to adsorb during microscopy. This bacteriophage particle was not observed before stable adsorption and appeared to have diffused into focus while it was attaching. Thus, we did not obtain information about the events that occurred before stable attachment. This limitation exists, in general, unless the thermal motion of the bacteriophage particles is restricted to a thin enough zone of solution so that the particles are tracked before and during adsorption.
Discussion
Genes for both bacteriophage-encoded DNA translocation and other functions non-essential in the laboratory
We have found that (1) a DNA translocation operon is in the bacteriophage 0305φ8-36 genome and (2) this DNA translocation operon does not provide any selective advantage in the laboratory, the latter as judged by the absence of a significant effect of deleting this operon on either I or D. To our knowledge, a DNA translocation operon has not been previously documented for other characterized bacteriophages, although at least bacteriophage SPO1 has a similar operon, based on the analysis in Table 2 and Figure 2.
For the following reason, we conclude that DNA translocation does have a function in the wild. A previous study8 has shown that no 0305φ8-36 genes have characteristics that suggest disuse-derived deterioration. The projected functionality of all 0305φ8-36 genes leads to the following conclusion. All laboratory propagation generated/selected deletions are likely to compromise propagation in the environment.
This conclusion is potentially significant for the use of bacteriophages to manage bacteria, because similar laboratory propagation-generated deletions will occur, although more slowly, even in the absence of selection for stability to elevated temperature. For any given procedure of propagation, laboratory propagation-generated deletions are expected to occur more rapidly when the bacteriophage has (unlike 0305φ8-36) a permuted genome. The reason is that non-permutation has been previously found36 to cause laboratory propagation-generated selection for retention of DNA length and, therefore, selection for retention of genes non-essential in the laboratory. That is to say, genome non-permutation has an anti-deletion activity that does not exist when the genome is permuted.
These considerations suggest that bacteriophages with non-permuted genomes be preferred when selecting bacteriophages for managing bacteria, assuming that the “environment function only” genes, such as those observed here, are present. In addition, long-genome bacteriophages should be preferred, because they are likely to have more “environment function only” genes than bacteriophages with shorter genomes. Managing of bacteria is essentially never done in laboratory conditions and is, therefore, likely to be assisted by these genes.
Biofilms
The laboratory propagation-based isolation of a reduced aggregation mutant suggests that, like presence of the DNA translocation operon, the occurrence of propagation-associated extensive aggregation4 has a selective advantage in the wild that it does not have in the laboratory. The possibility exists that the selective advantage specific to the wild occurs while 0305φ8-36 is part of the framework of a biofilm (bacteriophage as biofilm component hypothesis), as suggested by thin sections of human dental plaque (Introduction). Possible alternative selective advantages of aggregation include protection against environmental damage, even in the absence of a biofilm.
The following previous observations suggest additional roles for bacteriophages in biofilms. During biofilm formation, recombination-dependent genomic DNA rearrangement is a mechanism for bacterial phase variation, i.e., for epigenetically changing the state of bacteria37 in a relatively short time period.38 The rearrangement is sometimes among DNA segments from one bacterial cell. But, it also is sometimes between the genome of a cell and an externally derived DNA molecule, such as a plasmid, a transposon or a viral genome, as found for the filamentous Pseudomonas aeruginosa bacteriophage Pf4.39 A more comprehensive review has been presented and theory has been developed to describe the distribution of bacterial states induced by DNA translocation, followed by recombination.40 Thus, the DNA translocation operon of 0305φ8-36 has potentially been selected, in part, for promoting biofilm formation-associated bacterial phase variation.
The possibility also exists that the genomes of aggregating, biofilm-forming bacteriophages are among the DNA molecules translocated from cell-to-cell. These bacteriophages might even propagate in a biofilm by DNA translocation, without the classical bacteriophage particle. Propagation of this type is made efficient by the contact to each other of host cells. This concept of propagation-by-translocation is foreshadowed by theory that defines a virus via its intracellular state, rather than its extracellular state41,42 and that also sometimes postulates the evolution of viruses from cells via parasitism that results in loss of functions.42 Thus, the bacteriophage as biofilm component hypothesis leads to the prediction that aggregating, biofilm-forming bacteriophages in general and 0305φ8-36 in particular have DNA translocation operons that do not provide the advantage in the laboratory that they provide in the wild and that are non-essential in the laboratory.
Indeed, the data in Figure 2 and Table 2 show that other bacteriophages have DNA translocation operons. The DNA integration/recombination function of the translocation operon-proximal genes of lysogenic homologs (Fig. 2, rightmost genes in the bottom three rows) further supports a linking of the roles of translocation and recombination for these bacteriophages. In the case of the lytic bacteriophage, 0305φ8-36, a separate, neighboring gene for recombination is not present for the DNA translocation operon. Recombination may not require a separate gene in the case of some long-genome lytic bacteriophages, as found for the well-studied, smaller lytic bacteriophages, T3 and T7.43 To test the possibility of biofilm formation-associated translocation/recombination linking for 0305φ8-36, we need to determine the functions of the various 0305φ8-36 genes in a laboratory biofilm-forming system.
Unusual aspects of the propagation of bacteriophage 0305φ8-36
In the absence of a mitigating factor, the implied biofilm-associated host/bacteriophage co-existence is problematic, given that 0305φ8-36 is both genotypically and phenotypically lytic.8 A possible mitigating factor is a quorum-sensing, infection-inhibiting event triggered by increase in the concentration of bacteriophage particles. The simplest possibility is that, as the bacteriophage concentration increases, the tail-mediated dimerization observed here increasingly inhibits infection by obscuring tail tip components that adsorb to the host cell. This inhibition of infection would also (1) provide time for some bacterial cells to evolve resistance to bacteriophage infection and (2) explain why bacteriophage 0305φ8-36 does not propagate sufficiently in liquid culture to cause visible lysis, even though 0305φ8-36 is a lytic bacteriophage (Introduction). Dimerization is possibly a stage in the more extensive aggregation4 that subsequently occurs for the wild type bacteriophage.
The agarose-instigated increase in the level of 0305φ8-36 host cell clearing (Introduction) is additional evidence of environmental signaling of bacteriophage lytic activity, possibly via raising of the critical concentration for tail tip-tail tip dimerization. A bacteriophage adaptation to hydrated polymer makes sense in that environmental, hydrated polymer is likely to emerge whenever rain converts a polymer-rich, natural environment (a cattle pen in the case of 0305φ8-36) from a dry, hot (summer soil temperature usually reaching 49–62°C in the case of 0305φ8-36) condition to a wet condition in which both bacteriophages and hosts can propagate. Thus, hydrated polymer can, in theory, be a signal to a bacteriophage to become more aggressive in propagating. An objective for the future is further testing of the effects of various polymers on bacteriophage propagation, with potential application to managing environmental bacteria. The bacteriophage as biofilm component hypothesis discussed above can be subjected to further testing either by further developing a bacteriophage/host-based laboratory biofilm-forming system or by characterizing additional environmental biofilms by electron microscopy of thin sections, as done in reference 13.
Islands of non-essential genes
The closeness to each other of the various genes for DNA translocation [Δ(118,397–124,982) mutant] and the closeness to each other of the two genes for metal insertion [Δ(181,412–189,531) mutant] are explained by co-selection for common function. In the case of the metal insertion function of the Δ(181,412–189,531) mutant, we do not have data that further constrain the function, non-essential in the laboratory. However, we propose that, in a not-known way, metals are less available for protein insertion in the environment than they are in the laboratory. Parenthetically, both deletions include an orf that encodes an AAA ATPase (orfs 201 and 66), for reasons, if any, that are unknown.
However, the island-forming agglomeration of apparently unrelated non-essential genes, as observed here, is in need of additional explanation. One hypothesis is that gene insertions via horizontal transfer are most readily made in non-essential, multi-gene islands. By this hypothesis, the multi-gene islands are sites at which the probability of lethal damage is minimized during initial gene insertion and also during subsequent selection for optimization of the insertion. The edges of our deletions could have been generated either by an accident of the deletion process or by the presence of genes at the deletion edge that are essential with the current host. The above hypothesis predicts that the islands, once nucleated, will be self-propagating in the sense that genes introduced by horizontal transfer will be preferentially introduced in a previously initiated island of non-essential genes. This hypothesis is independent of the position within the genome of the various deletable segments.
Thus, we conjecture that islands of unrelated, non-essential genes constitute a theme, variations on which will be found among most, if not all, long-genome bacteriophages. The presence of a DNA translocation operon in bacteriophage genomes is already a theme, as established here.
Materials and Methods
Bacteriophage propagation and purification
Bacteriophage 0305φ8-36 was preparatively propagated in a 0.1% agarose overlay, as previously done,7,9 by use of stationary phase plating bacteria, grown with aeration in liquid culture. The medium was 10 g Bacto tryptone, 5 g KCl in 1000 ml water supplemented post-autoclaving with 0.002 M autoclaved CaCl2. The agarose overlay was then removed from Petri plates (plate lysate). Bacteriophage particles were extracted by shaking and, then, the preparation was clarified by relatively low-speed centrifugation. Bacteriophages were concentrated by higher-speed centrifugation. The procedures of reference 7 were used. Subsequently, concentrated, resuspended bacteriophage particles were purified either by ultracentrifugation in a cesium chloride step gradient8 or by revised procedure developed here to avoid the tail tip-initiated tail sheath contraction and loss of infectivity observed after purification by centrifugation in a cesium chloride density gradient.8
The revised procedure of purification was the following. A sucrose density gradient was pre-poured in a centrifuge tube for the Beckman SW41 rotor. The gradient had the following composition: (1) 1.0 ml of 65% sucrose at the bottom, (2) 9.3 ml of a 10–35% linear sucrose gradient above the bottom layer of 65% sucrose. The buffer for the sucrose solutions was 0.01 M Tris–HCl, 0.01 M MgCl2, pH 7.4. A clarified, concentrated plate lysate of infected cells was layered on at the top of the density gradient, as illustrated in Figure 5 (left). Then, centrifugation was performed at 33,000 rpm, 5°C, for 60 min. After centrifugation, the centrifuge tube was placed in a blackened aluminum holder with a hole at the bottom. A photograph was taken of the light scattering during fluorescent lamp, white light illumination from the bottom. During centrifugation, the bacteriophage particles sedimented through the 35%/65% sucrose interface and produced a band of light scattering when illuminated (Fig. 5). The gradient was then fractionated by pipeting from the top.
Figure 5.
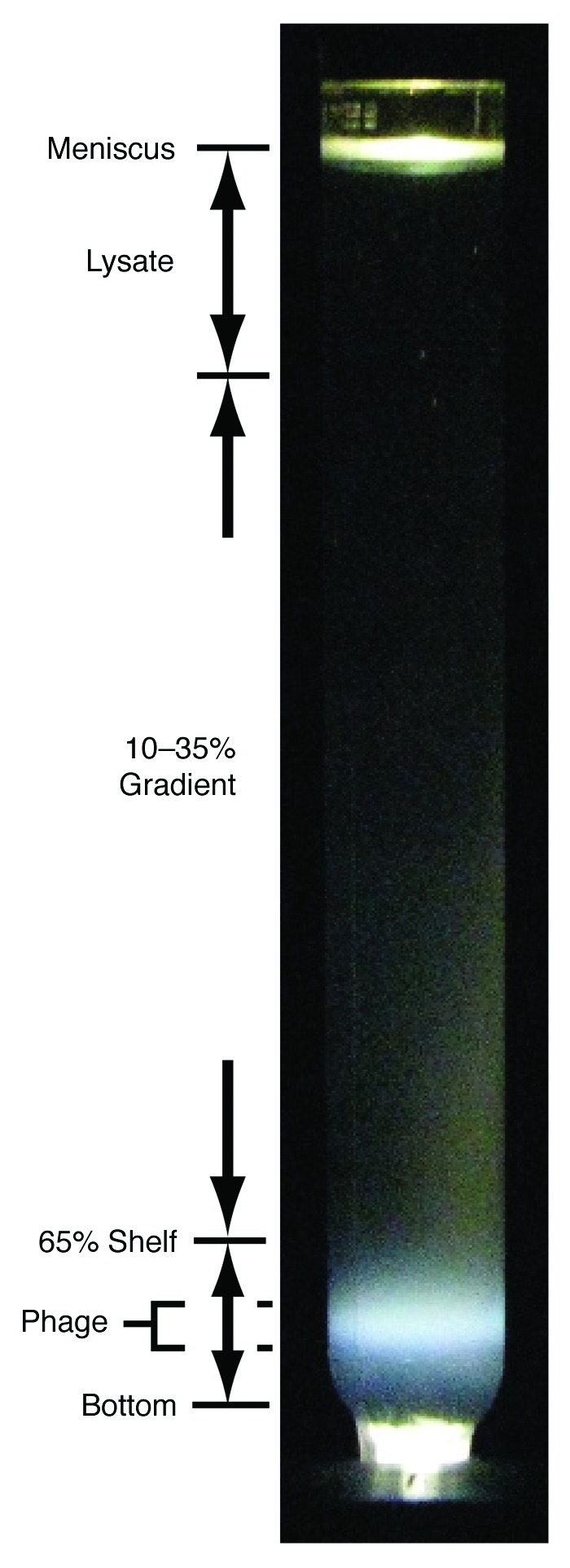
Revised purification of bacteriophage 0305φ8-36. After fractionation of a lysate of 0305φ8-36-infected cells in a sucrose density gradient, the post-centrifugation light scattering from the centrifuge tube was photographed. The light scattering from bacteriophage particles is indicated at the left near the tube bottom by “phage.” The pre-centrifugation composition of the tube contents is also indicated at the left. The “65% shelf” is the top of the 65% sucrose layer; the “10–35% sucrose gradient” was poured above the shelf; the lysate was layered above the 10–35% sucrose gradient. Light scattering from both the meniscus and the bottom of the centrifuge tube are indicated.
Isolation and propagation of deletion mutants
To select for deletion mutants, a clarified, concentrated, wild type plate lysate was diluted by at least 1:200 into buffer that contained 0.1 M NaCl, 0.01 M Tris-Cl, 0.002 M EDTA. The temperature was raised to 52°C for 10 min. The result was loss of titer by a factor of 1 × 106. The surviving bacteriophages were propagated as single plaques. Bacteriophages from a single plaque were then subjected to the same inactivation; the loss of titer was determined by dilution and traditional plating44 in a plaque-supporting 0.1% agarose overlay. If a plaque yielded bacteriophage particles significantly more temperature-resistant than the wild type bacteriophage, then two additional single-plaque purifications were performed and the elevated temperature-resistance of the more purified mutant was again tested. Two deletion mutants were isolated and, characterized, Δ(118,397–124,982), which was more resistant to inactivation by a factor of ~106 and Δ(181,412–189,531), which was also more resistant by a factor of ~106 than the wild type bacteriophage.
To compare D and I for a mutant to D and I for wild type bacteriophage, plaques were formed by dilution and traditional plating44 in a plaque-supporting 0.1% agarose overlay. Procedure, host preparation and media were the same for mutants as they were for wild type bacteriophage. The plates (10 for each bacteriophage) were incubated overnight, at room temperature (25 ± 2°C) in thermal contact with each other, so that the temperature did not vary by more than 0.2°C among the 30 plates. The values of plaque diameter were measured from digital photographs of the plates. The average (i.e., D) and standard deviation are reported after measurement of a total of 100 plaques for each bacteriophage.
To determine I, 0.2 ml of each of 10 plaques was removed from the center of a plaque and diluted in growth medium for plating and plaque formation in a 0.1% agarose supporting gel. The average (i.e., I) and standard deviations are reported for the number of plaque-formers per ml of plaque pipeted.
Extraction and restriction endonuclease analysis of bacteriophage DNA
To extract bacteriophage 0305φ8-36 DNA, we made clarified, concentrated plate lysates. We then digested host DNA and, next, purified bacteriophage DNA via phenol extraction, by use of procedures previously described.8 These procedures do not include purification of the bacteriophage particles.
To approximately map deletions, purified bacteriophage DNA was digested with the following restriction endonucleases: BamH1, BstN1, SpeI. DNA fragments in a digest were fractionated by electrophoresis through a horizontal, submerged 0.7% agarose gel (Seakem, LE agarose, Lonza). The electrophoresis buffer used was 0.09 M TRIS-acetate, pH 8.4, 0.001 M EDTA. The region of 0305φ8-36 genome covered by each restriction endonuclease-generated DNA fragment was determined from the 0305φ8-36 sequence and the fragment length. We approximately located the ends of a deletion by observing which bands were missing. We then located the deletion more precisely by accounting for the new bands that were produced by the digestion of all restriction endonucleases used.
To obtain single-base resolution, we designed primers and performed PCR across the suspected site of a deletion and compared the length of the PCR fragment and its digestion products to the corresponding lengths obtained from the wild type DNA. If this result confirmed deletion, we then Sanger sequenced the PCR fragment to identify the site of the deletion with single-base resolution.
Interpretation of DNA sequences
To advance beyond the original annotation8 of bacteriophage 0305φ8-36, we constructed extended protein families by iterative application of PsiBlast and SAM and an updated database.45,46 We also used HHpred47,48 to address a common problem with iterative family building methods, in which an unnamed member of a distant family is included at an early round with poor confidence, causing the inclusion of named members of the distant family at a later round with high confidence. In this case, the E value reported, when the named member of the distant family appears, exaggerates the confidence in the link between the query protein and the distant family. In cases where we suspected this problem, we verified the weak step of the linkage as follows. Sequences clearly related to the query protein within a conservative E value were aligned and converted to an HHpred-style HMM. Sequences clearly related to the named member of the target family were similarly aligned and converted to an HMM. Both HMMs were inserted in the ~100,000 member HHpred library. The E value for the weak link joining the two families was then evaluated by HHpred during its search of this library with each family HMM used as the query.
Electron microscopy
Bacteriophages purified by centrifugation in a sucrose gradient were dialyzed against the following buffer: 0.01 M Tris-Cl, 0.01 M MgSO4, pH 7.4. The dialyzed bacteriophages were then prepared for electron microscopy by negative staining with 1.0% sodium phosphotungstate, pH 7.6, as previously done.9 Specimens were observed with a JEOL100CX electron microscope in the Department of Pathology at The University of Texas Health Science Center at San Antonio.
Fluorescence microscopy
To prepare a specimen for in situ (in-plaque) fluorescence microscopy, part of the clear region of a 0305φ8-36 plaque was excised and then mixed with a ~1:20× amount of β-mercaptoethanol and a ~1:10× amount of 15 μg/ml DAPI (4′,6-diamidino-2-phenylindole). Then, this mixture was incubated for at least 1 min at room temperature and 0.5–1.0 μl was placed between cover glass and microscope slide, cleaned as described in reference 4. Specimens were visualized by use of an Olympus BX 60 upright fluorescence microscope with a short arc mercury vapor lamp and an oil immersion lens. The filter set is described in reference 4. Real time analog video was recorded and digitized as previously described.4
To prepare a specimen for observation of the contents of a fraction of a sucrose gradient, 5 μl of a fraction of a sucrose gradient was mixed with 0.2 μl of 15 μg/ml DAPI and 0.2 μl of β-mercaptoethanol. This mixture was placed between a cover glass and microscope slide cleaned as described in the previous paragraph. Specimens were visualized by use of an Olympus BX60 upright fluorescence microscope with a short arc mercury vapor lamp and an oil immersion lens. The filter set was the NU set from Chroma Technology Corp.
Acknowledgments
This work was supported by grants from The Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation and the Welch Foundation (AQ-764).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/19546
References
- 1.Clokie MRJ, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers WC. In the beginning…. Bacteriophage. 2011;1:50–1. doi: 10.4161/bact.1.1.14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serwer P, Wang H. Single-particle light microscopy of bacteriophages. J Nanosci Nanotechnol. 2005;5:2014–28. doi: 10.1166/jnn.2005.447. [DOI] [PubMed] [Google Scholar]
- 4.Serwer P, Hayes SJ, Lieman K, Griess GA. In situ fluorescence microscopy of bacteriophage aggregates. J Microsc. 2007;228:309–21. doi: 10.1111/j.1365-2818.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- 5.Pais-Correia AM, Sachse M, Guadagnini S, Robbiati V, Lasserre R, Gessain A, et al. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med. 2010;16:83–9. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- 6.Jolly C. Cell-to-cell transmission of retroviruses: Innate immunity and interferon-induced restriction factors. Virology. 2011;411:251–9. doi: 10.1016/j.virol.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serwer P, Hayes SJ, Thomas JA, Demeler B, Hardies SC. Isolation of novel large and aggregating bacteriophages. Methods Mol Biol. 2009;501:55–66. doi: 10.1007/978-1-60327-164-6_6. [DOI] [PubMed] [Google Scholar]
- 8.Thomas JA, Hardies SC, Rolando M, Hayes SJ, Lieman K, Carroll CA, et al. Complete genomic sequence and mass spectrometric analysis of highly diverse, atypical Bacillus thuringiensis phage 0305phi8-36. Virology. 2007;368:405–21. doi: 10.1016/j.virol.2007.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serwer P, Hayes SJ, Thomas JA, Hardies SC. Propagating the missing bacteriophages: a large bacteriophage in a new class. Virol J. 2007;4:21. doi: 10.1186/1743-422X-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutherland IW, Hughes KA, Skillman LC, Tait K. The interaction of phage and biofilms. FEMS Microbiol Lett. 2004;232:1–6. doi: 10.1016/S0378-1097(04)00041-2. [DOI] [PubMed] [Google Scholar]
- 11.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 12.Abedon ST. Bacteriophages and Biofilms: Ecology, Phage Therapy, Plaques. Nova Science Publishers 2011, Hauppauge, New York. [Google Scholar]
- 13.Brady JM, Gray WA, Caldwell MA. The electron microscopy of bacteriophage-like particles in dental plaque. J Dent Res. 1977;56:991–3. doi: 10.1177/00220345770560082901. [DOI] [PubMed] [Google Scholar]
- 14.Fujisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–45. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 15.Son M, Hayes SJ, Serwer P. Concatemerization and packaging of bacteriophage T7 DNA in vitro: determination of the concatemers’ length and appearance kinetics by use of rotating gel electrophoresis. Virology. 1988;162:38–46. doi: 10.1016/0042-6822(88)90392-3. [DOI] [PubMed] [Google Scholar]
- 16.Studier FW, Rosenberg AH, Simon MN, Dunn JJ. Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol. 1979;135:917–37. doi: 10.1016/0022-2836(79)90520-5. [DOI] [PubMed] [Google Scholar]
- 17.Rhoades M, Schwartz J, Wahl JM. New deletion mutant of bacteriophage T5. J Virol. 1980;36:622–6. doi: 10.1128/jvi.36.2.622-626.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Se´chaud J, Streisinger G, Emrich J, Newton J, Lanford H, Reinhold H, et al. Chromosome structure in phage T4, II. Terminal redundancy and heterozygosis. Proc Natl Acad Sci U S A. 1965;54:1333–9. doi: 10.1073/pnas.54.5.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, et al. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–74. doi: 10.1016/0022-2836(92)90469-Z. [DOI] [PubMed] [Google Scholar]
- 20.Nu´ñez B, De La Cruz F. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol Microbiol. 2001;39:1088–99. doi: 10.1046/j.1365-2958.2001.02308.x. [DOI] [PubMed] [Google Scholar]
- 21.Peed L, Parker AC, Smith CJ. Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. J Bacteriol. 2010;192:4643–50. doi: 10.1128/JB.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frost LS, Ippen-Ihler K, Skurray RA. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol. 2008;70:89–99. doi: 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- 25.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–85. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vollmer W. The prokaryotic cytoskeleton: a putative target for inhibitors and antibiotics? Appl Microbiol Biotechnol. 2006;73:37–47. doi: 10.1007/s00253-006-0586-0. [DOI] [PubMed] [Google Scholar]
- 27.Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol. 2010;192:1671–84. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frost LS, Koraimann G. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 2010;5:1057–71. doi: 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- 29.Villafane R, Zayas M, Gilcrease EB, Kropinski AM, Casjens SR. Genomic analysis of bacteriophage epsilon 34 of Salmonella enterica serovar Anatum (15+) BMC Microbiol. 2008;8:227. doi: 10.1186/1471-2180-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brok-Volchanskaya VS, Kadyrov FA, Sivogrivov DE, Kolosov PM, Sokolov AS, Shlyapnikov MG, et al. Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res. 2008;36:2094–105. doi: 10.1093/nar/gkn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen MJ, Locker J, Weiss SB. The physical mapping of bacteriophage T5 transfer tRNAs. J Biol Chem. 1976;251:536–47. [PubMed] [Google Scholar]
- 32.Wilson JH. Function of the bacteriophage T4 transfer RNA’s. J Mol Biol. 1973;74:753–7. doi: 10.1016/0022-2836(73)90065-X. [DOI] [PubMed] [Google Scholar]
- 33.Snider J, Houry WA. MoxR AAA+ ATPases: a novel family of molecular chaperones? J Struct Biol. 2006;156:200–9. doi: 10.1016/j.jsb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Snider J, Thibault G, Houry WA. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 2008;9:216. doi: 10.1186/gb-2008-9-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–70. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray NE. The impact of phage lambda: from restriction to recombineering. Biochem Soc Trans. 2006;34:203–7. doi: 10.1042/BST20060203. [DOI] [PubMed] [Google Scholar]
- 37.Henderson IR, Owen P, Nataro JP. Molecular switches--the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–32. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 38.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–68. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:8066–73. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chia N, Woese CR, Goldenfeld N. A collective mechanism for phase variation in biofilms. Proc Natl Acad Sci U S A. 2008;105:14597–602. doi: 10.1073/pnas.0804962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forterre P. Giant viruses: conflicts in revisiting the virus concept. Intervirology. 2010;53:362–78. doi: 10.1159/000312921. [DOI] [PubMed] [Google Scholar]
- 42.Bândea CI. A new theory on the origin and the nature of viruses. J Theor Biol. 1983;105:591–602. doi: 10.1016/0022-5193(83)90221-7. [DOI] [PubMed] [Google Scholar]
- 43.Pajunen MI, Elizondo MR, Skurnik M, Kieleczawa J, Molineux IJ. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J Mol Biol. 2002;319:1115–32. doi: 10.1016/S0022-2836(02)00384-4. [DOI] [PubMed] [Google Scholar]
- 44.Adams M. Bacteriophages. Interscience Publishers, Inc. 1959, Woods Hole, Mass. [Google Scholar]
- 45.Hughey R, Krogh A. Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput Appl Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- 46.Hughey R, Krogh A. SAM: Sequence alignment and modeling software system. Technical Report UCSC-CRL-99-11, University of California, Santa Cruz, CA. http://www.soe.ucsc.edu/research/compbio/papers/sam_doc/sam_doc.html
- 47.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 48.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]