Abstract
We present an empirical method for identification of distinct structural motifs in proteins on the basis of experimentally determined backbone and 13Cβ chemical shifts. Elements identified include the N-terminal and C-terminal helix capping motifs and five types of β-turns: I, II, I′, II′ and VIII. Using a database of proteins of known structure, the NMR chemical shifts, together with the PDB-extracted amino acid preference of the helix capping and β-turn motifs are used as input data for training an artificial neural network algorithm, which outputs the statistical probability of finding each motif at any given position in the protein. The trained neural networks, contained in the MICS (motif identification from chemical shifts) program, also provide a confidence level for each of their predictions, and values ranging from ca 0.7–0.9 for the Matthews correlation coefficient of its predictions far exceed that attainable by sequence analysis. MICS is anticipated to be useful both in the conventional NMR structure determination process and for enhancing on-going efforts to determine protein structures solely on the basis of chemical shift information, where it can aid in identifying protein database fragments suitable for use in building such structures.
Keywords: artificial neural network, backbone chemical shift, helix capping, β-turn, CS-Rosetta, MCC score, protein structure prediction, Rosetta, secondary structure prediction
Introduction
The most common elements of secondary structure in proteins include β-sheet, α-helix and 310 helix. However, many other small structural motifs exist and in particular N-terminal and C-terminal capping motifs have long been identified (Presta and Rose 1988; Richardson and Richardson 1988; Harper and Rose 1993; Aurora et al. 1994), as has a wide range of different turn types (Richardson 1981; Rose et al. 1985; Sibanda et al. 1989; Hutchinson and Thornton 1994). It is well recognized that such structural motifs, mostly containing specific H-bond patterns, play a key role in stabilizing protein structure and are likely to be important in the protein folding process (Dyson et al. 1988; Becker and Karplus 1997; Baldwin and Rose 1999). Extensive efforts have focused on identification of such motifs from the protein's amino acid sequence (Bystroff and Baker 1998; Chou 2000; Kaur and Raghava 2003; Fuchs and Alix 2005; Petersen et al. 2010), but considering the enormous variety of sequences that can form such motifs, the success rate of even the most advanced programs remains very limited beyond identification of β-sheet and α-helix.
Reliable prediction of structural motifs holds strong potential for enhancing protein structure prediction programs such as Rosetta (Rohl et al. 2004; Das and Baker 2008), which assembles structures of low empirical energy from small fragments taken from a large structural database. These fragments are selected to mimic the secondary structure prediction probabilities for each small segment of the query protein. When NMR chemical shifts are available for the query protein, this allows selection of database fragments that are much more likely to match the structure of the segment in the query protein than would be possible on the basis of amino acid sequence only. Indeed, improved fragment selection provides much of the basis for improved performance of the chemical-shift Rosetta (CS-Rosetta) program over the regular Rosetta method (Shen et al. 2008; Shen et al. 2009; Sgourakis et al. 2011).
An empirical relation between protein backbone structure and deviations of chemical shifts from random coil values, so-called secondary shifts Δδ, has long been recognized (Saito 1986; Pastore and Saudek 1990; Williamson 1990; Spera and Bax 1991; Asakura et al. 1995; Iwadate et al. 1999). Most significantly, upfield 1Hα and downfield secondary 13Cα secondary chemical shifts are commonly associated with α-helix, whereas negative Δδ13Cα together with positive Δδ1Hα values point to β-sheet (Wishart et al. 1991; Wishart and Sykes 1994). However, both computational and empirical analyses indicate that this correlation is mostly an indirect consequence of the local secondary structure, and that these secondary shifts relate more directly to the backbone torsion angles (Spera and Bax 1991; Pearson et al. 1997; Case 1998; Cornilescu et al. 1999; Vila et al. 2007; Vila et al. 2008). Thus, the correlation applies equally to residues with a given local backbone conformation, regardless of whether they are engaged in the H-bond pattern associated with helix or sheet. For nuclei other than 13Cα, the relation between chemical shift and local structure tends to be more complex. For example, for 15N the chemical shift is known to be a function not only of local backbone torsion angles, but also is impacted by H-bonding, electric field effects, and sidechain torsion angles (de Dios et al. 1993). Similarly, the amide 1HN chemical shift can be strongly impacted by ring current, susceptibility, electrostatic, and H-bonding effects (Asakura et al. 1995; Moon and Case 2007). Several computational approaches have been put forward in recent years which capitalize on these known relations to predict chemical shifts for proteins of known structure (Wishart et al. 1997; Meiler 2003; Neal et al. 2003; Shen and Bax 2007; Shen and Bax 2010; Han et al. 2011). Inversely, other programs aim to predict local backbone geometry for proteins of unknown structure but with experimentally determined chemical shifts. Such methods include purely empirical approaches such as the popular program TALOS (Cornilescu et al. 1999), which simply searches a database of previously assigned proteins of known structure for tripeptides fragments with similar backbone chemical shift and residue type, and its recent successor TALOS+, which adds an artificial neural network component to filter TALOS results, while at the same time identifying α-helix and β-sheet secondary structures (Shen et al. 2009). Whereas TALOS and TALOS+ only focus on very short fragments, other programs such as PREDITOR (Berjanskii et al. 2006) additionally are able to take advantage of sequence homology with proteins of known structure, yielding both accurate backbone and sidechain torsion angles.
Numerous methods exist for secondary structure prediction aided by chemical shifts. The most popular method is known as CSI, or chemical shift index, which makes a "consensus estimate" based on appropriately weighted 13Cα, 13Cβ, 1Hα, and 13C' secondary shift values (Wishart and Sykes 1994). Wang and Jardetzky's PSSI method additionally takes 1HN chemical shifts and nearest neighbor effects into account (Wang and Jardetzky 2002), whereas Hung and Sumudrala included an artificial neural network analysis, potentially taking better advantage of correlated changes in secondary shifts to predict secondary structure (Hung and Samudrala 2003). Other, more recent programs such as PECAN (Eghbalnia et al. 2005), 2DCSI (Wang et al. 2007), and TALOS+ (Shen et al. 2009) also simultaneously consider the chemical shifts of adjacent residues for predicting α-helices and β-strands, reaching prediction accuracies that approach the uncertainty limit associated with identification of secondary structure in proteins with known atomic coordinates.
Apart from α-helix and β-sheet, very few programs have focused on identification of other distinct elements in protein structures, although the potential of chemical shifts to reveal such motifs has been long recognized. For example, Gronenborn and Clore showed that the helical N-cap box motif can be recognized on the basis of a negative Δδ13Cα (ca −1 to −2 ppm) together with a positive, ca 1–4 ppm, Δδ13Cβ secondary shift for the N-cap residue, followed by a string of residues with an α-helical chemical shift signature (Gronenborn and Clore 1994). Other structural motifs, including helical C-caps, and the various types of β-turns have proven to be more difficult to identify on the basis of chemical shifts. This is due in part to less distinct chemical shift patterns for such motifs, but also to the fact that the database of proteins of accurately known structure and fully assigned chemical shifts remains relatively small. The latter makes it difficult to draw statistically warranted conclusions, in particular when considering that a wide range of different amino acid types is often found in such motifs, and it remains uncertain whether the backbone atoms of all residue types are subject to the same secondary chemical shift perturbation when located at any given position in such a motif.
Here, we describe an empirical approach, based on trained artificial neural network algorithms, to identify small secondary structure elements, including the N-cap and C-cap motifs, and the various types of β-turns. Our method takes advantage a carefully pruned NMR chemical shift database of proteins with accurately known structures and chemical shifts, recently enlarged for the program PROMEGA which aims to predict the cis or trans nature of peptide bonds preceding Pro residues (Shen and Bax 2010). This 580-protein database derives from the much larger collection of assigned chemical shift data, contained in the BMRB database (Doreleijers et al. 2005). Analogous to our recent programs TALOS+ and SPARTA+, our new method for structural motif identification from chemical shifts, named MICS, is based on an artificial neural network or ANN algorithm. The advantage of a properly trained ANN over other, more direct probabilistic methods, is that it can combine in an optimally weighted manner the wide range of input parameters, including the residue types at the different positions in the motif and the secondary chemical shifts from six different types of nuclei. MICS yields good results for identification of both N-caps and C-caps, as well as the most common types of β-turns, including I, I', II, II' and VIII. It has been implemented as a webserver program and can be accessed at http://spin.niddk.nih.gov/bax/nmrserver/mics/
Methods
Preparation of the protein database
The chemical shift patterns of the different helix capping motifs and β-turns in proteins were explored using the protein database recently developed for the Promega program (Shen and Bax 2010). This database, referred to as the chemical shift database, contains 580 proteins for which both a high-resolution X-ray structure and (nearly) complete backbone chemical shifts (δ15N, δ13C', δ13Cα, δ13Cβ, δ1Hα and δ1HN) are available. The preparation of this chemical shift database, including the calculation of the secondary chemical shifts, the chemical shift re-referencing, exclusion of residues with large B-factors in the X-ray reference structure, exclusion of chemical shift outliers, and 2H isotope effect correction, followed the same procedure as that used for precursors of this database, originally developed for the program TALOS (Cornilescu et al. 1999).
A second database, referred to as the structure database and containing 9446 proteins (2,468,258 residues) for which a high-resolution (≤2.5Å) X-ray structure was available, was constructed and used to explore the sequence and structure-related patterns of different helix capping motifs and different types of β-turns. This database was originally used by the CS-Rosetta program for its hybrid protocol (Shen et al. 2008; Shen et al. 2009). For each residue in the above two databases, a three-state secondary structure classification was assigned according to its DSSP-identified secondary structure (Kabsch and Sander 1983), determined from the X-ray atomic coordinates: H (Helix; DSSP classification of H or G), E (Extended strand; E or B) and L (Loop; I, S, T or C).
Classification of helix capping motifs
At a first stage, the helix capping motifs are identified based on their original definitions (Harper and Rose 1993; Aurora et al. 1994; Aurora and Rose 1998). As discussed below, the original definitions are not well suited as training input for an artificial neural network, and will be modified to identify essentially the same sets of residues, but associate a score with each such motif that indicates how closely it resembles the idealized motifs.
For capped helices extending from residues N1 through C1, which make canonical backbone H-bonds and have regular helical backbone torsion angles, the flanking residues are labeled as follows:
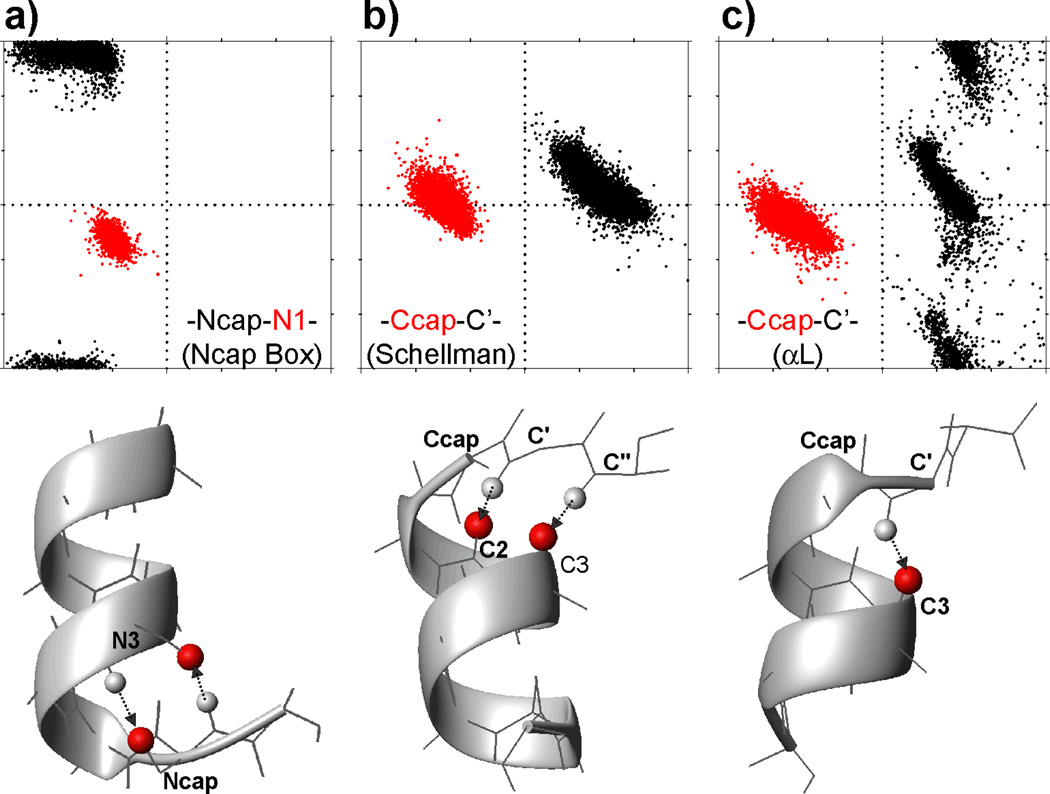
Thus, the Ncap and Ccap residues are always located immediately adjacent to the helix. They make H-bonds with the helical residues but have backbone torsion angles that deviate significantly from ideal helical values. Residues N″, N′, C′ and C″ do not participate in the helix hydrogen bonding network and/or do not have regular helical backbone torsion angles. An ideal N-terminal helix capping box (often referred as an Ncap box motif) contains two reciprocal backbone to side-chain H-bonds between the Ncap and the N3 residues: bb(i)→sc(i+3) and sc(i)←bb(i+3), where sc refers to sidechain and bb to backbone, and residue i is the Ncap residue, with the arrow denoting the donor to acceptor direction. In this study, a more generous definition for the N-cap is used, where only one of the two H-bonds is required. In our sequence database, which includes 57413 helices with a length of at least 7 residues, 2808 Ncap motifs are observed with a single bb(i)→sc(i+3) H-bond, and 17255 N-caps with a single sc(i)←bb(i+3) H-bond. For 6017 helices, 11%, an ideal Ncap box with both reciprocal H-bonds is present. The distributions of the backbone ϕ/ψ torsion angles for each of the six residues are listed in Table 1 and shown in Fig. 1 and Supplementary Information (SI) Fig. S1. In the 580-protein chemical shift database, 223 ideal Ncap boxes, 411 sc(i)←bb(i+3) Ncaps, and 66 bb(i)→sc(i+3) Ncaps are present. Amino acid sequence preferences for each position in the N- motif (SI Table S1) show a preference for Ser, Thr, Asp and Asn for the Ncap residue, allowing it to accept an H-bond from the backbone amide of residue N3. The N-cap motif also shows an elevated presence of hydrophobic residues in positions N' and N4, enabling hydrophobic interactions between the sidechains of these residues.
Table 1.
Average backbone ϕ/ψ torsion angles and H-bond energies for residues in helix capping motifs.
| Torsion Anglesa | Nb |
H-bond Energy [kcal/mol]c |
||||||
|---|---|---|---|---|---|---|---|---|
| i−1 | i | i+1 | i+2 | i+3 | i+4 | |||
| N’ | Ncap | N1 | N2 | N3 | N4 | |||
| Ncap Box | ||||||||
| <ϕ> | - | −92±24 | −59±5 | −64±5 | −64±5 | −64±5 | 6017 | E1: −2.0± 0.7 |
| <ψ> | - | 167±8 | −39±6 | −41±5 | −41±5 | −42±6 | E2: −2.0±0.6 | |
| i | i+1 | i+2 | i+3 | i+4 | ||||
| C3 | C2 | C1 | Ccap | C’ | ||||
| Ccap (Schellman) | ||||||||
| <ϕ> | −62±6 | −65±6 | −62±6 | −90±11 | 73±19 | - | 12718 | E1: −2.4±0.6 |
| <ψ> | −42±7 | −43±7 | −31±9 | 5±10 | 25±18 | - | E2: −1.6±0.5 | |
| Ccap (αL) | ||||||||
| <ϕ> | −64±6 | −65±7 | −79±12 | −99±21 | 75±22 | - | 4640 | E3: −1.9±0.7 |
| <ψ> | −42±7 | −39±9 | −38±11 | −19±16 | - | - | ||
| Ccap (Schellman + αL)d | ||||||||
| <ϕ> | −63±6 | −64±6 | −65±11 | −91±14 | 74±19 | 98±13e | 15793 | |
| <ψ> | −42±7 | −43±7 | −32±10 | −1±14 | 24±20 | |||
The positional mean ϕ and ψ torsion angles for each of the residues in all hexapeptides in the structure database forming an Ncap box motif, an (ideal) Schellman Ccap motif or an (ideal) αL Ccap motif (see Methods).
The numbers of the hexapeptides in the sequence database forming an Ncap box motif, an ideal Schellman Ccap or an ideal αL Ccap motif.
The electrostatic interaction energy (Kabsch and Sander 1983) of the characteristic H-bonds in the Ncap box and two Ccap motifs. For all hexapeptides forming an Ncap box motif, the average energy of bb(i)→sc(i+3) and sc(i)←bb(i+3) H-bonds is referred to as E1 and E2, respectively. For all Schellman Ccaps, the average energy of bb(i)←bb(i+5) H-bond [E1] and bb(i+1)←bb(i+4) H-bond [E2] is provided. For αL Ccaps, the average energy of bb(i+1)←bb(i+5) H-bond [E3] is listed.
The combined ideal Schellman Ccap and ideal αL Ccap torsion angle information, excluding αL motifs with a ψ angle of the C’ residue falling outside +/− 80°.
The average value of ϕ+ψ for the C’ residue.
Fig. 1.
ϕ/ψ torsion angle distributions for residues in three helix capping motifs. Plots of the backbone torsion angle ψ vs ϕ are shown for two of the six residues in (a) all N-terminal helix capping box (Ncap box) motifs, and (b) Schellman and (c) αL helix capping motifs in our structural database. Only ideal Schellman and αL helix capping motifs are considered (see Methods). ϕ/ψ angles of the Ncap residue are in black, and angles for the N1 residue in red. For the Ccaps, the C-cap residue angles are in red, and the C' residue in black. All ϕ/ψ torsion angle plots cover a range of −180° to 180° for both ϕ and ψ. The typical backbone conformation for each motif is shown below their respective ϕ/ψ plot. For simplicity, only CO atoms (red balls) and amide protons (small light gray balls) involved in the characteristic intra-motif H-bonds (marked by arrows; Table 1) are shown, and Ala is used for all residues, except for a Ser for the Ncap/N3 residues in the Ncap box, and Gly for the C' residue of the two Ccap motifs.
A Schellman C-terminal capping motif, or Schellman Ccap motif, is defined as a six-residue fragment (from residues C3 [i] to C” [i+5]) which locates at the end of an α-helix and exhibits a double H-bond pattern: bb(i)←bb(i+5) between the N-H of C″ (i+5) and C=O of C3 (i), and bb(i+1)←bb(i+4) between the N-H of C′ and C=O of C2. The αL C-terminal capping motif (αL Ccap motif) contains a single bb(i)←bb(i+4) H-bond between the N-H of residue C′ and the C=O of C3. For both the Schellman and αL Ccap motifs the backbone ϕ angle of the C′ residue (residue i+4) has a positive sign. A total of 15173 and 11453 helices in the protein sequence database are observed to be ended with a Schellman motif and αL motif, respectively. Interestingly, 2455 (16.2%) of those ending with a Schellman motif also exhibit a αL-like bb(i)←bb(i+4) H-bond, and 6813 (59.5%) of the helices ending with an αL motif also contain one of the Schellman-like bb(i)←bb(i+5) or bb(i+1)←bb(i+4) H-bonds. This mixed character of the H-bonding patterns and the high similarity in backbone angles makes it difficult to unambiguously distinguish the Schellman and αL motifs by empirical methods. For 12718 (83.8%) of the helices in the structure database ending with a Schellman C-cap and 4640 (40.5%) helices capped by an αL motif, no such mixed H-bonding pattern is observed, and these are referred to as ideal Schellman and ideal αL Ccap motifs, respectively. The ϕ/ψ torsion angle distributions of these ideal Ccap motifs observed in the structure database are presented in Fig. 1 and Table 1. The number of helix capping motifs in the chemical shift database is far smaller than in the structure database (totals of 355 [310 ideal] Schellman Ccaps and 253 [129 ideal] αL Ccaps) but follows the same distribution. Amino acid sequence preferences in C-cap motifs (SI Table S1) show a strong preference for Gly in the C' position of C-cap motifs, as expected on the basis of its required positive backbone angle, ϕ.
Classification of turn motifs
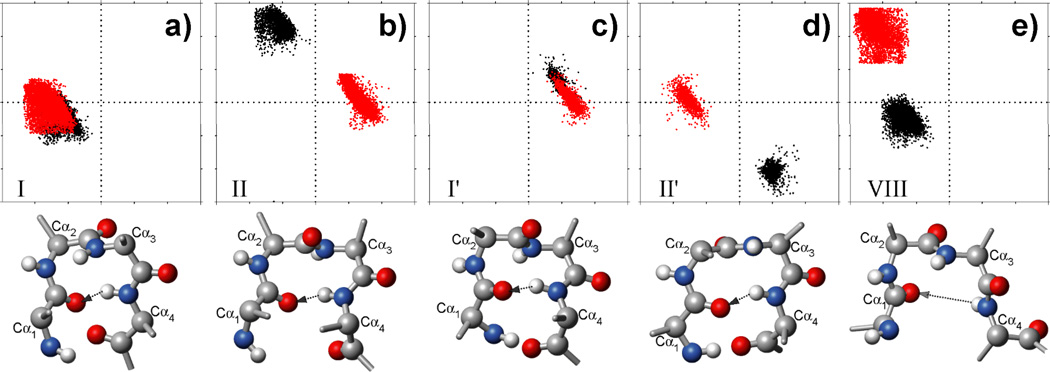
A β-turn is formed by four consecutive residues which are not part of an α-helix and where the Cα distance between the first (i) and the last (i+3) residue, referred to as the Cα(i,i+3) distance, is shorter than 7 Å (Richardson 1981; Rose et al. 1985). The backbone ϕ/ψ torsion angles of the two center residues (i+1 and i+2) are then used to define five different types of β-turns (Wilmot and Thornton 1990; Hutchinson and Thornton 1994), i.e., type I, II, I’, II’ and VIII. The standard definition for distinguishing these turn types requires that at least three of the four backbone torsion angles for the center two residues must fall within 30° of their ideal value, whereas one torsion angle is allowed to deviate by up to 45°. When using these definitions, ideal torsion angles for each of these turns, as defined by Hutchinson and Thornton (1994), agree closely with the average values observed in our structure database (Table 2). Besides the above five types of β-turns, there are four other types, including VIa1, VIa2 and VIb, for which the third position must be a cis-Proline, and a miscellaneous type IV, which includes all other β-turns with a Cα(i,i+3) < 7 Å, but backbone ϕ/ψ torsion angles for the two center residues that fall outside the ranges specified for the other turns (Richardson 1981; Hutchinson and Thornton 1994). Due to the rarity of the types VIa/b and the wide structural variety of type IV, these are not evaluated in our study. Moreover, only isolated β-turns, which have no overlapped residues with other β-turns, are considered first and used to derive their structural (Table 2) and chemical shift (Table S3) patterns. In total, 34337, 15690, 5238, 3522 and 14879 isolated β-turns with type I, II, I’, II’ and VIII, respectively, are observed in the sequence database. The distributions of their ϕ/ψ torsion angles, Cα(i,i+3) distances, and bb(i)←bb(i+3) intra-turn H-bond energy, as well as the positional amino acid preference (Tables 2 and S2; Figs 2, S1, S2 and S3) closely match values expected for these five types of turns. For the chemical shift database, the numbers of isolated β-turns observed for type I, II, I’, II’ and VIII are 914 (out of in total 1235 with at least three chemical shifts for each of four residues), 432 (460), 175 (198), 107 (110) and 309 (354), respectively.
Table 2.
Characteristic ϕ/ψ torsion angles, Cα distances and intra-turn hydrogen bonds for five types of β-turns.
|
Type (k) |
Torsion Anglesa |
Nb |
Cα(i,i+3) distance [Å] |
H-bond (i←i+3 /(i→i+3)c [%] |
Energy (i←i+3)d [kcal/mol] |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ϕi+1,k | ψi+1,k | ϕi+2,k | ψi+2,k | (ϕ+ψ)i+1,k | (ϕ+ψ)i+2,k | |||||
| Ie | (−64) −65±12 |
(−27) −24±13 |
(−90) −93±16 |
(−7) −3±17 |
−89±14 | −95±19 | 34337 | 5.5±0.4 | 80/10 | −1.7±0.6 |
| II | (−60) −61±13 |
(131) 136±11 |
(84) 80±16 |
(1) 5±20 |
75±14 | 85±11 | 15690 | 5.7±0.4 | 90/4 | −1.8±0.6 |
| I’ | (55) 53±7 |
(38) 40±10 |
(78) 77±12 |
(6) 5±15 |
93±7 | 83±9 | 5238 | 5.4±0.3 | 95/84 | −1.8±0.5 |
| II’ | (60) 60±9 |
(−126) −126±11 |
(−91) −93±13 |
(1) 5±16 |
−66±14 | −88±12 | 3522 | 5.5±0.4 | 95/64 | −2.0±0.6 |
| VIII | (−72) −76±15 |
(−33) −32±14 |
(−123) −131±20 |
(121) 131±23 |
−108±16 | 0±25 | 14879 | 6.5±0.5 | 0.6/0.2 | −0.7±0.2 |
The average ϕ/ψ torsion angles of the two center residues (residues i+1 and i+2) of the isolated β-turns observed in the structure database (SI Fig. S1 D–H). The ideal ϕ/ψ torsion angles of the two center residues for five types β-turns (Hutchinson and Thornton 1994) are given in parentheses.
Number of isolated β-turns in the structure database, which are not overlapped with other β-turns.
Percentage of the isolated β-turns in the structure database with bb(i)←bb(i+3) and bb(i)→bb(i+3) H-bonds between the first and the last residue.
Electrostatic interaction energy (Kabsch and Sander 1983) of the bb(i)←bb(i+3) H-bond between the C=O of residue i and N-H of residue i+3 in all isolated β-turns in the structure database.
For type I turns, the sum of ψi+1 and ϕi+2 is also used in Eq 5, with a value of −118+20° degree (see Methods).
Fig. 2.
ϕ/ψ torsion angle distribution for the two center residues (residue i+1 in black; residue i+2 in red) in five types of isolated β-turns in our structural database. All turns are identified using the original literature definitions (see Methods). All plots have a range of −180° to 180° for both ϕ (horizontal) and ψ angles. A typical conformation of each type of β-turn is shown as a ball-stick cartoon below their respective ϕ/ψ plot. Only backbone heavy atoms (N: blue balls; Cα/C′: gray; CO: red) and amide protons (small light gray balls) are shown, and Ala is used for all four residues of each β-turn, and idealized ϕ/ψ values for these five turn types are used (Table 2). The Cα atoms are numbered according to their position in the β-turn; the potential intra-turn H-bond from the HN of the last residue to the CO of the first residue is marked by an arrow. Note that for type VIII β-turns the long HN-O=C distance and often unfavorable H-bond angles usually result in vanishing H-bond energy.
In our study, we aim to train an artificial neural network algorithm to recognize the various types of helix capping and β-turn motifs on the basis of the experimental chemical shifts. In principle, this can be done by assigning a value of 1 to elements in the database that meet the criteria for a given motif, and the remainder a value of 0. However, such a binary distinction is not optimal for purposes of training an artificial neural network because for motifs that borderline fall within or outside the cut-off limits the difference in chemical shifts will be small. Moreover, as can be seen in Fig. 2 and SI Fig. S1, the original definitions for the various turn types result in angular ranges for the backbone torsion angles of turn types I, II, and VIII that show "truncation" behavior, meaning that turns which are geometrically very similar to e.g. a turn type I but fall just outside the allowed ϕ/ψ limits are assigned a 0, while other turns that are just within the tolerance limits are assigned a 1. A better approach therefore assigns a numerical value between 1 and 0 to the motif, depending on how closely it mimics ideal values in terms of backbone angles. It turns out that the backbone-backbone H-bonding patterns observed in β-turns, as well as the Cα(i,i+3) distances, are closely correlated with the backbone torsion angle values. Indeed we show below that a scoring function can be developed based only on backbone torsion angles which results in virtually the same classifications as those obtained from the original definitions.
Scoring of helix capping motifs
For helix capping, we find that the characteristic backbone angles associated with ideal Ncap and Ccap motifs can be stabilized by a substantial array of different H-bond pairings (see Results), with virtually indistinguishable chemical shift patterns. For the helix capping motifs, we therefore remove the requirements for specific H-bonds from the definition, and simply aim to identify motifs with backbone angles that overlap with those of the classic motifs.
The implementation of a scoring function is first illustrated for the case of the helical N-cap. For any given hexapeptide i, containing residues i−1 to i+4, the Ncap score, SNcap, is defined as a function of its backbone angles according to:
| [1] |
which is derived from a sigmoid normalization of the χ2 distribution of deviations of the ϕ/ψ torsion angles of five residues, i to i+4, from the mean ϕ/ψ torsion angles of the five residues, Ncap to N4, respectively, in an Ncap box motif:
| [2] |
where the average Ncap box values for ϕj and ψj together with their standard deviations, σ, are listed in Table 1, and the value for σ is set to 10° for cases where the experimental distribution is less than 10°. Note that the ϕ/ψ angles of the i−1 residue, N', are not considered as they vary widely among different Ncaps. The Ncap score SNcap is calculated for all available hexapeptides in the structure database, and this process is repeated for different values of the constants in the exponent of Eq. 1 (final values shown are 3 and 2.5), in order to find an SNcap scoring function that most effectively allows discrimination of Ncap motifs from other structural elements. Using the scoring function of Eq. 1, all hexapeptides corresponding to an Ncap box, with its characteristic pair of H-bonds, and Ncaps with a single sc(i)←bb(i+3) or bb(i)→sc(i+3) H-bonds, yield an SNcap score > 0.3 (Fig. 3a). The hexapeptides with an Ncap box motif yield the highest Ncap scores, mostly > 0.95, while nearly all other (non-Ncap) hexapeptides yield SNcap scores ≤0.1. A modest number of hexapeptides with Ncap-like backbone torsion angles but lacking the characteristic sc(i)←bb(i+3) or bb(i)→sc(i+3) H-bonds show high scores too (Fig. 3a), most of which are stabilized by different H-bonds (see Results section), but which are also included as Ncaps in this study.
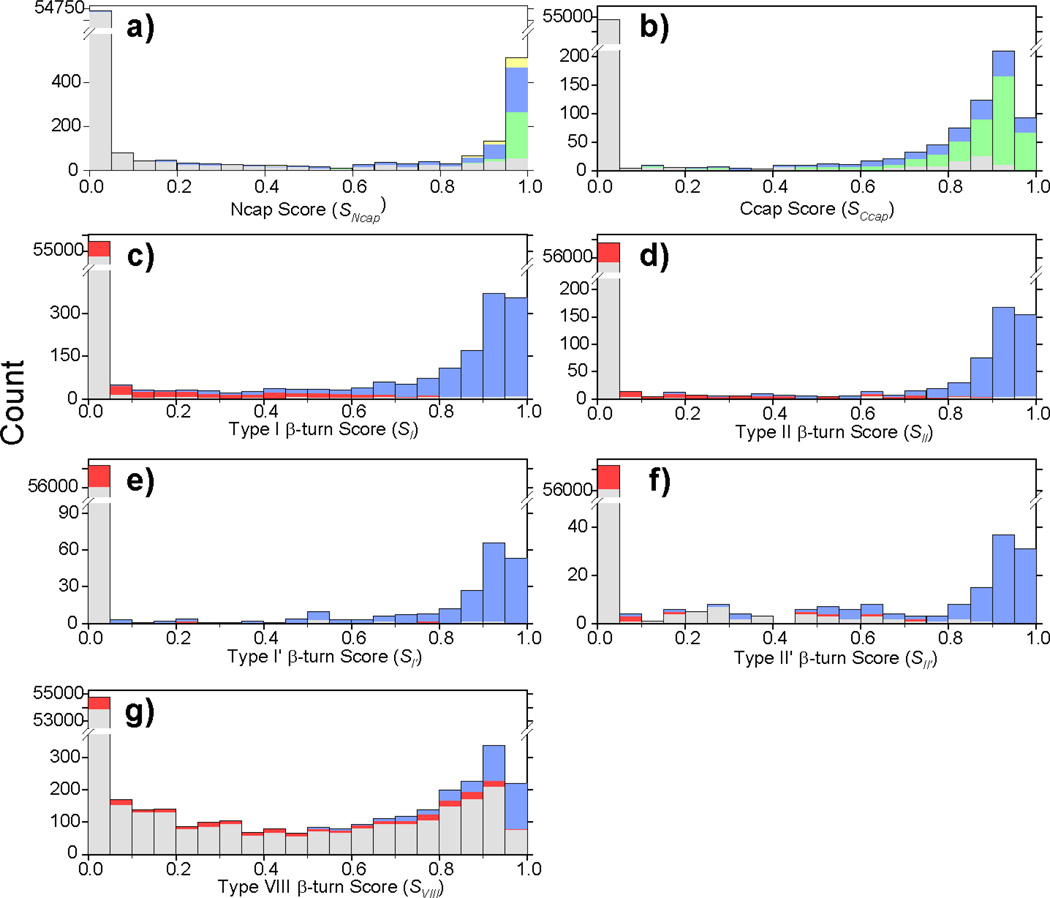
Fig. 3.
Histograms of empirical helix capping and β-turn scores. (a) Ncap score (SNcap, Eq. 1) calculated for all hexapeptides in the chemical shift database. Green, blue and yellow colors, respectively, correspond to the Ncap box motif, the sc(i)←bb(i+3) Ncap motif and the bb(i)→sc(i+3) Ncap motif. All other hexapeptides (incl. non-Ncap) are shown in gray. (b) Histogram of Ccap scores (SCcap, Eq. 3) calculated for all hexapeptides in the chemical shift database, with Schellman Ccap motifs in green, αL in blue, and all others in gray (c–g) Histograms of β-turn scores (Sk, Eq. 5) calculated for all tetrapeptides in the chemical shift database. For each type of β-turn, the number of β-turns [(type I (c), II (d), I' (e), II' (f) and VIII (g)] identified by using the original definitions (see Methods) are shown in blue, the number of β-turns for which the original definition assigns a miscellaneous type IV are in red, and the number of all other tetrapeptides in gray.
Considering the very similar backbone ϕ/ψ angles in Schellman and αL Ccap motifs (Table 1; Figs. 1 and S1) and backbone chemical shifts (Fig. 4b), as well as their mixed patterns of hydrogen bonds, we combine them into a single category, hereafter simply referred to as the Ccap motif. A single Ccap score, SCcap, is then defined for any hexapeptide i (residues i to i+5) to report its similarity to a Ccap motif in terms of its backbone torsion angles:
| [3] |
which again is derived from the χ2 distribution of the deviation of the ϕ, ψ, and ϕ+ψ angles of the first five residues, C3 to C' (or i to i+4) from their mean ϕ/ψ angles in a Ccap box motif (Table 1):
| [4] |
Note that this χ2 function includes an additional term related to the sum (ϕ+ψ) of the C' residue. As can be seen from the distribution of the C' ϕ/ψ angles in Ccaps, these values are strongly correlated and the variance in their sum is considerably smaller than in their individual values.
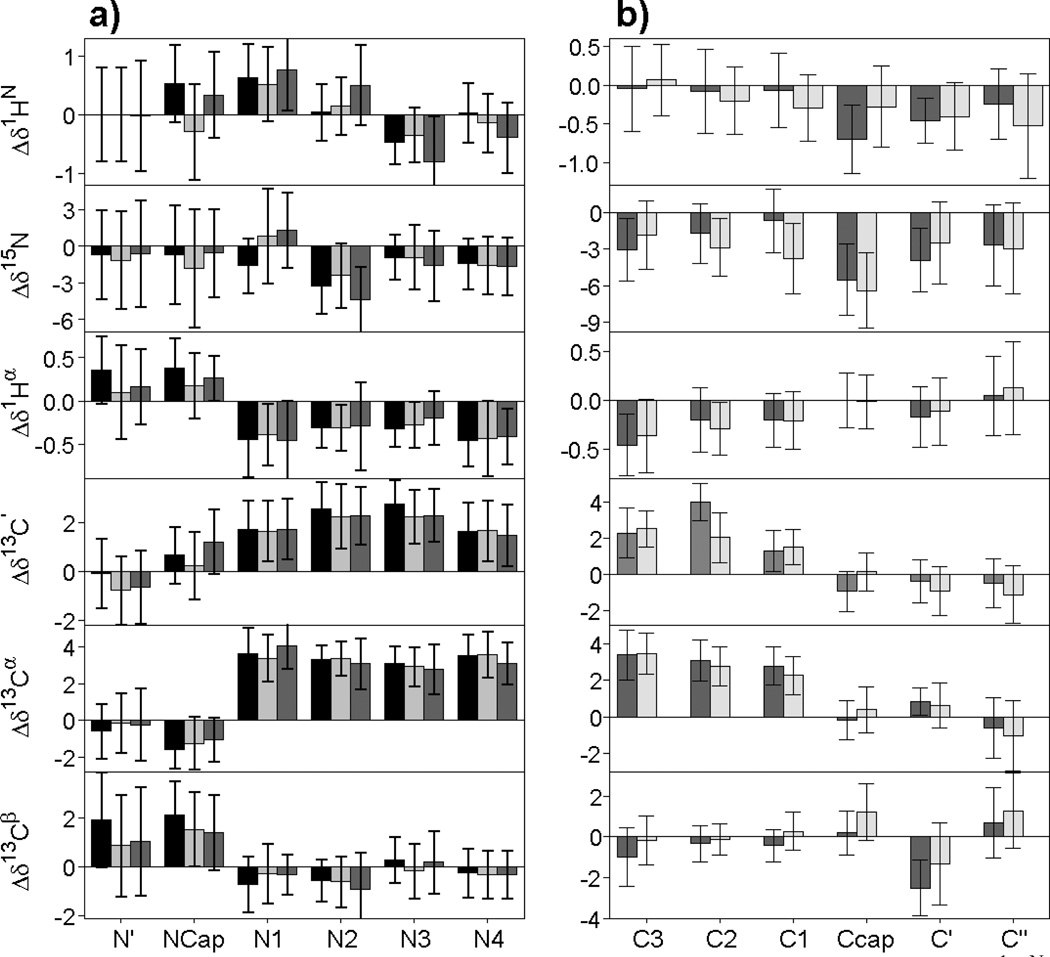
Fig. 4.
Chemical shift patterns of helix capping motifs. (a) The average secondary 1HN, 15N, 1Hα, 13C’, 13Cα and 13Cβ chemical shifts are plotted for each of the six consecutive residues (N′, Ncap, N1, N2, N3 and N4, respectively, with the Ncap box motif in black, the bb(i)→sc(i+3) Ncap motif in dark gray, and the sc(i)←bb(i+3) Ncap motif in light gray. (b) Average secondary chemical shifts for each of the six consecutive residues (C3, C2, C1, Ccap, C′, and C″, respectively) involved in a Schellman (dark gray) or αL (light gray) C-terminal helix capping motif.
Evaluation of the Ccap score, SCcap, over the chemical shift database (Fig. 3b), indicates that nearly all Schellman and αL Ccap motifs have SCcap > 0.3. A number of hexapeptides lacking the Schellman or αL H-bonding pattern have a high score too, however. Nearly all of these show at least one of two Schellman H-bonds, i.e., bb(i)←bb(i+5) or bb(i+1)←bb(i+4) H-bonds (see Results), and we therefore include this group as members of the generic Ccap class.
Scoring of β-turn motifs
Analogous to the helix capping motifs, we introduce a score of β-turn motifs, Sk (k=I, II, I′, II′, and VIII), for tetrapeptides simply on the basis of the backbone ϕ/ψ angles of their two center residues. The requirement of a short Cα(i,i+3) distance (≤7 Å) and a frequently observed intra-turn H-bond between the first and the last residue is not included in the scoring functions as they are closely correlated with the ϕ/ψ torsion angles of the two center residues (see Results). The scoring functions for β-turns are of the form
| [5] |
where hα,j,k is a penalty function which evaluates whether a given backbone torsion angle α (α = ϕ, ψ or ϕ+ψ) of residue j (j = i+1, i+2) in the query tetrapeptide has backbone torsion angles characteristic of a type k β-turn. The term fhelical,j is a Boolean number which accounts for the β-turn requirement that the two center residues (i+1 and i+2) are not part of a regular helix (as defined by the DSSP program (Kabsch and Sander 1983)), i.e., fhelical,j=0 if residue j (j=i+1, i+2) is not helical, and 1 otherwise. Analogous to Eqs. 1–4, the terms ha,j,k are sigmoid functions of the χ2 distribution of the deviation of the ϕ, ψ and ϕ+ψ angles (αj,k) from the mean ϕ/ψ/ϕ+ψ angles (<αj,k>) in a type k β-turn (Table 2):
| [6] |
| [7] |
where σα,j,k is the database standard deviation for torsion angle α of residue j in β-turn type k, with wa,j,k and ca,j,k being constants that define the steepness and the center of the sigmoid normalization function. With a typical value of 2 and 3, respectively, for wa,j,k and ca,j,k, the sigmoid function of Eq. 6 corresponds to a 50% penalty when the angle α deviates by 3 σα,j,k from the corresponding ideal angle, 88% for 4σα,j,k, and 98% for 5σα,j,k. The wa,j,k and ca,j,k constants are adjusted for the different turn types, k, to best match the original β-turn definitions (Table S4; see Results). For type I β-turns, the summed angle term of ψi+1+ϕi+2 (see footnote to Table 2) is also included in Eq. 5, and serves to minimize the inclusion of “open” type I β-turns (with Cα(i,i+3) distance > 7Å).
The β-turn score Sk is calculated for all available tetrapeptides in the chemical shift database, and the distribution of β-turn scores for the five types of β-turn studied in this work, I, II, I′ and II′ and VIII, shows that they can be accurately distinguished by their Sk value (Fig. 3c–g). For β-turns of type I, II, I′ and II′, a β-turn score Sk cutoff of 0.2–0.3 ensures that all these tetrapeptides are recognized as type k β-turns. However, a small fraction of tetrapeptides is found to yield a high score Sk, despite not being a type k β-turn. Closer inspection of those tetrapeptides shows that all of these either are type IV β-turns (represented by red bars in Fig. 3c–g, a miscellaneous category that contains all β-turns which are not of type I, II, I′, II′ or VIII (or three minor types VIa1, VIa2 and VIb, which are not considered in this work) according to the ϕ/ψ angles of the two center residues, or they correspond to β-turn like structures with a Cα(i,i+3) distance slightly above the 7Å cut-off (gray bars in Fig. 3c–g, or black bars in Fig S2). The type IV β-turns with a high score Sk actually have ϕ/ψ angles close to those of a β-turn type k for the two center residues but cannot be classified as β-turn type k due to the hard cutoff (30°/45°, see above) of the ϕ/ψ angles in the original definitions. For type VIII β-turns (Fig. 3g), a β-turn score SVIII >0.5 is observed for all type VIII β-turns, while a considerably number of the “non-β-turns” (as defined by Cα(i,i+3) distance of ≥7Å) have a high SVIII score too. Inspection indicates that those “non-β-turns” with a high SVIII score have slightly larger Cα(i,i+3) distances, ranging up to ~8.5 Å (Fig. S2; Table S6), and can be classified as slightly more "open" β-turns. Considering that those open type VIII β-turns have backbone ϕ/ψ torsion angle patterns very similar to the standard type VIII β-turns, they here are included in the type VIII β-turn classification. In other words, the Cα(i,i+3) distance requirement of ≤7 Å for a type VIII β-turn is now relaxed to ≤8.5 Å when considering a SVIII score (>~0.25).
Neural network architecture and training
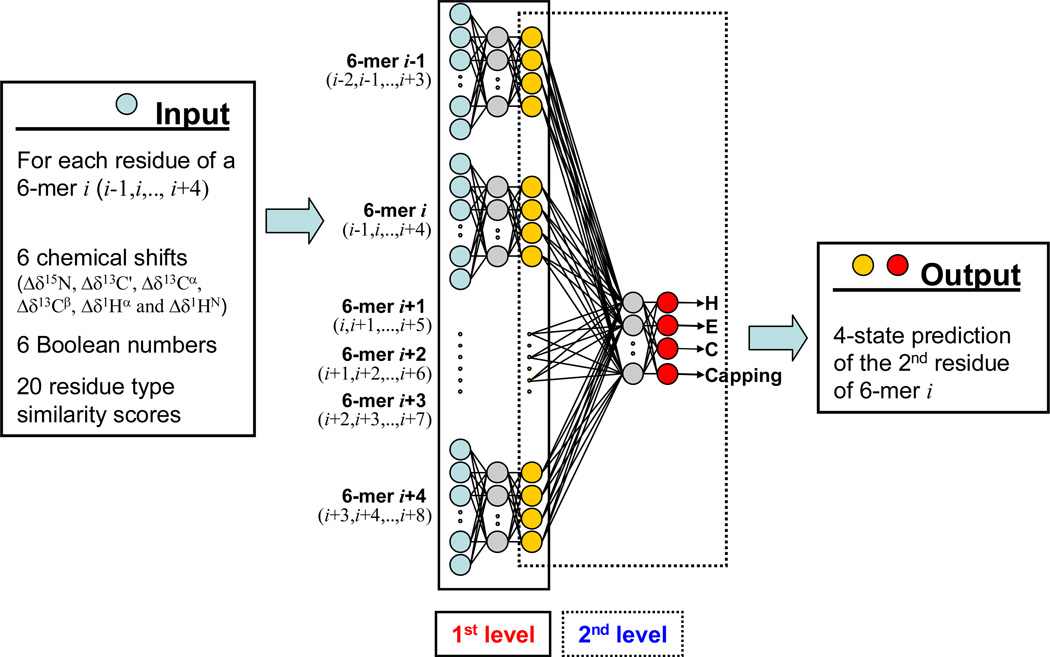
We use two-level feed-forward multi-layer artificial neural networks (ANN) to correlate the backbone chemical shifts and amino acid sequence patterns with the various helix capping and β-turn motifs (SI Table S5). The trained networks are then used to predict these motifs on the basis of their amino acid sequence and experimental chemical shifts.
The architecture of the two-level artificial neural network will first be illustrated for the Ncap motif, but is very similar for the other motifs. The input signals (aqua circles, Fig. 5) to the first layer of the Ncap ANN consist of hexapeptide parameter sets derived from the above chemical shift database. Each hexapeptide i (denoted as 6-mer i in Fig. 5), comprising residues i−1 to i+4, has 192 nodes, representing the six secondary chemical shift values Δδx,j (x=15N, 13C', 13Cα, 13Cβ, 1Hα and 1HN), six Boolean numbers bx,j and twenty amino acid type similarity scores, taken from the BLOSUM62 matrix (Henikoff and Henikoff 1992), for each residue j (j = i−1,…,i+4). The Boolean number bx,j is set to 1 if the chemical shift Δδx,j exists, otherwise, bx,j is set to 0 and an average (near zero) chemical shift <Δδx> in the database is assigned to Δδx,j. Only residues with at least three available experimental chemical shift assignments are used for ANN training purposes. In the hidden layer of the network, where each node receives the weighted sum of the input layer nodes as a signal, 60 such nodes (or hidden neurons; grey, Fig. 5) are used. The output of the hidden layer is obtained through a nodal transformation function; here a standard sigmoid function is used (see Eq. 8).
Fig. 5.
Architecture of the two-level feed-forward artificial neural network used to predict the presence of an Ncap motif (Ncap ANN). The Ncap ANN calculates the probability for each hexapeptide in a protein to be an Ncap motif with the second residue in the Ncap position. The Ncap ANN uses as input for the first level feed-forward prediction the known parameters characterizing each of the six residues. The ANN is trained on the 580-protein chemical shift database to predict the known output state. Besides the six chemical shifts and six Boolean numbers representing the chemical shifts, input parameters for each residue of the hexapeptide also include a 20-dimensional vector, consisting of the coefficients of its row in the BLOSUM62 matrix. A total of 192 input parameters (aqua) per hexapeptide are used to predict the probability for it to be an Ncap (yellow), which is then used as input for the second level of the ANN. 60 hidden nodes (grey) are used for the first level of the ANN. The ANN output of the first level for six overlapped hexapeptides is used to refine the final prediction of the four elements of the output vector (red), using a hidden level consisting of six nodes (grey).
The TALOS+ program (Shen et al. 2009b) uses a three-state secondary structure classification: helix (H), strand (E) and loop (L). An ANN similar to the one used in the present study predicted the secondary structure with an overall correctness of ~89% on the basis of inputs from tripeptides. In the present study, the Ncap ANN uses the input from hexapeptides, and an additional ANN target/output number representing the Ncap structure motif is used to complement the conventional three-state secondary structure identifiers (H, E and L). Thus, a four-state structure classification of the second residue of each hexapeptide i in the database will be assigned as the target/output of the first level network: [1 0 0 0]i for helix (H), [0 1 0 0]i for strand (E) and [0 0 1-SNcap SNcap]i for those identified as loop (L) by the DSSP secondary structure classification of residue i. The 1-SNcap and SNcap terms in the target vector indicates whether the second residue (i) is the Ncap residue in an Ncap motif, where SNcap is the Ncap score of Eq. 1. Below, we will refer to the fourth element of the P1×4 output vector simply as P4. Each output value has one node with a linear activation function [f2(x)=x] (see Eq. 8). The same procedure previously was used for the TALOS+ and SPARTA+ programs (Shen et al. 2009; Shen and Bax 2010). The empirical relationship between the four-state structure classification and NMR chemical shift data received by the first level network is given by:
| [8] |
with f1(x) = 1/(1+e−x), and f2(x) = x. X1×192 is the input data vector consisting of 192 elements; W(1) and b(1) are the weight matrix and bias, respectively, for the connection between the nodes in the input and the hidden layer; W(2) and b(2) are the weight matrix and bias, for the connection between the nodes in the hidden and output layer; p1×4 is the training target or the output vector of the first level of the neural network (yellow circles, Fig. 5), indicating the four-state structure classification (H, E, L and Ncap) of the second residue (residue i) in a given hexapeptide i.
The second level of the Ncap neural network is used to smoothen the prediction by accounting for commonly observed patterns in proteins, and follows its use in the TALOS+ program and several widely used sequence-based secondary structure prediction programs (Rost and Sander 1993; Jones 1999). The two-level artificial neural network, referred as a 4-4 ANN model, uses the input information from six sequential residues (from i−1 to i+4) for both the first and the second level. The input layer for the second level comprises the parameter set of the four-state structure classification predicted by the first level of the network for each available hexapeptide in the database, i.e., each hexapeptide set i (or 6-mer i, Fig. 5) of the input layer for the second level has 24 nodes, containing the four-state structure classification of each residue (from residues i−1 to i+4) predicted by the first level network (for hexapeptide sets i−1 to i+4, respectively). The hidden layer contains 6 nodes, and the four-state structure classification of the second residue (i) of the corresponding hexapeptide in the database is used in the output layer and as the target of the neural network. The equation used for the second level of the neural network is similar to Eq. 8:
| [9] |
where p1×24 is the input vector containing the 24 nodes and the definitions of weights, biases, and activation functions are the same as those in Eq. 8. Eqs. 8 and 9 of this two-level network, with their optimized weights and biases obtained from the training dataset, are then used to predict the four-state structure classification for residues in any protein of unknown structure. The Eq. 9 network output vector, P1×4, represents the ANN scores for the query residue i, or the second residue of the query hexapeptide i, to be within each of the four states: helix, strand, loop and Ncap.
Similarly, a two-level artificial neural network with a 4-4 ANN model, referred as Ccap ANN, is used to predict the probability for a hexapeptide to adopt a Ccap motif (Schellman or αL motif). The input signals to the first layer again consist of hexapeptide parameter sets derived from the chemical shift database. Each hexapeptide set i (or 6-mer i, SI Fig. S4a), now consists of residues i-3 to i+2, and again has 192 nodes, analogous to the Ncap ANN. In the output layer, a four-state structure classification of the fourth residue of each hexapeptide i in the database is assigned as the target/output of the first level network, i.e., [1 0 0 0]i for helix (H), [0 1 0 0]i for strand (E), and [0 0 1-SCcap SCcap]i, for those identified as loop (L) by DSSP, where the 1- SCcap and SCcap terms in the target vector are taken from the SCcap score of Eq. 3. The architecture and implementation of the second level of the Ccap ANN (SI Fig. S4a) are the same as for the Ncap ANN.
For each of the five types of β-turns (I, II, I′, II′ and VIII) considered in our study, again a two-level ANN, referred to as a β-turn ANN, is used to predict the probability of a four-residue fragment to adopt a specific type β-turn. For the first level, the two-level β-turn ANN uses input information from four sequential residues, and for the second level it uses the input from ten sequential residues. It therefore is referred to as a 4–10 ANN model (SI Fig. S4b). The input signals to the first layer of the first level ANN comprise the tetrapeptide parameter sets derived from the chemical shift database. Each tetrapeptide set i (or 4-mer i), consisting of residues i−1 to i+2, has 128 nodes, representing the six secondary chemical shift values Δδx,j (x=15N, 13C', 13Cα, 13Cβ, 1Hα and 1HN), six Boolean numbers bx,j (which have the same definition as for Ncap and Ccap ANNs) and twenty amino acid type similarity scores for each residue j (j = i−1,…,i+2). The first level β-turn ANN contains 40 nodes in its hidden layer, and four nodes in its output layer. In the output layer of the first level network, a four-state structure classification of the second residue of each tetrapeptide i is used as the target/output, i.e., [1 0 0 0]i for helix (H), [0 1 0 0]i for strand (E) and [0 0 1-Sk Sk]i for those identified as loop (L) by DSSP, where the 1-Sk and Sk terms in the target vector are taken from the Sk score of Eq. 5, and Sk (k = I, II, I’, II’ and VIII) is the β-turn score of residue i. The input layer for the second level uses the output of the four-state structure classification predicted by the first level of the network, but uses this information from 10 sequential residues, such that the ANN can take advantage of the fact that β-turns are commonly found in between elements of regular secondary structure. Thus, the input layer for the second level has 40 nodes, containing the four-state structure classification of each of the 10 residues (i−4 to i+5) predicted by the first level network (for tetrapeptide sets i−4 to i+5, respectively. The hidden layer contains 10 nodes, and the four-state structure classification of residue i is used as the target of the neural network. The connections between the three layers of the two levels of the network (SI Fig. S4) are to the same as those in the Ncap/Ccap ANN. The empirical formulas of the first and second levels of neural network are:
| [10] |
| [11] |
Neural network training
The weights and bias terms of each of the Ncap/Ccap/β-turn ANNs were determined by training of the network, using the chemical shift and sequence information of the 580-protein chemical shift database, described above. To prevent over-training, a three-fold training and validation procedure was performed for each neural network model by dividing the input training dataset into three input subsets followed by separate training of the corresponding neural networks. For each of these three network optimizations, one input subset was excluded from the training dataset but then used to evaluate the performance of the neural network during the training. Thus, this validation subset was not used to calculate the weight changes in this network. Training of the network was terminated when the performance of the network on the validation dataset, represented by the mean squared errors (MSE) between the predicted values and targets, began to degrade. This procedure was repeated three times for each network, each time with a different one-third of the database entries assigned to the validation set, and the average of the three P1×4 output vectors generated by each of the three separately trained networks is used by MICS to generate the final prediction.
Neural network testing and validation
The predicted Ncap/Ccap score SNcap/SCcap, as represented by the fourth number P4 of the output vector P1×4 (Eq. 9) of the Ncap/Ccap ANN, is used to decide if a given residue i is predicted to be an Ncap/Ccap residue in an Ncap/Cap motif. Similarly, the predicted β-turn score P4k, corresponding to the fourth element of the P1×4 output vector (Eq. 11) of the ANN for predicting β-turn k (k=I, II, I′, II′ and VIII), is used to decide if a given tetrapeptide i is a type k β-turn or not. The actual probability of a hexapeptide to represent an Ncap/Ccap motif, or of a tetrapeptide to be a type k β-turn, subsequently will be derived from these P4k scores by establishing an empirical relation between the P4k and the accuracy of the prediction (see Results).
To inspect the network prediction performance, an accuracy score Qpred is used to report the percentage of the total number of positive Ncap/Ccap/β-turn predictions (NP) that are correct (true positives; NTP):
| [12] |
A sensitivity score Qobs is used to report the percentage of the total number of Ncaps/Ccaps/β-turns present in the database (NT) that are correctly predicted:
| [13] |
Finally, a Matthews correlation coefficient MCC is used as a measure of the overall quality of prediction:
| [14] |
where NTP, NFP, NTN and NFN are the number of true positives, false positives, true negatives and false negatives, respectively (Matthews 1975; Baldi et al. 2000). The MCC score is generally recognized as a balanced measure for prediction accuracy as it takes into account both true and false positives and negatives. Therefore, it can be used even if the classes are of very different sizes, as for example applies in our study of the β-turn/non-β-turn classification. A MCC value of +1 indicates a perfect prediction, 0 an average random prediction, and −1 an inverse prediction; MCC values <~0.4 were obtained for the early β-turn/non-β-turn predictions performed by various bioinformatics methods, with the best empirical programs now approaching MCC values of 0.5 but being unable to accurately predict the type of β-turn (Kirschner and Frishman 2008; Petersen et al. 2010).
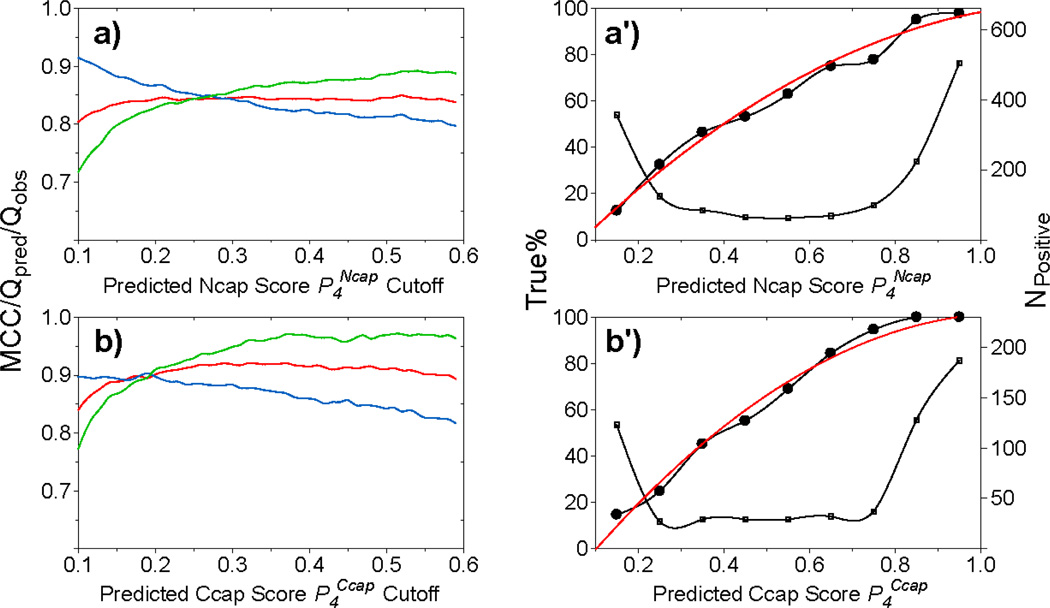
In order to estimate the true probability of a prediction from its predicted Ncaps/Ccaps/β-turns score, P4, a distribution of the prediction accuracy Qpred is generated as a function of its predicted Ncap/Ccap/β-turn score P4 (Table S5; Fig. 6).
Fig. 6.
MICS prediction accuracy for Ncap (a and a') and Ccap (b and b') helix capping motifs, as calculated for all available hexapeptides in the validation datasets when using the trained ANN parameter sets (see Methods). (a,b) Matthews correlation coefficient (MCC, red), Qpred (green) and Qobs (blue), as a function of the threshold value P4 for a positive prediction of a Ncap or Ccap motif. These scores are used to select a best threshold (see SI Table S5) for a positive Ncap/CCap prediction. (a',b') Numbers of hexapeptides with a positively predicted Ncap or CCap motif (small open squares; right y-axis) as a function of the (binned) P4Ncap (a') or P4Ccap (b') value. The fraction of true Ncaps or Ccaps (i.e., with an SNcap or SCcap >0.3) over the number of Ncap/Ccap predictions as a function of the P4Ncap (a') or P4Ccap (b') value (left y-axis, bold line with filled circle). This fraction corresponds to the actual probability that a hexapeptide with a given P4Ncap (P4Ccap) score is an actual Ncap (Ccap) motif, from which fitted polynomial equations (Eqs. S1–S2) are derived (red solid lines) and used by MICS to calculate the actual probability for any given prediction from the P4 score.
In addition to the above three-fold training and validation procedure, a second validation was carried out for a set of eleven proteins not contained in the database. This set of eleven proteins also has nearly complete chemical shifts, high quality reference structures, and no homologous proteins (<30% sequence identity) in the 580-protein database. This same set of eleven proteins previously also was used to validate the SPARTA+ method (Shen and Bax 2010).
Results and Discussion
Neural network prediction of Ncap motifs
In its original definition, the Ncap motif, comprising residues N’-Ncap-Nl-N2-N3-N4 and numbered i−1,…,i+4, is characterized by backbone-sidechain H-bonds between the Ncap residue, i, and the third residue (N3, i+3) of the α-helix, with the ideal N-cap box showing both sc(i)←bb(i+3) and bb(i)→sc(i+3) H-bonds (Harper and Rose 1993). The polypeptide backbone torsion angles of the Ncap residue then cluster around distinct values of ϕ=−92±24° and ψ=167±8° (Table 1; Fig. 1; SI Fig. S1). The calculated electrostatic interaction energy (Kabsch and Sander 1983), averaged over all Ncap motifs in our database, suggests that the sc(i)←bb(i+3) H-bond is slightly more stable than the bb(i)→sc(i+3) H-bond (Table 1). This difference is also reflected in the observation that we find more than twice as many sc(i)←bb(i+3) than bb(i)→sc(i+3) H-bonds in the database Ncap motifs. The sidechains of Ser, Thr, Asp and Asn residues can easily accept a H-bond from a backbone amide, and hence are favored at the Ncap position, whereas the sidechains of Glu, Asp and Gln are preferred at the N3 position (Table S1). Moreover, the juxtaposition of residues N’ and N4 in an Ncap motif favors hydrophobic residues, such as Ile, Leu, Met, Val and Ala at these two positions (SI Table S1) (Aurora and Rose 1998).
The average secondary chemical shift, , and its standard deviation, , for each backbone atom x (x=15N, 13C', 13Cα, 13Cβ, 1Hα and 1HN) at each position j within the Ncap hexapeptide are shown in Fig. 4 and SI Table S3. The chemical shift patterns of the Ncap box, the sc(i)←bb(i+3) Ncap, and the bb(i)→sc(i+3) Ncap motifs are observed to be very similar (Fig. 4A), making it impossible to distinguish these three motifs from their backbone chemical shifts. All three types of Ncaps show the expected large positive Δδ13 Cα and Δδ13C' values expected for the helical residues N1-N4, and the distinct negative Δδ13Cα and positive Δδ13Cβ values highlighted previously by Gronenborn and Clore as being characteristic of the N-cap residue (Gronenborn and Clore 1994). A modest upfield Δδ1HN shift for the N3 residue may reflect the observation that in many of the Ncaps the amide of N3 cannot form a very stable H-bond. Other characteristic features include downfield Δδ1HN for N1 and upfield Δδ15N secondary shifts for N2, but again these features are very similar for the three Ncap motifs. Therefore, for purposes of our computational analysis, the Ncap box, the sc(i)←bb(i+3) Ncap, and the bb(i)→sc(i+3) Ncap are grouped together in a single Ncap motif.
The scoring function, SNcap, of Eq. 1 evaluates how closely the positional ϕ/ψ torsion angles of any given hexapeptide resemble an Ncap motif. A histogram of all hexapeptides in the chemical shift database shows that the SNcap values for Ncap motifs mostly are larger than 0.9 (Fig. 3a), with the Ncap box motif yielding the highest scores.
It is interesting to note that there are a substantial number of hexapeptides in the database that match the backbone angles of an Ncap motif, but lack both the sc(i)←bb(i+3) and the bb(i)→sc(i+3) H-bonds. So, even while the backbone of these hexapeptides is geometrically very close to that of true Ncap motifs, they lack the requisite H-bonds. Inspection shows that these Ncap-like motifs are stabilized by alternate H-bonds, most commonly bb←bb(i+3) (23%), sc←bb(i+2) (21%) and sc→sc(i+3) (4%) between the Ncap residue and its flanking residues in the helix Table S7). Considering the close structural similarity and nearly indistinguishable chemical shifts relative to true Ncaps, we include these Ncap-like motifs in the Ncap classification.
The Ncap neural network (Ncap ANN) is trained by using the amino acid type and chemical shift data present in the chemical shift database as input, and the four-state structural classification [helix, strand, loop, Ncap]. So, as training target for the ANN we use 0 or 1 for the first two elements of this vector, based on the residue's three-state DSSP secondary structure classification [helix, strand, loop]. For the third element of residues identified by DSSP as loop, a modified loop score is used, 1- SNcap, and SNcap is used as the value of the fourth element (see Methods). The trained Ncap ANN performs well in terms of reproducing the four-state training target in the validation datasets, in particular the Ncap score. In total, 84% (Qobs) of the observed Ncaps in the validation datasets (with SNcap score ≥0.3) are positively predicted, i.e., yield a predicted score ≥0.3. Importantly, the fraction of hexapeptides for which the ANN predicts an Ncap in the validation set and which indeed corresponds to an Ncap based on the ϕ/ψ-derived SNcap (cf Eq. 1) is 86% (Qpred), i.e. at the SNcap = 0.3 threshold, 86% of the predictions are correct (Table S5). As shown in Fig. 6a, Qobs and Qpred are a function of the value of the SNcap threshold used, even though the MCC score of 0.85 is relatively insensitive to this threshold value.
Neural network prediction of Ccap motifs
As with the Ncap motifs, distinction of the different types of capping motifs, in particular Schellman and αL, can be problematic due to the mixed H-bonding patterns noted above. Moreover, very similar secondary chemical shifts are also observed for the Schellman and αL Ccap motifs (Fig. 4b), foreshadowing the difficulty to distinguish these Ccap motifs on the basis of their chemical shifts. Due to the requirement of a positive ϕ torsion angle for the residues at the C’ position, Gly and Asn are the two residues most commonly observed in this position. Analogous to the Ncap, hydrophobic interaction between residues C3 and C″ often bracket the helix capping motif (Aurora et al. 1994; Aurora and Rose 1998), resulting in an increased propensity for Leu, Ala and Met in the C3 position, and Ile, Leu and Val at C″ (SI Table S1). By contrast, the hydrophilic residues Glu, Lys, Arg and Gln are more often observed at positions C2 and C1 (SI Table S1).
As expected, the Ccap motif (C3-C2-C1-Ccap-C’-C”; numbered i,…,i+5) exhibits the Δδ13Cα and Δδ13Cβ values characteristic of α-helix for residues C3, C2 and C1. The backbone torsion angles of the Ccap residue itself cluster around ϕ=−92±15 and ψ=−1±16 (Table 1; Fig. 1b,c). Despite these angles being close to α-helical values, the 13Cα, 13Cβ, 13C’ and 1Hα Δδ values are close to zero (SI Table S3; Fig. 4b). By contrast, Δδ15N of both the αL and Schellman Ccap residue shows a substantial upfield shift of ca −6 ppm. Although, with the exception of the Ccap Δδ15N, the Δδ values for the Ccap, C' and C" residues are small in magnitude, their values are weakly correlated with one another (SI Fig. S7), a feature which is automatically taken advantage of during the training of the ANN. So, even while the secondary shifts for the Ccap motifs at first sight appear to be close to random coil, together with the Ccap amino acid propensities they suffice for training the ANN to identify the Ccap motifs. However, the differences between Schellman and αL Ccaps are insufficiently distinct to allow them to be distinguished from one another by the ANN, and we therefore combine them into a single Ccap motif.
Occasionally, α-helices are terminated by a Pro residue, where a steric collision between the pyrrolidine ring and the preceding residue prevents continuation of the α-helix. About 4800 and 200 such helices are found in the structure database and chemical shift database, respectively, and 144 and 3 of them can be classified as being a praline Ccap motif, which must have a Pro at C' and a three-center H-bond linking the C=O at the Ccap position to the N-H in C‴ and C′‴ positions, i.e., bb(Ccap)←bb(C‴) and bb(Ccap)←bb(C′‴) H bonds (Aurora and Rose 1998). If the requirement of a three-center H-bond is relaxed to include either a bb(Ccap)←bb(C‴) H-bond or a bb(Ccap)←bb(C′‴) H-bond, 1704 and 70 helices can be recognized as terminated by a proline Ccap motif, respectively. The backbone conformation and chemical shift patterns for this extended proline Ccap motif are essentially the same as those containing the three-center H-bond, and we will therefore use this more generous definition instead. Of the 1704 helices terminated by such a proline Ccap motif, the ϕ/ψ torsion angles of the Pro residue cluster in two regions, centered at −60±8°/−23±12° (1434) and at - 58±8°/137±12° (270), respectively; the residues at the Ccap position also exhibit a bimodal ϕ/ψ distribution with its two main clusters centered at −135°/75° and −75°/130° (SI Fig. S5). However, there appears no direct correlation between the ϕ/ψ torsion angles of the Ccap residue and the Pro at C'. Consequently, the Δδ values of the Ccap residues in the 70 proline Ccaps in the chemical shift database show large dispersion (Table S3), making it difficult to identify the Pro Ccap motifs on the basis of chemical shifts, and proline Ccaps are therefore not included in our analysis.
The Ccap score, SCcap, calculated using Eq. 3 for all available hexapeptides in the chemical shift database shows that the vast majority of Ccaps yield a score higher than 0.6, with the majority clustered between 0.9 and 0.95 (Fig. 3b). The Ccap ANN is trained in a manner very similar to that used for the Ncap ANN, and the trained network shows excellent performance to reproduce the four-state structural classification [helix, strand, loop, Ccap]. Despite lacking very distinct chemical shifts, except for the Δδ15N of the Ccap residue, the Ccap ANN is remarkably effective at identifying Ccap motifs on the basis of chemical shifts and amino acid composition. Using a prediction score cutoff value of 0.3, 94% of its selected Ccap hexapeptides represent true Ccap motifs (Qpred = 0.94) and 88% of the true Ccaps are identified by the ANN (Qobs = 0.88), for a total MCC value of 0.92 (SI Table S5). As with the N-cap motifs, the values of Qpred and Qobs scale with the prediction threshold value used, but the MCC value remains relatively constant (Fig. 6b). Evaluation of the trained Ccap ANN on the separate validation dataset of 11 proteins not used in any of the ANN training yields Qpred and Qobs values very similar to those observed for the chemical shift database (Table 3).
Table 3.
MICS prediction performance for 11 proteins which are not present in the training database
| BMRB/PDB | Ncap | Ccap | β-turn I | β-turn II | β-turn I′ | β-turn II′ | β-turn VIII |
|---|---|---|---|---|---|---|---|
| dinI/1ghh | 1/1/1a | - | 2/2/2 | - | - | - | 3/2/2 |
| 5589/1nxi | 2/2/2 | 3/3/3 | 2/2/1 | - | - | - | - |
| 16146/1enfA | 4/3/3 | 1/1/1 | 4/6/4 | 2/0/0 | 2/1/1 | - | 3/3/2 |
| 16321/1wzvA | - | 0/1/0 | 7/6/6 | - | - | - | 6/5/5 |
| 16362/1gwyA | 2/2/1 | 1/1/1 | 2/1/1 | 1/0/0 | 1/0/0 | - | 2/1/1 |
| 16447/1phpA | 3/3/2 | 6/5/5 | 5/8/5 | 2/2/2 | - | 1/2/1 | 7/7/6 |
| 16537/2etlA | 3/2/2 | 2/1/1 | 4/4/2 | 2/1/1 | 1/1/1 | - | 3/2/2 |
| 16572/3hn9A | - | - | 4/6/4 | 3/2/2 | - | 1/1/1 | 3/4/3 |
| 16656/3ipfA | - | - | 2/2/2 | - | - | - | 2/2/2 |
| 16661/3gzmA | 4/4/4 | 3/2/2 | 1/1/1 | - | - | - | - |
| 16684/3l48C | - | - | 3/3/3 | 3/2/2 | 1/1/1 | - | 1/1/1 |
| overall | 19/17/14 | 16/14/13 | 36/41/31 | 13/7/7 | 5/3/3 | 2/3/2 | 30/27/24 |
number of the observed, predicted, and correctly predicted elements. Only those observed N-caps, C-caps and β-turns with sufficient observed chemical shift data, i.e., with at least three backbone and 13Cβ chemical shifts per residue, are counted. The thresholds of the predicted score used to assign a positive prediction are listed in SI Table S5.
Neural network prediction of β-turns
Tight β-turn motifs are highly abundant in proteins. They play an important role in protein folding. In addition to other tight turns, such as δ-, γ-, α-, and π-turns, they enable direct contacts between elements of regular secondary structure (α-helix and β-strand) by reversing the direction of the polypeptide chain. When intervening between two segments of regular secondary structure, a β-turn invariably is associated with important stabilizing interactions, such as pairing of β–strands and α-helix packing. Depending on the ϕ/ψ torsion angles of their two center residues, β-turns are further sub-categorized into types, I, II, VIII, I′, II′, VIa1, VIa2 and VIb, with the remainder being assigned to a "miscellaneous" type IV. In our study we aim to identify five of the most common types: I, II, I′, II′ and VIII. The ϕ/ψ torsion angles show large variations for the first and last residues in a β-turn, but tight clustering for the center two residues in each turn type (Fig. 2 and SI Fig. S1).
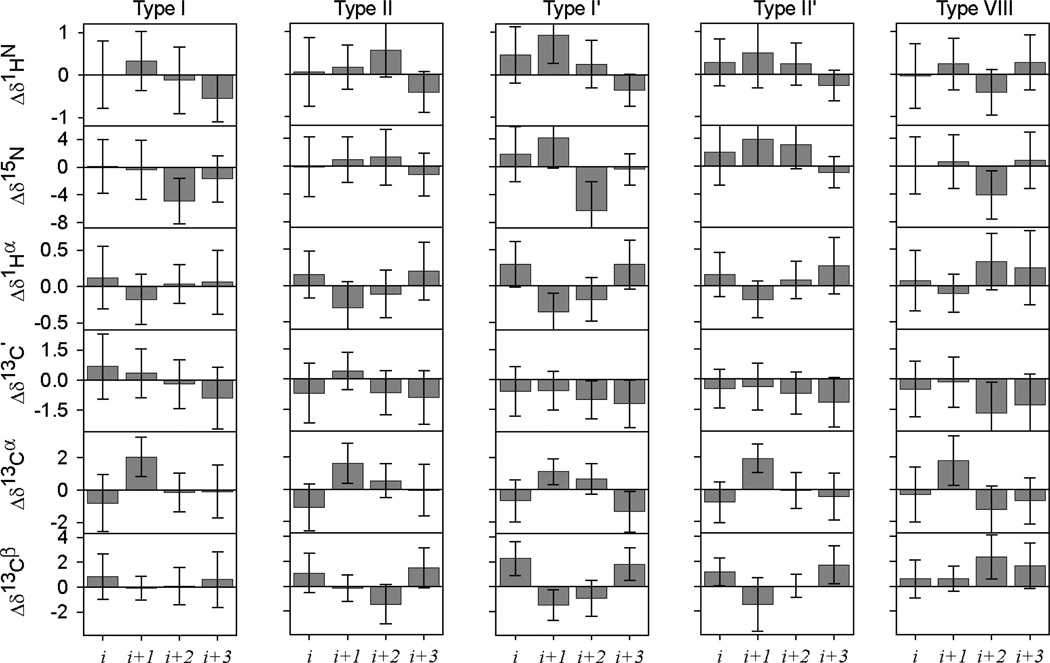
A total of 914, 432, 175, 107 and 309 isolated β-turns are observed in the chemical shift database with type I, II, I’, II’ and VIII, respectively. The secondary chemical shifts observed for each of these turn residues show substantial scatter and relatively few distinguishing features (Fig. 7). For most turn nuclei, the Δδ value differs by less than one standard deviation from zero. However, as was noted for the Ccap residues, the values within a given turn again are weakly correlated with one another (SI Fig. S8). It is difficult to take advantage of this feature in regular chemical shift analysis, but the correlated nature of the Δδ values is automatically included in the training process of the ANN. So, even while the secondary shifts for the different β-turn types at first sight appear to be very similar to one another, together with the amino acid propensity in different turn types these values contain sufficient information for the trained ANN to distinguish between the different turn types.
Fig. 7.
Chemical shift patterns for five types of β-turns. For all tetrapeptides in the chemical shift database identified as isolated type I, II, I′, II′ or VIII β-turns (according to the original definitions; see Methods), the average secondary 1HN, 15N, 1Hα, 13C′, 13Cα and 13Cβ chemical shifts are plotted together with their standard deviations for each of the four residues.
Of the different β-turns, types I, II, I′ and II′ exhibit similar Cα(i,i+3) distance distributions, with types I' and II' showing the tightest clustering and shortest average Cα(i,i+3) distance (Table 2; SI Fig. S2). Type VIII β-turns tend to have longer Cα(i,i+3) distances (Table 2) and appear contiguous with the more “open” turns, which frequently have type-VIII-like ϕ/ψ torsion angles, but Cα(i,i+3) > 7 Å (SI Fig. S2). About 75% of the type I, I', II and II' β-turns are expected to show a bb(i)←bb(i+3) H-bond from the last to the first residue (Hutchinson and Thornton 1994), whose presence correlates with the short β-turn Cα(i,i+3) distance (<7 Å). In our protein structure database, more than 80–90% of these turns include such an intra-turn H-bond (Table 2), exhibiting comparable distributions for the computed H-bond energy (Table 2; SI Fig. S3). In contrast, the bb(i)←bb(i+3) H-bond is rarely observed in type VIII β-turns, an observation that can be linked to their relative long Cα(i,i+3) distance. A large fraction (60–80%) of the type I′ and II′ β-turns additionally exhibit bb(i)→bb(i+3) H-bonds (Table 2). O t her intra-turn H-bonds, including bb(i)→sc(i+3), bb(i)←sc(i+3) and sc(i)←bb(i+3) are less common, and are observed with frequencies of (5%, 2%, 12%) for type I, (13%, 10%, 0.5%) for type II, (0.5%, 3%, 1%) for type I′, (2%, 4%, 0%) for type II′, and (1%, 1%, 0%) for type VIII.
The amino acid sequence preference of β-turns, and the association of those favored amino acids with the stability of β-turns has been studied extensively (Wilmot and Thornton 1988; Hutchinson and Thornton 1994). The amino acid sequence also serves as the input to a wide array of bioinformatics algorithms to predict the presence of β-turns (Wilmot and Thornton 1990; Chou 2000; Kirschner and Frishman 2008; Petersen et al. 2010). However, using sequence information alone it proves difficult to distinguish the different β-turn types, and often all β-turns are grouped into either a single class or two classes, I and II (Petersen et al. 2010). The positional preference observed for the various turn types in our protein structure database is very similar to those reported earlier, showing a high occurrence of short and hydrophilic amino acids, such as Asp, Asn and Gly, at the two center positions (Table S2), which are responsible for reversing the direction of the polypeptide chain. The polar character of these residues relates to the fact that most turns are located on the protein surface and therefore exposed to solvent.
Type VIII β-turns, added by Wilmot and Thornton (1990) to complement the six categories (types I, II, I′, I I′, VIa and VIb) defined by Richardson (1981), are characterized by a second residue with an αR and a third residue with a β backbone conformation. As mentioned above, they tend to be more “open” in terms of Cα(i,i+3) distance and they lack intra-turn H-bonds. Moreover, in contrast to the other β-turns, the number of type VIII turns rapidly increases when the Cα(i,i+3) distance cutoff is slightly increased (SI Fig. S2). This makes it difficult to draw a sharp distinction on the basis of backbone torsion angles alone, as reflected in the broad and contiguous distribution of β-turn score values SVIII (Fig. 3G). The broader distribution of geometries represented in type VIII turns is the likely reason that bioinformatics predictions of these turns show much lower success ratios compared to predictions of the other turn types (Shepherd et al. 1999; Kirschner and Frishman 2008; Kountouris and Hirst 2010).
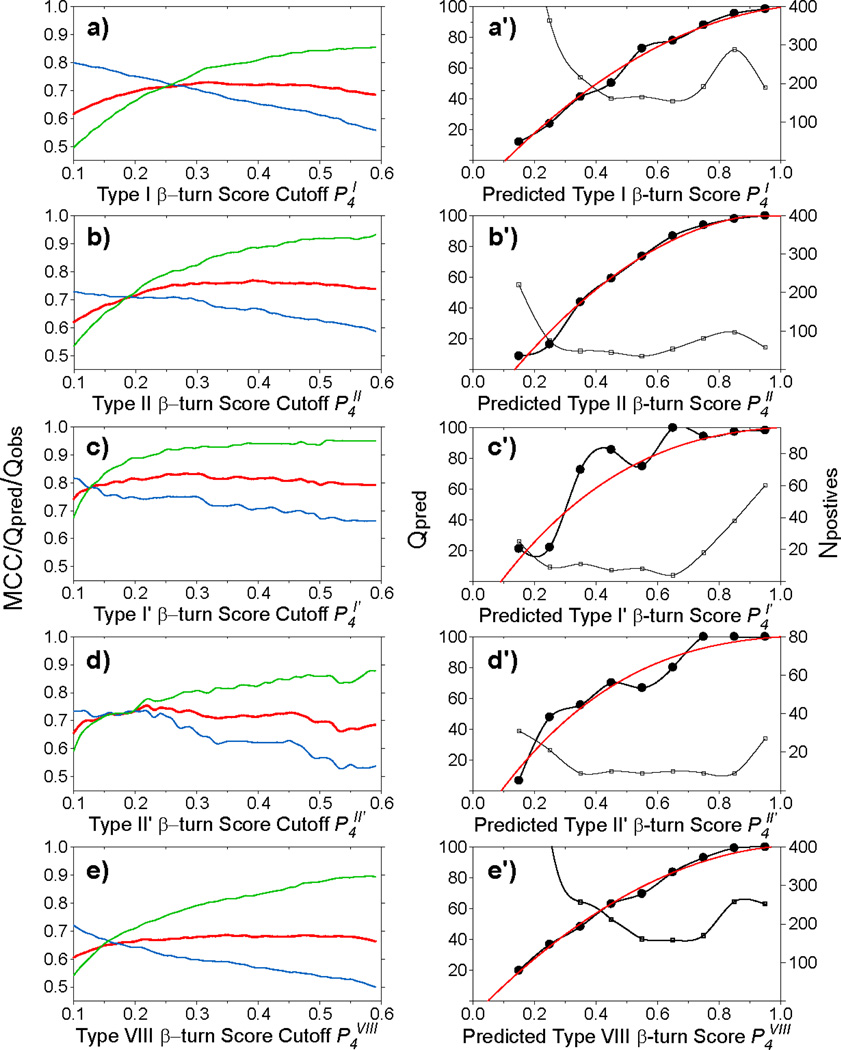
The empirical β-turn score, Sk (Eq. 5), represents a numerical value reflecting how closely any given turn mimics the idealized turn type in terms of backbone angles. β-turn scores, Sk (k = I, II, I′, II′ and VIII), are calculated for each tetrapeptide in the chemical shift database. With the exception of the type VIII β-turn, the calculated β-turn scores are shown to be highly effective at identifying each of these five types of β-turns (Fig. 3c–f). Almost all type I, II, I′, II′ β-turns have a corresponding calculated score Sk > ~0.4, while nearly all non β-turns and non type k β-turns have a very low score Sk < 0.05. Type IV β-turns are the only exception, and a number of these exhibit elevated Sk scores between 0.1 and 0.5. These type IV β-turns with high Sk values actually are very similar to type k β-turns in their ϕ/ψ angle pattern, but are excluded by the hard cutoff (30°/45°; see Methods) used for the ϕ/ψ angles in the original definitions. We use a Sk score cutoff of ~0.2–0.3 (Table S5) to assign type I, II, I′, II′ and VIII β-turns, such as to yield optimal overlap between turns identified by Eq. 5 and those of the original literature definitions.
As described above for the helix capping motifs, extending the three-state secondary structure classification [helix, strand, loop] by a fourth class, the specific β-turn character at any given position in the sequence, provides a suitable avenue to train the various specific β-turn ANNs. Five separate β-turn ANN are trained for the five types of β-turns considered in our study. Using the chemical shifts and amino acid sequence information contained in our chemical shift database, the trained networks are proving to be highly effective at identifying type I, II, I′ and II′ β-turns, and at a somewhat lower level of accuracy also for the less distinct type VIII β-turn. The values obtained for Qpred and Qobs again scale with the value chosen for the score cutoff (Fig. 8), whereas the MCC value is less sensitive to the score cutoff. Final Sk score cut-off values (Table S5) are selected to optimize the overall β-turn ANN prediction. Performance of the ANNs for both the validation datasets used during training (Table S5) and the separate set of eleven additional validation proteins (Tables 3, S8) show that ca 75% of the β-turns actually present are positively identified by their specific ANN, with relatively few false positives among these predictions, in particular when the prediction yields a high P4k value (Table S5). MCC scores range from 0.67 for the type VIII β-turn to 0.83 for type I'. The best performance is seen for type I', and correlates with the unique backbone conformation of its two center residues, which both have positive ϕ angles.
Fig. 8.
MICS prediction accuracy for five types of β-turns. Prediction performance scores for type I (a and a'), II (b, b'), I′ (c,c'), II′ (d, d') and VIII (e, e') β-turns for all available tetrapeptides obtained by the trained ANN in the validation datasets (see Methods). (a–e) Matthews correlation coefficient (MCC, red), Qpred (green) and Qobs (blue), as a function of the P4k threshold value used for a positive prediction of a type k β-turn. These scores are used to select the threshold (see SI Table S5) for a positive type k β-turn prediction. (a'–e') The numbers of the positively predicted β-turns are plotted (right y-axis, thin line with open squares) as a function of the binned P4k value; the ratio of true type k β-turns relative to the number of predictions (Qpred) (left y-axis, bold line with filled circles) shows the probability that any given tetrapeptide is correctly identified as a type k β-turn, from which fitted polynomials (Eqs. S3–S7) are derived (red solid lines) and used by MICS to convert the P4 scores into actual probabilities.
The statistics reported in Table 3 and SI Tables S5 and S8 suggest that there is a non-negligible fraction of false positive β-turn predictions, and the same applies for positive Ncap/Ccap predictions. Of these false positives, many yield a high predicted score and it appears that in many cases the appearance of a false positive may be caused by actual differences between the reference structure, studied in the crystalline state, and the structure present in solution. The vast majority of β-turns locate on the protein surface, which frequently exhibit increased backbone mobility and sometimes adopt different conformations when crystallized in a different space group For example, the tetrapeptide V83-N84-G85-H86 adopts a type IV β-turn (ϕi+2/ψi+2=−116°/52°; SI′ = 0.04) in reference structure 2ZJD (solved at a crystallographic resolution of 1.56 Å, and used by our chemical shift database), and as a type I′ β-turn (SI′ = 0.99) in reference structure 2Z0E (solved at 1.9 Å). MICS predicts this element to be a type I′ β-turn with a predicted P4I′ score of 0.9. Another reason for false positive predictions, in particular for type I β-turns, is that when such turns partially overlap with one another, these adjacent type I β-turns are often classified as 3–10 helices by the DSSP program. Interestingly, some of the 3–10 helical segments identified by DSSP are designated as loops or β-turns by other secondary structure assignment programs, such as STRIDE (Heinig and Frishman 2004). In this respect it is important to note that even for the same set of atomic coordinates different programs do not always agree in their designation of regular secondary structure, which therefore limits the extent to which such elements can be recognized from NMR chemical shifts (Wishart 2011).
Concluding remarks
Both the N- and C-terminal helix capping motifs as well as five specific types of β-turns can be predicted with quite good accuracy on the basis of amino acid type and backbone and 13Cβ chemical shifts. The trained ANNs report a score that is directly related to the likelihood for any given residue to be either the second residue in a four-residue β-turn, or the helical Ncap or Ccap residue. An empirical relation between these ANN scores and the likelihood that the ANN prediction is correct (red lines in Figs. 6a’–b’; 8a’–e’) is then used to convert the ANN scores into actual probabilities. As the ANN programs for recognizing the different elements of secondary structure, including α-helix, β-sheet, loop, turns, and helix caps have been trained separately, use of empirical correlations such as graphed in Figs. 6 and 8 potentially can result in total probabilities, summed over the ten different categories, that are higher than 100%. The probability output is therefore normalized such that its output cannot exceed 100%. The full MICS output file reports for each residue both the ten ANN output scores for it to be of type k (P1 for helix, P2 for sheet, P3 for loop and P4k for Ncap, Ccap and the five β-turns) and the actual probabilities, derived from these ANN output scores. MICS is available as a user-friendly webserver (http://spin.niddk.nih.gov/bax/nmrserver/mics). It accepts chemical shift input files that are either in BMRB or TALOS input format and presents a graphical display reporting the probabilities that each given residue is of type k. In addition, the full output file is emailed to the user.
We anticipate MICS to be useful both in conventional protein structure determination as well as in the chemical shift based structural modeling, as exemplified by the programs CHESHIRE and CS-Rosetta. For conventional protein structure determination, in particular for larger proteins where the number of NOEs per residue frequently is smaller, correct identification of local turns and helix capping motifs on the basis of NOE data can be difficult and the MICS output will serve as a guide for correct identification of these local structural elements. CHESHIRE and CS-Rosetta use as their input a large ensemble of short peptide fragments, selected from a protein structure database. These fragments are then used in a computationally intensive search to derive a structure that is of low empirical energy. Finding suitable input fragments is easiest for α-helical and β-sheet segments of a protein, but can be very difficult for other regions. We anticipate that the quantitative probabilities provided by MICS will aid in assembling collections of input fragments that better reflect the true structure, and therefore will result in improved convergence and potentially extend the size limit of proteins that can be studied by such methods.
The limitations of secondary structure prediction from amino acid sequence information alone are exemplified by a recent study where a single amino acid substitution can switch the protein structure from a mixed α/β structure to a 3-helix fold (Alexander et al. 2009). Clearly, chemical shifts which report on the local backbone torsion angles are invaluable in such cases and can unambiguously identify the correct secondary structure (Shen et al. 2010). Analogously, we here have demonstrated that the use of chemical shifts, in addition to sequence, can dramatically improve the accuracy at which specific β-turns can be identified (SI Table S8) over what can be achieved with some of the latest sequence-only programs (Fuchs and Alix 2005; Kountouris and Hirst 2010; Petersen et al. 2010). To the best of our knowledge, there are currently no webserver programs that predict helix-capping motifs from amino sequence alone. However, when we train our Ncap and Ccap ANN for identification of such motifs in the absence of any chemical shift information, the MCC scores for corresponding predictions are approximately two-fold lower than obtained by MICS (data not shown).
Supplementary Material
Fig. 9.
Output of the MICS program, displaying results of the helix capping motif and β-turn type predictions for ubiquitin. MICS also includes in its output the secondary structure prediction for α-helix (red bars) and β-sheet (cyan bars) as well as the chemical shift based RCI-S2 prediction (Berjanskii and Wishart 2005) (green dots and lines; upper panel). The predicted Ncap and Ccap motifs are marked by yellow arrows (second panel), and the predicted type I, II, I′, II′ and VIII β-turns (blue bars, with solid color for the two center residues and transparent for the first and last residues) are displayed in separate panels. Note that for ubiquitin no type II, II' or VIII turns are predicted, and those panels have therefore been deleted from the figure. The heights of the bars and arrows correspond to the normalized probabilities assigned by MICS.
Acknowledgments
We thank Frank Delaglio for helpful comments and suggestions. This work was funded by the Intramural Research Program of the NIDDK and by the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH.
Footnotes
Electronic supplementary material The online version of this article (doi:xxxxxxxxxx) contains supplementary material, which is available to authorized users.
Software availability
The MICS program can be downloaded from http://spin.niddk.nih.gov/bax/software. MICS can also be run in webserver mode at http://spin.niddk.nih.gov/bax/nmrserver/mics.
References
- Alexander PA, He Y, Chen Y, Orban J, Bryan PN. A minimal sequence code for switching protein structure and function. Proc. Natl. Acad. Sci U.S.A. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T, Taoka K, Demura M, Williamson MP. The relationship between amide proton chemical shifts and secondary structure in proteins. J. Biomol. NMR. 1995;6:227–236. doi: 10.1007/BF00197804. [DOI] [PubMed] [Google Scholar]
- Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora R, Srinivasan R, Rose GD. Rules for α-helix termination by glycine. Science. 1994;264:1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- Baldi P, Brunak S, Chauvin Y, Andersen CAF, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- Baldwin RL, and Rose GD. Is protein folding hierarchic? II. Folding intermediates and transition states. Trends Biochem. Sci. 1999;24:77–83. doi: 10.1016/s0968-0004(98)01345-0. [DOI] [PubMed] [Google Scholar]
- Becker OM, Karplus M. The topology of multidimensional potential energy surfaces: Theory and application to peptide structure and kinetics. J. Chem. Phys. 1997;106:1495–1517. [Google Scholar]
- Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjanskii MV, and Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- Bystroff C, Baker D. Prediction of local structure in proteins using a library of sequence-structure motifs. J. Mol. Biol. 1998;281:565–577. doi: 10.1006/jmbi.1998.1943. [DOI] [PubMed] [Google Scholar]
- Case DA. The use of chemical shifts and their anisotropies in biomolecular structure determination. Curr. Opin. Struct. Biol. 1998;8:624–630. doi: 10.1016/s0959-440x(98)80155-3. [DOI] [PubMed] [Google Scholar]
- Chou KC. Prediction of tight turns and their types in proteins. Anal. Biochem. 2000;286:1–16. doi: 10.1006/abio.2000.4757. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- de Dios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts - an ab initio approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- Doreleijers JF, Nederveen AJ, Vranken W, Lin JD, Bonvin A, Kaptein R, Markley JL, Ulrich EL. BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. J. Biomol. NMR. 2005;32:1–12. doi: 10.1007/s10858-005-2195-0. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. Folding of immunogenic peptide fragments of proteins in water solution. 1. Sequence requirements for the formation of a reverse turn. J. Mol. Biol. 1988;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- Eghbalnia HR, Wang LY, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- Fuchs PFJ, Alix AJP. High accuracy prediction of beta-turns and their types using propensities and multiple alignments. Proteins-Structure Function and Bioinformatics. 2005;59:828–839. doi: 10.1002/prot.20461. [DOI] [PubMed] [Google Scholar]
- Gronenborn AM, Clore GM. Identification of N-terminal helix capping boxes by means of 13C chemical shifts. J. Biomol. NMR. 1994;4:455–458. doi: 10.1007/BF00179351. [DOI] [PubMed] [Google Scholar]
- Han B, Liu YF, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper ET, Rose GD. Helix stop signals in proteins and peptides - the capping box. Biochemistry. 1993;32:7605–7609. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500–W502. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino-acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LH, Samudrala R. Accurate and automated classification of protein secondary structure with PsiCSI. Protein Sci. 2003;12:288–295. doi: 10.1110/ps.0222303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EG, Thornton JM. A revised set of potential for β-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwadate M, Asakura T, Williamson MP. C-alpha and C-beta carbon-13 chemical shifts in proteins from an empirical database. J. Biomol. NMR. 1999;13:199–211. doi: 10.1023/a:1008376710086. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kaur H, Raghava GPS. Prediction of beta-turns in proteins from multiple alignment using neural network. Protein Sci. 2003;12:627–634. doi: 10.1110/ps.0228903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner A, Frishman D. Prediction of beta-turns and beta-turn types by a novel bidirectional Elman-type recurrent neural network with multiple output layers (MOLEBRNN) Gene. 2008;422:22–29. doi: 10.1016/j.gene.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Kountouris P, Hirst JD. Predicting beta-turns and their types using predicted backbone dihedral angles and secondary structures. Bmc Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Comparison of predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Meiler J. PROSHIFT: Protein chemical shift prediction using artificial neural networks. J. Biomol. NMR. 2003;26:25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- Moon S, Case DA. A new model for chemical shifts of amide hydrogens in proteins. J. Biomol. NMR. 2007;38:139–150. doi: 10.1007/s10858-007-9156-8. [DOI] [PubMed] [Google Scholar]
- Neal S, Nip AM, Zhang HY, Wishart DS. Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. J. Biomol. NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- Pastore A, Saudek V. The relationship between chemical shift and secondary structure in proteins. J. Magn. Reson. 1990;90:165–176. [Google Scholar]
- Pearson JG, Le HB, Sanders LK, Godbout N, Havlin RH, Oldfield E. Predicting chemical shifts in proteins: Structure refinement of valine residues by using ab initio and empirical geometry optimizations. J. Am. Chem. Soc. 1997;119:11941–11950. [Google Scholar]
- Petersen B, Lundegaard C, Petersen TN. NetTurnP - Neural Network Prediction of Beta-turns by Use of Evolutionary Information and Predicted Protein Sequence Features. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta LG, Rose GD. Helix signals in proteins. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- Richardson JS. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using rosetta. Meth. Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv. Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70 percent accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Saito H. Conformation-dependent C13 chemical shifts - A new means of conformational characterization as obtained by high resolution solid state C13 NMR. Magn. Reson. Chem. 1986;24:835–852. [Google Scholar]
- Sgourakis NG, Lange OF, DiMaio F, Andre I, Fitzkee NC, Rossi P, Montelione GT, Bax A, Baker D. Determination of the Structures of Symmetric Protein Oligomers from NMR Chemical Shifts and Residual Dipolar Couplings. J. Am. Chem. Soc. 2011;133:6288–6298. doi: 10.1021/ja111318m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J. Biomol. NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J. Biomol. NMR. 2010;46:199–204. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. SPARTA plus : a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bryan PN, He YN, Orban J, Baker D, Bax A. De novo structure generation using chemical shifts for proteins with high-sequence identity but different folds. Protein Sci. 2010;19:349–356. doi: 10.1002/pro.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+ : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd AJ, Gorse D, Thornton JM. Prediction of the location and type of beta-turns in proteins using neural networks. Protein Sci. 1999;8:1045–1055. doi: 10.1110/ps.8.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Blundell TL, Thornton JM. Conformation of b-hairpins in protein structures: A systematic classification with applications to modelling by homology, electron density fitting and protein engineering. J. Mol. Biol. 1989;206:759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- Spera S, Bax A. Empirical correlation between protein backbone conformation and Ca and Cb 13C nuclear magnetic resonance chemical shifts. Journal of American Chemical Society. 1991;113:5490–5492. [Google Scholar]
- Vila JA, Aramini JM, Rossi P, Kuzin A, Su M, Seetharaman J, Xiao R, Tong L, Montelione GT, Scheraga HA. Quantum chemical C-13(alpha) chemical shift calculations for protein NMR structure determination, refinement and validation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14389–14394. doi: 10.1073/pnas.0807105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila JA, Villegas ME, Baldoni HA, Scheraga HA. Predicting C-13(alpha) chemical shifts for validation of protein structures. J. Biomol. NMR. 2007;38:221–235. doi: 10.1007/s10858-007-9162-x. [DOI] [PubMed] [Google Scholar]
- Wang CC, Chen JH, Lai WC, Chuang WJ. 2DCSi: identification of protein secondary structure and redox state using 2D cluster analysis of NMR chemical shifts. J. Biomol. NMR. 2007;38:57–63. doi: 10.1007/s10858-007-9146-x. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP. Secondary structure dependent chemical shifts in proteins. Biopolymers. 1990;29:1428–1431. doi: 10.1002/bip.360291009. [DOI] [PubMed] [Google Scholar]
- Wilmot CM, Thornton JM. Analysis and prediction of the different types of b-turn in proteins. J. Mol. Biol. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- Wilmot CM, Thornton JM. Beta-turns and their distortions - a proposed new nomenclature. Protein Eng. 1990;3:479–493. doi: 10.1093/protein/3.6.479. [DOI] [PubMed] [Google Scholar]
- Wishart DS. Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The C-13 chemical shift index - a simple method for the identification of protein secondary structure using C-13 chemical shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Watson MS, Boyko RF, Sykes BD. Automated (1)H and (13)C chemical shift prediction using the BioMagResBank. J. Biomol. NMR. 1997;10:329–336. doi: 10.1023/a:1018373822088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.