Abstract
Background & Aims
Intestinal dysbiosis and bacterial translocation is common in patients with advanced liver disease, and there is strong evidence that the translocation of bacteria and their products across the epithelial barrier drives experimental liver disease progression. The aims of our study were to investigate dynamics of bacterial translocation and changes in the enteric microbiome in early stages of liver disease.
Methods
Cholestatic liver injury was induced by ligation of the common bile duct (BDL) and toxic liver injury by injection of carbon tetrachloride (CCl4) in mice.
Results
Increased intestinal permeability and bacterial translocation occurred one day following liver injury in both disease models. This was accompanied by decreased intestinal expression of the tight junction protein occludin. Although BDL resulted in a rapid onset of intestinal bacterial overgrowth, bacterial overgrowth was observed in mice injected with CCl4 only in advanced stages of liver fibrosis. To further assess the qualitative changes in the intestinal microbiome, massively parallel pyrosequencing of 16S rRNA genes revealed minor microbial changes following BDL, while CCl4 administration resulted in a relative abundance of Firmicutes and Actinobacteria compared with oil injected mice. Four different liver disease models (cholestasis, toxic, alcohol, obesity) show few similarities in their intestinal microbiome.
Conclusions
Acute liver injury is associated with an early onset of increased intestinal permeability and bacterial translocation that precede changes in the microbiome. The enteric microbiome differs with respect to the etiology of liver disease.
Keywords: bacterial overgrowth, microbiome, dysbiosis, bacterial translocation, 16S rRNA sequencing
Introduction
Acute liver injury results in hepatocyte damage and acute inflammation, while chronic liver injury of any origin exhausts the regenerative capacity of the liver and results in fibrosis, which is characterized by an excessive deposition of extracellular matrix proteins. Cirrhosis occurs with the development of regenerating nodules of hepatocytes. Patients with decompensated liver cirrhosis have a poor prognosis and liver transplantation is often necessary [1, 2]. Increased mortality in patients with liver cirrhosis is most often attributed to direct complications resulting from the loss of liver function, variceal hemorrhage as a sequela of portal hypertension, and the development of hepatocellular carcinoma. However, a significant percentage of patients also succumb to bacterial infections with an infection-attributed mortality of 30% to 50% [3–5]. Bacterial translocation, defined as the passage of viable endogenous bacteria or their products from the intestinal tract through the epithelial barrier to the mesenteric lymph nodes, systemic circulation, or extraintestinal organs, is considered an important mechanism for the incidence of infections including spontaneous bacterial peritonitis and sepsis in cirrhotic patients [6].
Patients with liver cirrhosis have increased intestinal permeability as compared to healthy controls, and it was more apparent in patients with overt clinical complications [7]. Direct evidence for increased bacterial translocation comes from studies demonstrating increased viable bacteria in the mesenteric lymph nodes [8], increased bacterial DNA [9, 10] and increased endotoxin (lipopolysaccharide or LPS) in the plasma of patients with cirrhosis [11–13]. Endotoxemia has been associated with decreased survival rates in patients with liver cirrhosis [14]. LPS-induced local and systemic inflammation is associated with cirrhosis and predicts progression to end-stage liver disease in patients with HBV or HCV infection [15]. In addition to bacterial translocation, patients with advanced liver dysfunction of various etiologies show intestinal bacterial overgrowth [16–19].
Changes in the gut microflora and microbiome may also affect bacterial translocation. Increases in plasma endotoxin and bacterial DNA have been associated with small intestinal bacterial overgrowth in cirrhotic patients [14, 19]. Furthermore, small intestinal bacterial overgrowth was an independent and major risk factor for the presence of bacterial DNA in cirrhotic patients. In addition, patients with bacterial overgrowth of the small intestine develop spontaneous bacterial peritonitis more frequently than patients without bacterial overgrowth [16, 20]. Interestingly, selective intestinal decontamination decreased translocation to the mesenteric lymph nodes to the level of non-cirrhotic patients [8]. Thus, intestinal bacterial overgrowth might predispose patients with liver disease to bacterial translocation.
Most studies assessing bacterial translocation and changes in the intestinal microflora have been conducted in patients with end-stage liver disease. There are currently no patient data or experimental studies assessing bacterial translocation and the enteric microbiome in early, non-cirrhotic stages of liver disease. We therefore took an unbiased approach to investigate the dynamics of these two events in early stages of liver disease using complementary mouse models of liver injury and fibrosis.
Material and Methods
Animal models of liver disease
Male wild-type BALB/c mice were obtained from Charles River or bred in our specific pathogen free vivarium. TLR4 mutant mice on a BALB/c genetic background were purchased from Jackson Laboratory. All animals received humane care in compliance with institutional guidelines. Mice had access to water and irradiated chow diet (PicoLab Rodent Diet 20 5053) ad libitum. Ligation of the common bile duct (BDL) or sham operations as control have been described [21]. One, five, or ten days after BDL or sham operation, mice were sacrificed for analysis. A second complementary model of carbon tetrachloride (CCl4)-induced liver injury and fibrosis was used. Corn oil as control or CCl4 (2 μl CCl4/g mouse body weight; the initial two injections were 1:100 and 1:50 dilutions, followed by 1:4 dilutions with corn oil; for mice that received only one injection, a dilution of 1:4 was used) are injected intraperitoneally every third day. Injections were repeated for a total of one, four, twelve and twenty-four times. For selective intestinal decontamination, mice were started on poorly absorbable antibiotics (polymyxin B (Bedford Laboratories; 0.4 mg/ml), neomycin (Sigma; 0.4 mg/ml)and rifaximin (Salix; 0.2 mg/ml) [22]) in the drinking water one week prior to the experimental procedure and were kept on antibiotics for the entire length of the experiment. The intragastric feeding model of continuous ethanol infusion in mice has been described [23]. Sequences from our previous publication [23] were used to compare microbial changes between various liver disease models.
FITC-dextran permeability assay
Intestinal permeability was assessed by gastric gavage of FITC-dextran (3kDa; Sigma), a non-metabolizable macromolecule that is used as a permeability probe as described by us [24]. All mice were gavaged with 200μl FITC-dextran (100mg/ml) 4 hrs before sacrifice. FITC-dextran measurements were performed in plasma by fluorometry. To assess intestinal permeability of each gastrointestinal segment, we used an intestinal loop model as we have previously described [25, 26]. After anesthesia, a midline laparotomy incision was made. A 4cm long segment of the gastrointestinal tract (proximal, mid or distal small intestine, cecum or colon) was created with two vascular hemoclips without disrupting the mesenteric vascular arcades. The length of intestine between the two clips was injected with 50 μl FITC-dextran. After 1 hr, mice were sacrificed and fluorescence was measured in the plasma as described above.
16S rRNA sequencing
For 16S rRNA sequencing, male mice from one litter were used for the sham/BDL studies; two consecutive litters from the same parents were studied in the corn oil/CCl4 experiments. Control and experimental mice were housed in separate cages in the same specific pathogen free vivarium. DNA from cecal contents and mucosa was extracted as previously described [23]. Deep DNA pyrosequencing of the hypervariable V1–V3 region of prokaryotic 16S rRNA loci was performed to generate microbial community profiles using species-level (97% similarity) operational taxonomic unit (OTU)-based classification and analysis as described in our previous publication [23] with the following exceptions. For all OTU-based analysis other than the Venn diagram, we used 2600 randomly selected sequence reads from each mouse and analyzed statistical significance using Student’s t-test. Sample coverage was estimated using an implementation of Good’s coverage[27]. We used all good reads for construction of the Venn diagram: 15174 454 reads (86–93% coverage) from alcohol, 12116 reads (85–90% coverage) from BDL, 13651 reads (93–96% coverage) from CCl4, and 1118 Sanger reads (71–76% coverage) from ob/ob experimental groups, and 13328 454 reads (91–93% coverage) from alcohol, 9657 reads (86–87% coverage) from BDL, 20833 reads (93–95% coverage) from CCl4, and 552 Sanger reads (63–83% coverage) from ob/ob control groups. A total of 42059 sequences from experimental mice and 44370 sequences from control mice were used in constructing the Venn diagram. Three mice were used for each of alcohol, BDL and CCl4 experimental groups and control groups. Sequences from four ob/ob male offspring mice from Ley et al. were used as the experimental group and sequences from two +/+ male offspring were used as the control group (63–83% coverage)[28]. Homozygous ob/ob mice are deficient in the leptin gene that produces a stereotyped, fully penetrant obesity phenotype with fatty liver[28, 29].
Statistical analysis
Mann-Whitney rank sum test was used for statistical analysis except when stated otherwise. All data are presented as mean ± SEM except when stated otherwise. p<0.05 was selected as the level of significance.
Additional Material and Methods are described in the Supplementary Data section.
Results
Cholestatic liver injury results in early bacterial translocation and increased intestinal permeability
To characterize the dynamics of bacterial translocation in early cholestatic liver disease, the common bile duct was ligated in BALB/c mice for one, five, and ten days. Mice with sham operations served as controls. Plasma ALT levels, a common measure for liver injury, were increased in bile duct ligation (BDL) animals as compared to sham controls, with highest levels observed one day following surgery (Suppl. Fig. 1A). To assess fibrosis as a consequence of chronic liver injury, deposition of collagen was stained with Sirius red. Deposition of extracellular matrix proteins was not observed after one day following BDL. Periportal and bridging fibrosis occurred after five and ten days, respectively, following BDL (Suppl. Fig. 1B).
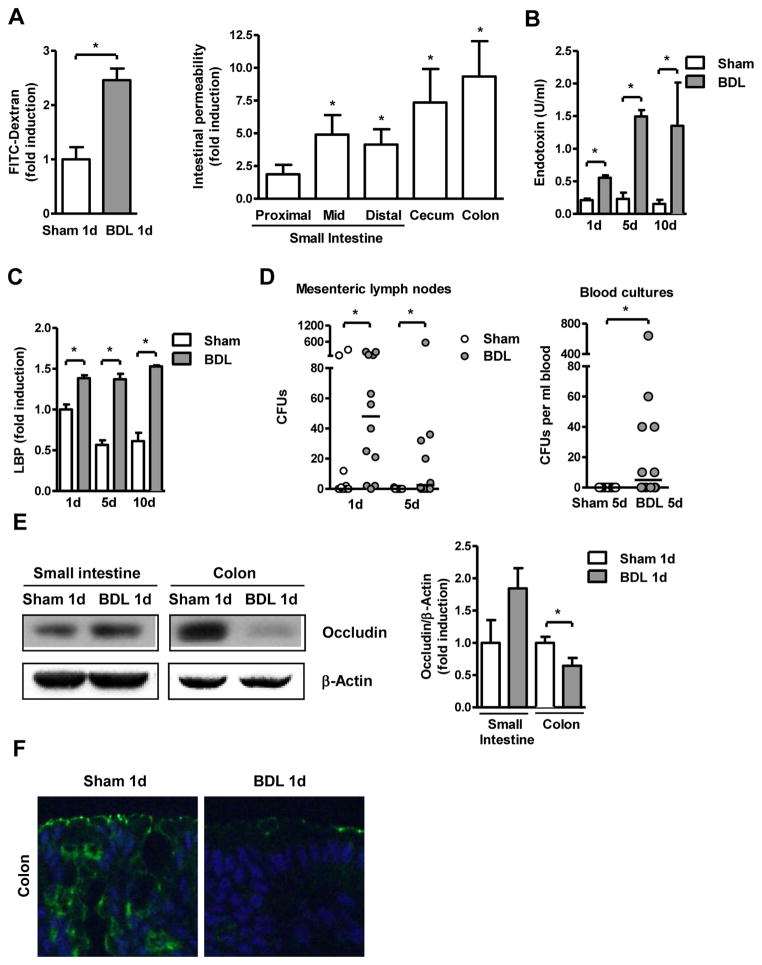
To directly assess intestinal permeability we used an in vivo method by measuring recovery of indigested dextran (molecular mass 3kDa) labeled with FITC. BDL resulted in an increase of fluorescence in the plasma indicative of increased intestinal permeability one day after BDL (Fig. 1A). To determine the gastrointestinal site of increased intestinal permeability in vivo, a ligated intestinal loop model was used. A 4-cm loop of the proximal, mid or distal small intestine, cecum or colon was ligated (without disrupting the mesenteric vascular arcades and blood supply) and FITC-dextran was injected in anesthetized mice that have undergone sham or BDL operations one day prior. At 1 hr, mice were sacrificed and fluorescence was measured in the plasma. Results of a sham and BDL mouse done in parallel were compared, and showed a significant increase in permeability in the mid and distal small intestine, cecum and colon following BDL. The highest increase in permeability was observed in the colon(Fig. 1A). To assess whether increased intestinal permeability directly translates into translocation of bacterial products, systemic endotoxin levels were investigated. Lipopolysaccharide (LPS), a component of the outer cell wall of Gram-negative bacteria, was higher in mice one day following BDL and continued to rise after five and ten days of BDL as compared to sham operated mice (Fig. 1B). Furthermore, plasma levels of lipopolysaccharide-binding protein (LBP) were similarly elevated after BDL (Fig. 1C). As an additional measure of bacterial translocation across the mucosal barrier, viable bacteria were cultured in mesenteric lymph nodes (the first organ encountered in the translocation route from the gastrointestinal tract) and in the systemic circulation following sham and BDL operation. BDL resulted in a significant increase in bacterial numbers in mesenteric lymph nodes one day and five days after BDL (Fig. 1D). Venous blood cultures of sham and BDL mice remained sterile one day after BDL (not shown); however, five days following BDL there was an increase in positive blood cultures of BDL as compared to sham operated mice (Fig. 1D). The identity of translocated bacteria was identified by sequencing and revealed that these organisms can be broadly categorized into three families, Enterobacteriaceae, Enterococcaceae and Bacillaceae (Supp. Table 1). Finally, bacterial DNA was detectable by PCR in whole blood of mice following BDL for one day, but not in sham operated mice (Suppl. Fig. 1C).
Figure 1. Early bacterial translocation following ligation of the common bile duct.
Mice underwent sham operation or bile duct ligation (BDL) for indicated time intervals. (A) Mice were gavage-fed FITC labeled dextran (200μl, 100mg/ml) one day following surgeries. Mice were sacrificed 4 hrs later, and fluorescence measured in the plasma (n=5; left panel). FITC-dextran (50μl, 100mg/ml) was injected into intestinal loops of mice which underwent sham or BDL operations one day prior. FITC was measured in the plasma 1 hr after injection (n=5; right panel). (B and C) Plasma endotoxin and lipopolysaccharide-binding protein (LBP) levels were measured (n=4–8). (D) Aerobic bacteria were cultured and quantified in mesenteric lymph nodes (n=10–13) and blood (n=12–13); data are presented as median. CFU = colony forming unit. (E) Occludin and β-actin protein expression in intestinal sections were analyzed by Western blot analysis. A representative Western blot image is shown. Densitometry of Western blot images was performed. Values are presented relative to sham operated mice (n=5 in each group). (F) Immunofluorescent staining for occludin (green) was performed on colonic sections from mice that underwent sham or BDL operation. Nuclei are stained with Hoechst (blue). *p<0.05.
Disruption of the epithelial barrier function is closely connected to a dysfunction of tight junctions [30]. Therefore, the integrity of tight junctions of small intestinal and colonic epithelial cells was investigated using western blotting and immunofluorescence for occludin, a component of tight junctions. Following one day of cholestatic liver injury, expression of occludin protein was diminished in the colon, but not the small intestine as compared to sham operated animals (Fig. 1E and F). Electron microscopy confirmed disrupted colonic tight junctions one day after BDL (Suppl. Fig. 1D). These findings are consistent with our intestinal loop model. Taken together, bacterial translocation occurs very early following cholestatic liver injury and is facilitated by a disruption of tight junctions.
Bacterial translocation occurs independently from changes in the microbiome and endotoxin
To determine whether bacterial translocation is dependent on quantitative changes in the microflora, we added a mixture of antibiotics with limitedoral bioavailability to the drinking water to reduce the intestinal bacterial burden. Mice receiving non-absorbable antibiotics for 7 days showed a reduction by 92% in luminal and adherent bacteria (Suppl. Fig. 2A). Despite a reduced quantitative amount of bacteria in the intestine, intestinal permeability (Suppl. Fig. 2B) and plasma endotoxin levels (Suppl. Fig. 2C) were unchanged following BDL for one day as compared to control mice not receiving antibiotics prior to BDL. This is consistent with another study showing that non-absorbable antibiotic treatment for 5 days after BDL did not decrease plasma LPS levels as compared to antibiotic treated sham animals. However, 21 days following BDL, mice on antibiotics had lower plasma LPS levels as compared to sham operations [21]. Thus, increased intestinal permeability and bacterial translocation is independent from quantitative changes in the microbiome.
As systemic LPS can cause an increase in intestinal permeability and bacterial translocation [31], we then asked whether LPS unresponsive TLR4 mutant mice on a BALB/c background show a difference in bacterial translocation. BDL for one day caused a significant increase in intestinal permeability as compared to sham operated TLR4 mutant mice (Suppl. Fig. 2D). Plasma endotoxin levels and bacterial translocation to mesenteric lymph nodes showed an increase in TLR4 mutant mice undergoing BDL as compared to sham mice (Suppl. Fig. 2E and F), which was not significantly different from wildtype BALB/c mice following BDL for one day (Fig. 1B and C). Bacterial DNA was detectable by PCR in whole blood of wildtype and TLR4 mutant mice following BDL for one day (Suppl. Fig. 2G).
Thus, from these experiments we conclude that an increase in intestinal permeability is independent of the LPS/TLR4 pathway in early stages of cholestatic liver disease. Our findings are consistent with a previous report showing the same elevated plasma LPS levels in wild type and TLR4 mutant mice following one injection of carbon tetrachloride (CCl4)[21].
Translocation of bacteria and their products in early stages of toxic liver injury
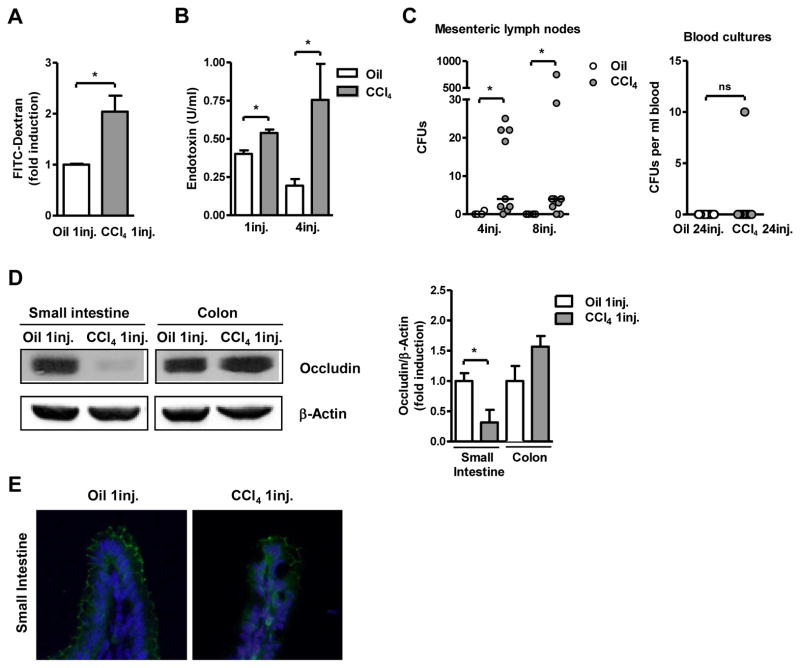
As bacterial translocation may result from the absence of bile acids in the gastrointestinal tract in cholestatic liver disease, a second model of toxic liver injury and fibrosis was studied. Repeated intraperitoneal injections of carbon tetrachloride (CCl4) caused liver injury as compared to mice injected with corn oil alone as vehicle control for CCl4. This is reflected by ALT levels, which were elevated with increasing numbers of CCl4 injections (Suppl. Fig. 3A). Minimal collagen deposition as a marker for liver fibrosis was seen after four injections of CCl4. CCl4 treated animals showed fibrotic septa extending from the portal tracts into the liver lobules and bridging fibrosis after 12 and 24 injections of CCl4, respectively (Suppl. Fig. 3B). One single dose of CCl4 resulted in an increase in intestinal permeability as assessed by the FITC-dextran assay (Fig. 2A). Similarly, endotoxin levels were increased after one single injection of CCl4 (Fig. 2B). Translocation of viable bacteria from the gastrointestinal tract to the mesenteric lymph nodes occurred after four and eight injections of CCl4 (Fig. 2C), which was not observed prior to the fourth injection (not shown) and is therefore relatively later as compared to BDL. Bacteria mainly from three families, Enterobacteriaceae, Bacillaceae and Lactobacillaceae, translocate to mesenteric lymph nodes (Supp. Table 1). Systemic blood cultures remained negative even after 24 injections of CCl4 (Fig. 2C). Expression of occludin as part of the tight junction complex was decreased in the small intestine, but not the colon after one injection of CCl4 as compared to oil injected mice (Fig. 2D and E). Thus, toxic liver injury causes early bacterial translocation. An increase of intestinal tight junction permeability might explain the early onset of bacterial translocation in both the cholestatic and toxic liver injury models.
Figure 2. Toxic liver injury is accompanied by early changes in intestinal permeability and bacterial translocation.
Mice were injected intraperitoneally with oil or carbon tetrachloride (CCl4) for indicated times. (A) 24 hrs following a single injection of oil or CCl4, mice were gavaged with 200μl of FITC labeled dextran (100mg/ml), and fluorescence in the plasma was determined 4 hrs later (n=3–5). (B) Plasma endotoxin levels were measured (n=4–7). (C) Colony forming units (CFUs) were counted on aerobic cultures of mesenteric lymph nodes (left panel) (n=4–9) and blood (right panel) (n=14–20); data are presented as median. (D) Occludin protein expression was analyzed in the intestine by Western blotting. β-actin was used as a loading control. Images are representative of one Western blot. Densitometry of Western blot images was performed. Values are presented relative to oil injected mice (n=5 in each group). (E) Immunofluorescent staining for occludin (green) was performed on colonic sections from mice that received one injection of oil or CCl4 one day prior. Nuclei are stained with Hoechst (blue). *p<0.05.
Bacterial overgrowth is an early event following bile duct ligation
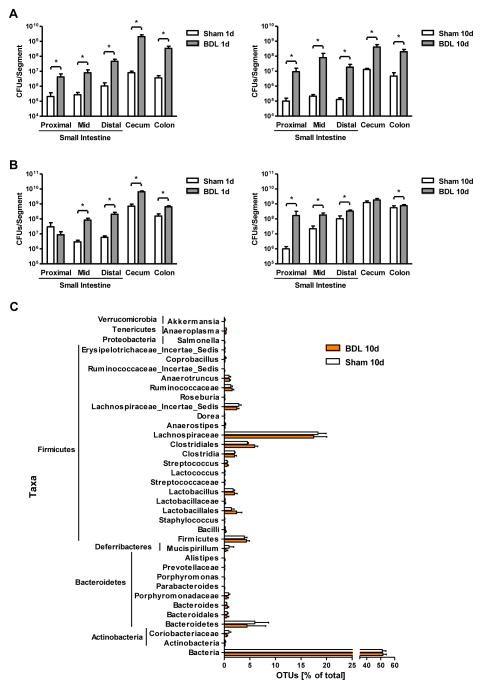
The effect of BDL or sham operations on bacterial contents of the small intestine (proximal, mid, and distal third) and large intestine (cecum and colon) was assessed by conventional culture techniques. The number of aerobic luminal and adherent bacteria increased across the gastrointestinal tract as early as one day following BDL, and continued to be higher ten days after BDL as compared to sham operated animals (Fig. 3A). Similarly, BDL caused bacterial overgrowth of anaerobic bacteria after one or ten days as compared to sham operation (Fig. 3B).
Figure 3. Bile duct ligation produces intestinal bacterial overgrowth.
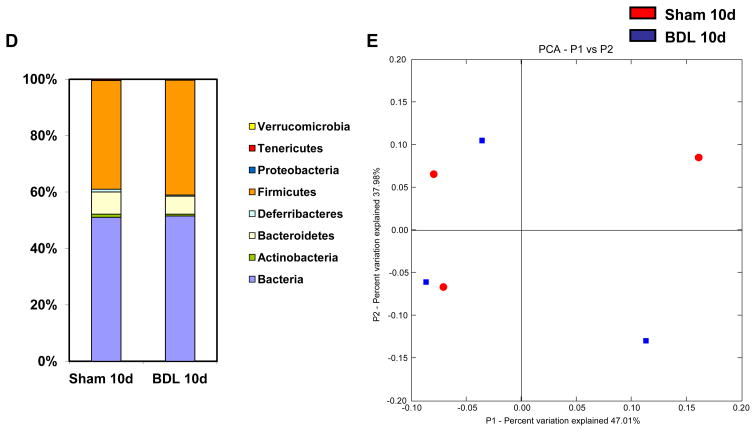
Mice underwent sham operation or bile duct ligation (BDL) for indicated time intervals. (A, B) Total aerobic bacteria (A) and anaerobic bacteria (B) were quantified by culture in the gastrointestinal tract (n=3–15). (C) 16S rRNA from the mouse cecum was sequenced using 454 Titanium technology. Experiment-specific operational taxonomic units (OTUs) representative sequences (97% identity) were classified using the Ribosomal Database Project (RDP) classifier and plotted. Orange bars indicate OTUs containing the BDL group (3 mice, distributed among 1264 OTUs) and white bars indicate OTUs containing the sham control group (3 mice, distributed among 1245 OTUs). (D) The graph demonstrates the percentages of each community contributed by the indicated phyla. (E) Scatter plot of Principal coordinates analysis (PCoA) on weighted and normalized P1 vs. P2 UniFrac distances. The BDL samples are in blue while the sham samples are in red. CFU = colony forming unit.*p<0.05.
As only a minority of enteric bacteria can be cultured by conventional culture techniques [32], qualitative changes in the intestinal luminal and adherent microbiome were assessed by massively parallel pyrosequencing. To rule out genetic, dietary, and environmental factors that influence the microbiome [33], we used littermates fed the same diet and housed in the same vivarium. We found that the mice in the BDL group at day 10 showed only minor microbial changes (Fig. 3C and D). To evaluate similarity among the samples, principle coordinates analysis (PCoA) was performed. The microflora of animals within the individual groups and in comparison to each other did not cluster separately by PCoA, suggesting that BDL did not have a significant effect on bacterial community composition (Fig. 3E). The microbial species richness, estimated by the Chao1 estimator [27, 34] was very similar between the control and experimental groups (median Chao1 of three control mice predicted 1162 OTUs, while the median Chao1 of three experimental mice predicted 1152 OTUs). Likewise, both the control and experimental groups had a median Shannon index [35]value of 5.3 and 5.1, respectively, indicating similar microbial species richness or evenness. Thus, the bacterial density increases very early following BDL, while the overall structure of the bacterial community is not altered suggesting that the selective pressures acting on the bacterial community are not significantly affected by cholestasis.
Carbon tetrachloride-induced liver injury is accompanied by intestinal bacterial overgrowth and dysbiosis
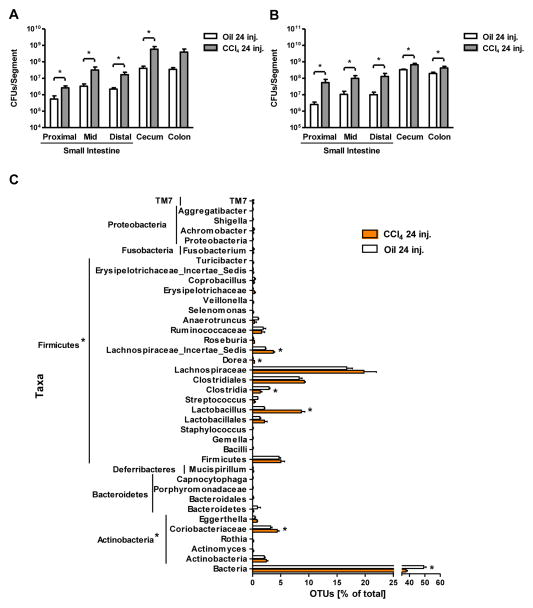
To confirm our cholestasis-induced bacterial overgrowth results in a second mouse model of liver fibrosis, mice were subjected to repeated intraperitoneal injections of CCl4 every third day for a total of 24 times resulting in bridging fibrosis. Fibrosis was associated with an increase in the luminal and adherent aerobic and anaerobic bacteria in the small and large intestine (Fig. 4A and B). Quantitative changes of the microflora were not observed at an earlier time point (data not shown).
Figure 4. Effects of carbon tetrachloride on microbial diversity of the mouse cecum.
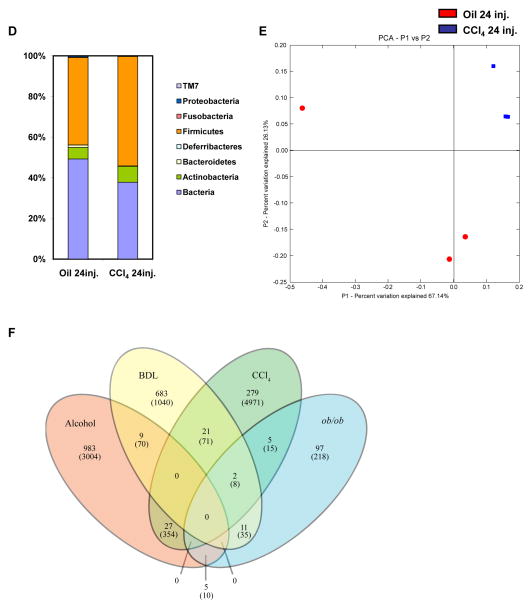
Mice were injected intraperitoneally with oil or carbon tetrachloride (CCl4) for a total of 24 times. (A and B) Luminal and adherent bacterial load was determined by aerobic (A) and anaerobic (B) cultures in various gastrointestinal segments (n=4–15). (C) 16S rRNA gene surveys show clustering of specific operational taxonomic units (OTUs) by treatments. Orange bars indicate OTUs containing the CCl4 group (3 mice, distributed among 823 OTUs) and white bars indicate OTUs containing the oil control group (3 mice, distributed among 563 OTUs). (D)The graph demonstrates the percentages of each community contributed by the indicated phyla. (E) Principal coordinates analysis (PCoA) was performed on weighted and normalized P1 vs. P2 UniFrac distances for the oil and the CCl4 group. The CCl4 samples are in blue while the oil samples are in red. CFU = colony forming unit. *p<0.05.(F) Venn Diagram of sequenced animals (cecal microbiome) is shown, which depicts the distribution of OTUs (and reads in parenthesis) that remain after subtracting any OTU containing a control group read. Mice were fed alcohol or an isocaloric diet as control via an intragastric feeding tube for 3 weeks [23], underwent BDL or sham operation for 10 days, or were injected 24 times with CCl4 or oil as control. Genetically obese ob/ob mice with fatty liver disease and their wildtype siblings were used as additional liver disease model [28].
Dysbiosis following the onset of toxic liver disease was analyzed by pyrosequencing after 24 injections ofCCl4. We found that the mice in the CCl4 treated group had significantly increased numbers of OTUs in the phylum Firmicutes, namely members in the genus Lactobacillus, Dorea and Lachnospiraceae_Incertae_Sedis. There was also a significant increase in the number of OTUs in the phylum Actinobacteria, particularly members in the genus Coriobacteriaceae in the CCl4 treated group (Fig. 4C and D). PCoA showed that the treated and control groups cluster separately and that the CCl4 treated mice cluster more tightly than do the oil treated samples (Fig. 4E). Thus, the quantitative and qualitative changes in the intestinal microflora following toxic liver injury are not an early event and occur after bacterial translocation has been observed. Unlike BDL treatment, the microbial species richness has decreased from a median Chao1 estimate of 779 OTUs, in control mice to 565 OTUs in CCl4-treated mice. This pattern is also reflected in the Shannon index: median of 5.21 for control mice and 3.91 for treated mice. Likewise, the Simpson index [36](where 0 = infinite diversity, 1 = no diversity) was six times higher in treated mice than control mice (median of 0.063 for treated and 0.011 for control mice), indicating lower diversity in the treated mice. This indicates that toxic liver injury, as modeled with CCl4, resulted in a reduction in cecal microbial diversity.
The enteric microbiome differs with respect to the etiology of liver disease
To determine similarities between the microbiome associated with various liver diseases, we compared sequencing data of mice treated with 24 injections of CCl4, subjected to BDL for 10 days, fed an alcohol diet via an intragastric feeding tube for 3 weeks (as published by us[23]), or genetically obese ob/ob mice with fatty liver disease [28]. For comparison we used a stringent analysis method by subtracting any OTU containing a control group read from any experimental group. The microbiome following CCl4 treatment shares 27 and 21 OTUs with the microbiome after alcohol feeding or BDL, respectively. The BDL-associated microbiome has 11 and 9 OTUs in common with the ob/ob microbiome and the microbiome following alcohol, respectively. Only few shared OTUs are observed between the ob/ob microbiome and the microbiome following alcohol or CCl4 administration. Interestingly, the four liver disease models do not share any common OTU (Fig. 4F). Thus, although experimental liver disease is dependent on gut-derived bacterial products in mice, there are no unique and common bacterial species dominating the microbiome associated with four different liver diseases.
Discussion
Although bacterial translocation and intestinal bacterial overgrowth are very well characterized phenomena in end-stage liver disease, there are no studies assessing bacterial translocation and changes in the microbiome in early stages of liver injury. As experimental studies suggest that liver disease progression, particularly fibrosis progression, is driven by gut-derived bacterial products [21], we took an unbiased approach and investigated dynamics of liver disease, bacterial translocation and changes in the intestinal microbiome in mice. Increased intestinal permeability and bacterial translocation is observed very early following liver injury. At a similar time, intestinal bacterial overgrowth of both aerobic and anaerobic bacteria is evident after one day following BDL, but only in mice with established liver fibrosis following CCl4-induced toxic liver injury. The microbiome following cholestasis is qualitatively similar to control mice, while dysbiosis associated with CCl4-induced bridging fibrosis is characterized by an increase of Firmicutes and Actinobacteria.
There is an evolving concept that gut-derived endotoxin (and possibly other, not yet characterized bacterial products) resulting from bacterial translocation plays a central role in the initiation of acute liver injury and progression to chronic liver disease. First, plasma endotoxin levels are increased in patients with liver disease, and the severity of liver disease correlates with the degree of endotoxemia [11]. Second, selective intestinal decontamination with antibiotics is beneficial for patients and prevents experimental liver injury [8, 37]. Third, mice with genetic deletions in the LPS signaling pathway are resistant to experimental liver injury and fibrosis [21, 38, 39]. Bacterial translocation as measured by systemic bacterial DNA was more frequently found in cirrhotic patients with small intestinal bacterial overgrowth as compared to patients without overgrowth [19]. Therefore, the question arises whether bacterial translocation is dependent on qualitative and/or quantitative changes in the intestinal microbiome. Interestingly, when bacterial overgrowth is induced in the small intestine experimentally, it results in hepatic injury mediated by translocated bacterial products [40]. Our results clearly suggest that bacterial translocation precedes qualitative and quantitative changes in the intestinal microflora in CCl4-induced toxic liver injury and alcoholic steatohepatitis [23]. Although bacterial translocation and intestinal bacterial overgrowth are observed at the same time following the onset of experimental cholestasis, non-absorbable antibiotics to decrease the intestinal bacterial burden, did not prevent increased intestinal permeability or bacterial translocation. Non-alcoholic fatty liver disease (NAFLD) in humans is similarly associated with increased gut permeability, which appears to be caused by disruption of intercellular tight junctions in the smallintestine [41]. In rats, acute ingestion of alcohol has been shown to alter the epithelial barrier in the colonic mucosa via ethanol oxidation into acetaldehyde by the enteric microflora with subsequent downstream activation of mast cells [42], while alcohol consumption in humans results in duodenal and jejunal mucosal injury [43]. Thus, based on these studies, increased intestinal permeability with subsequent translocation of bacterial products or bacteria occurs in the setting of acute liver injury independently from changes in the intestinal microflora. The dynamics of bacterial translocation are slightly different between cholestatic and toxic liver injury. Translocation of viable bacteria to the mesenteric lymph nodes occurs later in toxic liver injury than in cholestasis, and positive blood cultures are not observed even after 24 injections of CCl4. In addition, while the intestinal tight junction protein occludinis down-regulated in the colon following cholestatic liver injury, toxic liver damage results in a decreased expression of occludin in the small intestine. Using an intestinal loop model, intestinal permeability is elevated in the small and large intestine following acute cholestatic liver injury. Occludin is one of multiple tight junction proteins. Because we observed no change of occludin expression in the small intestine after the acute onset of cholestatis, other tight junction proteins might be affected resulting in increased small intestinal permeability. However, further studies are required to determine the contribution of individual tight junction proteins in the small and large intestine to increased permeability in different liver disease models.
Interestingly, all four etiologies of liver disease are accompanied by intestinal bacterial overgrowth [23, 41, 44]. The reason for overgrowth is likely multifactorial including absence or decreased intestinal levels of bile acids, changes in intestinal motility or feeding rates. There is evidence that conjugated bile acids regulate expression of host genes to promote innate defense against luminal bacteria. Conjugated bile acids bind to the nuclear receptor for bile acids, farnesoid receptor X (FXR), in enterocytes to induce the antimicrobial proteins angiogenin 1 and RNase family member 4 that prevent bacterial overgrowth and promote epithelial cell integrity [45]. Given a preexisting leaky gut barrier as evidenced by disrupted tight junctions, an increase in the intestinal bacterial burden elevates systemic levels of bacterial products to perpetuate later disease stages [46].
Technical advances have helped to characterize the deep biodiversity of the gastrointestinal microflora and its functional contribution to the host biology by examination of the 16S RNA genes used in taxonomical classification of bacteria. The microbiome of four liver disease models have now been described: high fat (diet)- and alcohol-induced steatohepatitis [23], and now cholestatic and toxic liver injury in the present study. Obesity-induced fatty liver disease is closely correlated with changes in the microbiome and metagenome. Studies in obese mice with a mutation in the leptin gene (ob/ob) and their lean wild-type littermates revealed that obesity is associated with a division-wide increase and decrease in the relative abundance of the Firmicutes and Bacteroidetes, respectively [28]. Furthermore, when wild-type germfree mice fed a standard chow diet are colonized with a microbiota harvested from ob/ob or lean donors, adiposity in recipients of the obese microbiota increased more than in recipients of a lean microbiota [47]. This change involves several mechanisms, which were mostly identified by metagenomic and metabolomic techniques. One is the fermentation of indigestible polysaccharides by the microbial flora into monosaccharides and short-chain fatty acids. More monosaccharides and short-chain fatty acids are subsequently absorbed from the gut lumen and transported to the liver, resulting in the induction of de novo hepatic lipogenesis [48]. In addition, fasting-induced adipocyte factor (Fiaf), a circulating lipoprotein lipase inhibitor, is selectively suppressed in the intestinal epithelium of the host resulting in deposition of triglycerides [48]. Metabolic profiling of genetic susceptibility to impaired glucose homeostasis and fatty liver disease demonstrated a disruption of choline metabolism. Conversion of choline into methylamines by microbiota in a susceptible mouse strain reduces the bioavailability of choline and mimics the effect of choline-deficient diets causing fatty liver disease [49]. This was confirmed in a recent human study, which demonstrated that variations between subjects in levels of only two bacterial classes, Gammaproteobacteria and Erysipelotrichi, were directly associated with changes in liver fat in each subject during choline depletion [50]. These data indicate an active role for the intestinal microflora in the development of fatty liver disease. We recently applied 454 massively parallel pyrosequencing of the gut microbiota to a mouse model of alcoholic steatohepatitis, and found that the mice in the alcohol treated group had reduced OTUs of Firmicutes, but an increase in the number of OTUs of Bacteroidetes and Verrucomicrobia, which is the opposite of the observed changes in the high fat diet-associated microbiome. Prebiotics are able to reverse bacterial changes and ameliorate alcoholic liver disease, suggesting a causative link between changes in the microflora and steatohepatitis [23]. Thus, both a high fat diet and alcohol induce dramatic changes in the enteric microbiome of mice that directly regulate their phenotypes. Our current study completes the project of liver disease-associated microbiome by analyzing qualitative changes in the enteric microflora following cholestatic and toxic liver injury. No major differences were observed between the composition of the intestinal microbiota following cholestasis. An increase of Firmicutes and Actinobacteria are found in the microbiome associated with toxic liver disease, which is similar to the changes observed in the high-fat diet model. However, a direct comparison of the microbiome associated with alcohol feeding, BDL, CCl4 treatment and obesity-induced fatty liver disease revealed few similarities between these four liver disease models. This might indicate that the relative abundance of microbial species between the individual treatment and the respective control group is important, although differences in diet, husbandry, mouse age and strain could contribute to variation of the microbial community. Based on our results, it is not conceivable that one unique microbial species is associated with liver disease and drives disease progression independent from its etiology. Currently we can only speculate how a dysbiotic microbiome contributes to alcoholic, cholestatic and toxic liver disease progression. Our study provides the groundwork and motivation for a humanized gut microbiome mouse model by colonizing germ-free animals with complex microbial communities from patients with chronic liver disease.
Taken together, liver disease results in qualitative (dysbiosis) and quantitative (bacterial overgrowth) changes of the intestinal microbiome. We also demonstrate that bacterial translocation is independent from changes in the microflora and occurs in the setting of acute liver injury. Our findings suggest that bacterial translocation is not a phenomenon in late stage liver disease and cirrhosis, and that it likely contributes to liver disease progression from a very early stage on. This novel insight will facilitate rationale attempts to manipulate the intestinal microbiome and the gut barrier in order to prevent and treat liver disease.
Supplementary Material
Acknowledgments
Financial support: The study was supported in part by NIH grants K08 DK081830(to BS) and RO1 DK072237(to DAB). This study was also supported by the UCSD Digestive Diseases Research Development Center (DK080506) (to BS), and by the AGA Fellowship to Faculty Transition Award (FFTA) (to BS).
We thank Dr. Keiko Iwaisako for assistance with intestinal loops, Drs. Alan Hofmann and Reiner Wiest for helpful discussion. We thank the Cellular & Molecular Medicine Transmission Electron Microscopy Core Facility at UCSD for assistance with electron microscopy.
Abbreviations
- ALT
Alanine Aminotransferase
- BDL
bile duct ligation
- CCl4
carbon tetrachloride
- CFU
colony-forming unit
- LBP
lipopolysaccharide-binding protein
- PCoA
principle coordinates analysis
- LPS
lipopolysaccharide
- OTU
Operational Taxonomic Unit
- TLR
Toll-like receptor
Footnotes
None of the authors has a financial, personal or professional conflict of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnabl B, Scholten D, Brenner DA. What is the potential role of antifibrotic agents for the treatment of liver disease? Nat Clin Pract Gastroenterol Hepatol. 2008;5:496–497. doi: 10.1038/ncpgasthep1200. [DOI] [PubMed] [Google Scholar]
- 3.Brann OS. Infectious complications of cirrhosis. Curr Gastroenterol Rep. 2001;3:285–292. doi: 10.1007/s11894-001-0051-2. [DOI] [PubMed] [Google Scholar]
- 4.Christou L, Pappas G, Falagas ME. Bacterial infection-related morbidity and mortality in cirrhosis. Am J Gastroenterol. 2007;102:1510–1517. doi: 10.1111/j.1572-0241.2007.01286.x. [DOI] [PubMed] [Google Scholar]
- 5.Wong F, Bernardi M, Balk R, Christman B, Moreau R, Garcia-Tsao G, et al. Sepsis in cirrhosis: report on the 7th meeting of the International Ascites Club. Gut. 2005;54:718–725. doi: 10.1136/gut.2004.038679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology. 2005;41:422–433. doi: 10.1002/hep.20632. [DOI] [PubMed] [Google Scholar]
- 7.Pascual S, Such J, Esteban A, Zapater P, Casellas JA, Aparicio JR, et al. Intestinal permeability is increased in patients with advanced cirrhosis. Hepatogastroenterology. 2003;50:1482–1486. [PubMed] [Google Scholar]
- 8.Cirera I, Bauer TM, Navasa M, Vila J, Grande L, Taura P, et al. Bacterial translocation of enteric organisms in patients with cirrhosis. J Hepatol. 2001;34:32–37. doi: 10.1016/s0168-8278(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 9.Frances R, Zapater P, Gonzalez-Navajas JM, Munoz C, Cano R, Moreu R, et al. Bacterial DNA in patients with cirrhosis and noninfected ascites mimics the soluble immune response established in patients with spontaneous bacterial peritonitis. Hepatology. 2008;47:978–985. doi: 10.1002/hep.22083. [DOI] [PubMed] [Google Scholar]
- 10.Such J, Frances R, Munoz C, Zapater P, Casellas JA, Cifuentes A, et al. Detection and identification of bacterial DNA in patients with cirrhosis and culture-negative, nonneutrocytic ascites. Hepatology. 2002;36:135–141. doi: 10.1053/jhep.2002.33715. [DOI] [PubMed] [Google Scholar]
- 11.Lin RS, Lee FY, Lee SD, Tsai YT, Lin HC, Lu RH, et al. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J Hepatol. 1995;22:165–172. doi: 10.1016/0168-8278(95)80424-2. [DOI] [PubMed] [Google Scholar]
- 12.Fukui H, Brauner B, Bode JC, Bode C. Plasma endotoxin concentrations in patients with alcoholic and non-alcoholic liver disease: reevaluation with an improved chromogenic assay. J Hepatol. 1991;12:162–169. doi: 10.1016/0168-8278(91)90933-3. [DOI] [PubMed] [Google Scholar]
- 13.Bode C, Kugler V, Bode JC. Endotoxemia in patients with alcoholic and non-alcoholic cirrhosis and in subjects with no evidence of chronic liver disease following acute alcohol excess. J Hepatol. 1987;4:8–14. doi: 10.1016/s0168-8278(87)80003-x. [DOI] [PubMed] [Google Scholar]
- 14.Bauer TM, Schwacha H, Steinbruckner B, Brinkmann FE, Ditzen AK, Aponte JJ, et al. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia. Am J Gastroenterol. 2002;97:2364–2370. doi: 10.1111/j.1572-0241.2002.05791.x. [DOI] [PubMed] [Google Scholar]
- 15.Sandler NG, Koh C, Roque A, Eccleston JL, Siegel RB, Demino M, et al. Host response to translocated microbial products predicts outcomes of patients with HBV or HCV infection. Gastroenterology. 2011;141:1220–1230. 1230 e1221–1223. doi: 10.1053/j.gastro.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morencos FC, de las Heras Castano G, Martin Ramos L, Lopez Arias MJ, Ledesma F, Pons Romero F. Small bowel bacterial overgrowth in patients with alcoholic cirrhosis. Dig Dis Sci. 1995;40:1252–1256. doi: 10.1007/BF02065533. [DOI] [PubMed] [Google Scholar]
- 17.Pande C, Kumar A, Sarin SK. Small-intestinal bacterial overgrowth in cirrhosis is related to the severity of liver disease. Aliment Pharmacol Ther. 2009;29:1273–1281. doi: 10.1111/j.1365-2036.2009.03994.x. [DOI] [PubMed] [Google Scholar]
- 18.Bauer TM, Steinbruckner B, Brinkmann FE, Ditzen AK, Schwacha H, Aponte JJ, et al. Small intestinal bacterial overgrowth in patients with cirrhosis: prevalence and relation with spontaneous bacterial peritonitis. Am J Gastroenterol. 2001;96:2962–2967. doi: 10.1111/j.1572-0241.2001.04668.x. [DOI] [PubMed] [Google Scholar]
- 19.Jun DW, Kim KT, Lee OY, Chae JD, Son BK, Kim SH, et al. Association Between Small Intestinal Bacterial Overgrowth and Peripheral Bacterial DNA in Cirrhotic Patients. Dig Dis Sci. 2010;55:1465–1471. doi: 10.1007/s10620-009-0870-9. [DOI] [PubMed] [Google Scholar]
- 20.Chang CS, Chen GH, Lien HC, Yeh HZ. Small intestine dysmotility and bacterial overgrowth in cirrhotic patients with spontaneous bacterial peritonitis. Hepatology. 1998;28:1187–1190. doi: 10.1002/hep.510280504. [DOI] [PubMed] [Google Scholar]
- 21.Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Ouyang Y, Guner Y, Ford HR, Grishin AV. Ubiquitin-editing enzyme A20 promotes tolerance to lipopolysaccharide in enterocytes. J Immunol. 2009;183:1384–1392. doi: 10.4049/jimmunol.0803987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan AW, Fouts DE, Brandl J, Stärkel P, Torralba M, Schott E, et al. Enteric Dysbiosis Associated with a Mouse Model of Alcoholic Liver Disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandl K, Rutschmann S, Li X, Du X, Xiao N, Schnabl B, et al. Enhanced sensitivity to DSS colitis caused by a hypomorphic Mbtps1 mutation disrupting the ATF6-driven unfolded protein response. Proc Natl Acad Sci U S A. 2009;106:3300–3305. doi: 10.1073/pnas.0813036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin Reg III gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin HZ, Yang SQ, Chuckaree C, Kuhajda F, Ronnet G, Diehl AM. Metformin reverses fatty liver disease in obese, leptin-deficient mice. Nat Med. 2000;6:998–1003. doi: 10.1038/79697. [DOI] [PubMed] [Google Scholar]
- 30.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 31.Deitch EA, Ma L, Ma WJ, Grisham MB, Granger DN, Specian RD, et al. Inhibition of endotoxin-induced bacterial translocation in mice. J Clin Invest. 1989;84:36–42. doi: 10.1172/JCI114164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- 35.Spellerberg IF, Fedor PJ. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Global Ecology & Biogeography. 2003;12:177–179. [Google Scholar]
- 36.Simpson EH. Measurement of Diversity. Nature. 1949;163:688. [Google Scholar]
- 37.Rutenburg AM, Sonnenblick E, Koven I, Aprahamian HA, Reiner L, Fine J. The role of intestinal bacteria in the development of dietary cirrhosis in rats. J Exp Med. 1957;106:1–14. doi: 10.1084/jem.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isayama F, Hines IN, Kremer M, Milton RJ, Byrd CL, Perry AW, et al. LPS signaling enhances hepatic fibrogenesis caused by experimental cholestasis in mice. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1318–1328. doi: 10.1152/ajpgi.00405.2005. [DOI] [PubMed] [Google Scholar]
- 39.Guo J, Loke J, Zheng F, Hong F, Yea S, Fukata M, et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lichtman SN, Sartor RB, Keku J, Schwab JH. Hepatic inflammation in rats with experimental small intestinal bacterial overgrowth. Gastroenterology. 1990;98:414–423. doi: 10.1016/0016-5085(90)90833-m. [DOI] [PubMed] [Google Scholar]
- 41.Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 42.Ferrier L, Berard F, Debrauwer L, Chabo C, Langella P, Bueno L, et al. Impairment of the intestinal barrier by ethanol involves enteric microflora and mast cell activation in rodents. Am J Pathol. 2006;168:1148–1154. doi: 10.2353/ajpath.2006.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bode C, Bode JC. Effect of alcohol consumption on the gut. Best Pract Res Clin Gastroenterol. 2003;17:575–592. doi: 10.1016/s1521-6918(03)00034-9. [DOI] [PubMed] [Google Scholar]
- 44.Wigg AJ, Roberts-Thomson IC, Dymock RB, McCarthy PJ, Grose RH, Cummins AG. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor alpha in the pathogenesis of non-alcoholic steatohepatitis. Gut. 2001;48:206–211. doi: 10.1136/gut.48.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 47.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 48.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association Between Composition of the Human Gastrointestinal Microbiome and Development of Fatty Liver With Choline Deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.