Abstract
Over the last decade, multiple functional genomic datasets studying chromosomal aberrations and their downstream effects on gene expression have accumulated for several cancer types. A vast majority of them are in the form of paired gene expression profiles and somatic copy number alterations (CNA) information on the same patients identified using microarray platforms. In response, many algorithms and software packages are available for integrating these paired data. Surprisingly, there has been no serious attempt to review the currently available methodologies or the novel insights brought using them. In this work, we discuss the quantitative relationships observed between CNA and gene expression in multiple cancer types and biological milestones achieved using the available methodologies. We discuss the conceptual evolution of both, the step-wise and the joint data integration methodologies over the last decade. We conclude by providing suggestions for building efficient data integration methodologies and asking further biological questions.
Keywords: data integration, copy number, gene expression, integrative analysis, cancer
INTRODUCTION
Human cancer genesis and progression are enabled by the aberrant function of genes that regulate aspects of cell proliferation, apoptosis, genome stability, angiogenesis, invasion and metastasis [1]. Even before the advent of functional genomic technologies, there was already a wide agreement that recurrent genomic abnormalities confer an underlying selection advantage by spanning across genes vital for tumor development and metastasis [2]. The importance of somatic copy number alterations (CNA) was particularly clear in the cases of oncogenes and tumor suppressor genes (TSGs) as the CNA resulted in altered expression of these genes compared with the physiological expression (dosage effect). There have been numerous examples in the literature of the genes identified using the dosage alterations resultant of focal or chromosomal arm-level amplification or deletions. Most notably amplified oncogenes include ERBB2 [3], MYC [4], CCND1 [5], CAD [6, 7], BCR−ABL [8] and AR [9], while deleted TSGs include PTEN [10], CDKN2A [11], RB1, BRCA1, BRCA2, PTPRJ and TP53 [12–15]. A recent work studying patterns of CNA across 26 cancer types, found a mean of 24 gains and 18 losses per tumor sample [2]. Discovery and functional assessment of oncogenes and TSGs is essential for understanding the biology of cancer and for clinical disease management.
In the last decade both CNA and gene expression (GE) profiles for multiple cancer types have been measured using microarray technologies in high-throughput manner. There are many experimental methods that provide information on CNA but they vary in terms of resolution (see Supplementary Data, Section 1 for more discussion). Both array comparative genomic hybridization (aCGH) and single-nucleotide polymorphism (SNP) microarrays have been used to obtain high-resolution information on CNA [16, 17]. With the availability of paired gene expression and CNA information from the same patients using high-throughput platforms, it is reasonable to expect that additional cancer related genes will be identified by assessing more recurrent abnormal regions and their corresponding dosage alteration [2]. The Cancer Genome Atlas project [18] (http://cancergenome.nih.gov) is generating multiple data types including gene expression and copy number (CN) data for the same set of patients. The critical challenge is in differentiating between alterations that drive the cancer growth and other seemingly random alterations that accumulate through instability induced by tumorigenesis. The availability of these paired data from same patients has facilitated this process. Although it is possible to carry out analysis even with unpaired data, the analysis becomes much more powerful when both types of data are derived from the same patients since the relationship can be inferred not just on averaged quantities but in each sample. The paired data structure allows for optimal power and a reduction in false positives [19, 20].
However, the data produced from the functional genomics platforms cannot be used without preprocessing. In fact, it is crucial to preprocess and normalize the data to effectively dissociate actual biological signal values from experimental noise [21–23] prior to integrating the GE and CNA signals from the microarray platforms. The analysis steps and the software tools for quantifying of GE levels and obtain CNA information from microarray data, are summarized in Supplementary Tables S1 and S2. In addition, replicate information would not be available in the case of patient samples making it more difficult to analyze the data. Finally, in tumor samples, ‘contamination’ of stromal cells is typically seen further complicating the analysis [24, 25]. The case of integrating aCGH profiles with gene expression information relatively straightforward, since a gene's expression is directly interrogated by gene-specific probes and the gene's CN is readily available for the same entity interrogated by the aCGH array. For the high-density single nucleotide polymorphism (SNP) arrays, the signal value refers to a SNP marker and the gene CN must be estimated [26]. The SNP arrays are denser and hold an advantage over aCGH by being able to simultaneously detect chromosomal loss of heterozygosity (LOH) and uniparental disomy (UPD) events, apart from the CNA.
QUANTITATIVE RELATIONSHIP BETWEEN CNA AND GE IN DIFFERENT CANCER TYPES
Before implementing integration methodologies, the extent of correlation between the CNA and the GE should be investigated. A number of studies have quantified this relationship across a wide range of DNA CNA like low-, mid- and high-level of focal and chromosome arm-level amplification and deletions. For example, Hyman et al. [27] used a cut-off to determine the unamplified and amplified samples. Pollack et al. [28] stratified the samples into five categories: deletion, no change and low-, medium- and high-level amplifications. These statistically arbitrary, yet intuitive cut-offs provided evidence for statistically significant correlation between CNA and GE data [27–34].
Transcriptional changes for 10–63% of genes in amplified regions and 14–62% in regions of loss, across multiple cancer types has been reported (Supplementary Table S1). Furthermore, a relative gain (or loss) in genomic content is shown to increase (or decrease) the expression levels averaged across all genes in the implicated regions [29, 32, 33]. In breast cancer, for example, a 2-fold change in DNA CN was found associated with a corresponding 1.5-fold change in mRNA levels on average [28]. A relative gain or loss of a chromosome or chromosomal arm usually resulted in a statistically significant increase or decrease, respectively, in the average expression level of all of the genes on the chromosome, even when many genes seemed to be unrelated to malignant progression or not expressed in a given cell type.
In the context of individual genes, however, the situation is often more complex as numerous regulatory mechanisms are all capable of controlling the mRNA transcription. Therefore, even in regions of large gains, one can expect to find significantly downregulated genes. For example, 14% of downregulated genes appeared within regions of DNA gain and 9% of upregulated genes appeared in regions of DNA loss [29]. Furthermore, even within a chromosomal arm that is amplified in its entirety, one may still find contiguous regions whose genes are expressed at levels similar to that of normal tissue [29]. These caveats not only caution the interpretation of some integrative analysis results, but also serve as a constant reminder that CNA and GE integration can only expose part of a complex biological picture.
SIMPLE CLASSIFICATION SCHEMES FOR THE AVAILABLE INTEGRATION METHODOLOGIES
Numerous methodologies capable of integrating genome-wide CNA information with GE profiles have been developed in the past decade (Table 1 and Supplementary Tables S2 and S3). Though each method is formulated uniquely, general trends can be deduced upon closer inspection. For example, all integration methods have a common input—the paired data in the form of sample by gene matrices. Most integrative methods can be categorized into three distinct classes based on their biological and methodological complexity. Initial stepwise methods designed for exploring the relationship between CN and GE employ relatively simple techniques to quantify this interaction on a global scale. Later stepwise methods take advantage of this established relationship to achieve well-defined biological endpoints. Finally, there is also a class of joint methods that are mathematically involved. Though some may still have routine biological endpoints, others can be more ambitious.
Table 1:
A representative list of available methodologies for CNA information with gene expression profiles for which software implementations are available
| Methodology/Reference | Integration type | Endpoints | Main statistical tools used |
|---|---|---|---|
| Ace-it [52] | S | Gene targets (dosage effect) | nPHT |
| Magellan [47] | S | Exploratory analysis; clustering | ES; nPHT; CA; GO |
| SODEGIR [26] | S | Gene targets (concomitant CN/GE alteration) | Own statistic; nPHT |
| edira [53] | S | Gene targets (dosage effect) | CA; nPHT |
| CNAmet [77] | S | Gene targets (concomitant CN/GE alteration) | Own statistic; nPHT |
| Berger et al. [62] | J | Gene targets (dosage effect) | SVD; gene shaving |
| SIGMA2 [75] | S; J | Exploratory analysis; gene targets (concomitant CN/GE alteration) | ES; CA; PHT |
| iCLUSTER [46] | J | Clustering | Latent variable Model; VS |
| Van Wieringen & van De Wiel [56] | S; J | Gene targets (CN-induced DEG) | Own statistic; BF; nPHT |
| CONNEXIC [38] | J | Gene targets (drivers) | BF; networking |
| remMap [78] | J | Gene targets (concomitant CN/GE alteration) | RA; VS |
| DR-Integrator [76] | J | Gene targets (correlated CN/GE) | CA; PHT |
Integration type: S, stepwise; J, joint. Main statistical tools used: ES, exploratory statistics; PHT parametric hypothesis test; nPHT, non-parametric hypothesis test; CA, correlation analysis; RA, regression analysis; GO, gene ontology; VS, variable selection; BF, Bayesian framework; SVD, singular value decomposition.
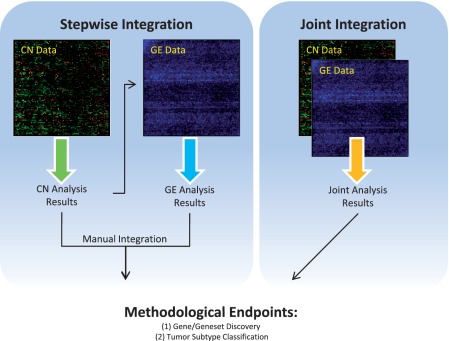
In terms of their objective and structure, methodologies can be grouped based on approach: stepwise or joint methodologies or endpoints: gene/gene-set discovery or subtype clustering (Figure 1). Gene/gene-set discovery methods aim to identify candidate genes or pathways [19, 35, 36], clusters of genes [19, 37] and candidate regulators involved in tumorigenesis [38–41]. Thus, they attempt to shed light on tumor biology and identify prognostic or therapeutic targets [42–45]. The subtype clustering methods are usually classification schemas designed to identify patient subgroups that may have similar prognosis or response to treatment [46–49], and therefore, improve on cancer risk and disease course prediction.
Figure 1:
Schematic overview of methods. Integrative methodologies can be grouped based on their integration structure and biological endpoints. Stepwise methods typically interrogate the CN data for regions of CNAs before results from a subsequent GE analysis are manually combined to complete the integrative procedure. Joint integration treats CN and GE as paired data entries. Thus, only one analysis is carried out in light of the pairing. Despite the contrasting approaches, most integrative methodologies arrive at the same biological endpoints of gene/geneset discovery or tumor subtype classification.
THE EVOLUTION OF SEQUENTIAL DATA INTEGRATION METHODOLOGIES
Following the intuitive blueprint that differential GE results from CNA in the DNA and aided by the notion that concordant amplification and overexpression are tell-tale signs of oncogenes and deletion along with the underexpression for TSGs, the ‘classical’ stepwise integration methodologies view integration as a two-step procedure. Typically, these methods identify aberrant chromosome regions before manually combining results from a separate expression analysis to arrive at their endpoints [27, 32, 34, 39, 42, 50, 51].
Not limited to quantifying the CNA and GE relationship, various exploratory statistical measures have been used for candidate gene identification. Methods aimed at exploring gene dosage effect have been the traditional hallmarks of stepwise approaches. ACE-it [52], for example, is a statistical tool intended to identify genes with concordant CNA/GE relationship. The overall implementation of the methodology involves stratifying samples into two groups based on CN gain or loss followed by implementing a one-sided Wilcoxon test to assess the concordant changes of GE values. Schafer et al. [53] also implemented a strategy to identify the driver genes related to disease development. To do so, they implement externally centered correlation coefficients to assess the degree of concordant CN and GE alteration.
There are several drawbacks common to these stepwise integration methods. The first drawback originates from the microarray platforms utilized to measure gene expression levels and CN profiles. Most integration methods base their analysis on the matched GE and CNA data of genes, and this requires steps of filtering, imputing or averaging features from one profile to the other since the probe sets from two platforms vary in chromosome loci and resolution. Many of the above mentioned methods use arbitrary thresholds to stratify CN and GE data. The use of the simple call data in downstream analysis may not be optimal, as calls do not fully account for the high degree of genetic heterogeneity amongst cancers [54]. Moreover, the data of some samples or genomic regions in samples are clearly noisier than others and thus, less confidence should be placed on such calls. Van de Wiel and van Wieringen [55] suggested that the uncertainty of the discrete CNs can be propagated in the test statistic for differential expression between CN groups or the call probabilities of ‘loss’, ‘normal’ and ‘gain’ regions can be considered, instead of the actual calls [56] for better data integration. Such call probabilities reflect both tumor cell heterogeneity and circumvent the loss of statistical power from which methods that discretize to hard calls suffer. The call probabilities also have a clear biological interpretation: the uncertainty with which a call is made. This difference in interpretation has two important consequences. First, CN profiles from different platforms can be compared directly when using the call probabilities. Second, the breakpoint nature of the CN data implies that neighboring probes (clones) share the same CN signature over samples. This will make it possible to borrow information across the genes within copy number extended regions [54, 55]. These drawbacks are also applicable to the joint integration methods.
The complexity associated with cancer cell–genomic environments also demand additional attention before meaningful biology can be extracted through such stepwise integration. For example, Garraway et al. [57] initially classified cancer samples based on chromosomal aberrations and analyzed aberration-based subgroups. By subsequently analyzing differential expression profiles between copy number derived sample clusters characterized by gain and no-gain, their method was capable of uncovering novel cancer biomarkers.
Alder et al. [36] have proposed a stepwise approach, Stepwise Linkage Analysis of Microarray Signatures (SLAMS), capable of uncovering transcriptional signature regulators that emerge due to CNAs. In this scenario, a prespecified gene expression signature was treated as the ‘phenotype’, while the CNAs of signature positive samples were labeled as the ‘genotype’. A similar stepwise analysis is then carried out on the observed CN changes to identify potential regulator genes.
JOINT INTEGRATION METHODOLOGIES
Unlike stepwise methods, joint integration techniques carry out one analysis by viewing CNA and GE as paired data entries. Thus, all sources of genomic information are treated as one coherent dataset instead of separate structures that require separate analysis. Typical to these approaches, consistent signals that emerge only as a result of combining both levels of evidence are used to conduct inference. As a result, joint methods are known to employ forms of correlation [19, 59, 60] or regression [31, 59, 61] analysis. These methods face two major challenges of high dimensionality and computational feasibility. The imbalance between the sample size and number of genes is the problem facing most genomic analysis methods, as it decreases the ability to differentiate between true signals and random noise. In an integrative setting, this problem is exacerbated as the additional data type doubles the number of existing features, whereas, the sample size stays the same. Therefore, data reduction methods have been commonly called upon to deal with such issue. Generalized Singular Value Decomposition (GSVD) is a popular regression framework used in joint analysis due to the added value of dimension reduction. Berger et al. [62] implemented this strategy to identify variation patterns between two biological inputs by iteratively projecting CNA/GE data onto different decomposition directions. Computation-wise, many joint methods have utilized correlation analysis as an approach to quantify the relationship between CNAs and GE [30, 63, 64].
Soneson et al. [65] also pursued a correlation-based approach to achieve integration. After using principal component analysis (PCA) to reduce dimensions, they employed Canonical Correlation Analysis (CCA) to identify highly correlated CNA/GE pairs. Similarly, Gonzalez et al. [66] implemented regularized CCA to explore the correlation structure between paired datasets with additional emphasis placed on the high dimensionality of the input data. Schafer et al. [53] also introduced a correlation approach that combines a bivariate analysis to assess the concordance of CN/GE abnormalities.
The Significant Overlap of Differentially Expressed and Genomic Imbalanced Regions (SODEGIR) [26] identifies discrete chromosomal regions of coordinated CN alterations and changes in transcriptional levels. Instead of utilizing all samples in one analysis, each tumor is sequentially studied for chromosome regions with concordant dosage effect. The results are then combined, elevating the analysis to the entire dataset of tumor specimens.
Many genes are coexpressed in the genome and CNAs occur simultaneously in multiple locations. This limits the precision in locating the interacting partners. To allow for an additional flexibility between CNA/GE relationships, Lee et al. [19] proposed a form of correlation analysis that allowed clusters of coexpressed genes to be simultaneously associated with CNAs throughout the genome [42]. By implementing a bi-clustering algorithm on the observed CNA/GE correlation matrix, their methodology identifies clusters of genes that are related to other clusters of CNAs. In addition, the methodological setup also accounted for downstream function analysis—a novelty at the time the method was proposed.
Altogether, many correlation-based methods employ measures (i.e. Pearson correlation) designed to identify features that vary linearly across both data types. However, in scenarios where the relationship is nonlinear, these techniques may lack the statistical power to pick out the interesting features. Furthermore, since detectable correlations, by nature, require data points to exhibit a certain degree of spread, the clustering of these points, even in the extreme regions of either data type, will pose its own set of issues. Thus, correlation-based methods may be less suited for integrative purposes in general. However, rank correlation or mutual information-based measures can be considered instead.
Specific to CNA and GE integration, the correlation structures refer to the within- and between-data correlation matrices (gene–gene and CN–GE correlation matrices). While correlation-based methods employ judicious assumptions to simplify their form, others may altogether ignore them. Therefore, a complete disregard or oversimplification of these structures can severely cripple the analysis despite the daunting task associated with their accurate formulation.
Dependency, as attested by such approaches may fail in situations where all profiles across the same feature exhibit abnormal levels. In such scenario, the lack of a decent spread will ultimately result in a correlation close to zero despite the strong inherent signal. Assuming no reference base is then used (common amongst these techniques), these techniques are essentially restricted for identifying features that: (i) exhibit a wide range of values and (ii) behave in the same direction. Furthermore, since these methods assume an existing linear relationship that cannot be guaranteed, they may actually be less suited for integration purposes. Thus, the specification and modeling process of the correlation structure inherent to multiple layers of genomic data becomes key if a correlation-based approach is indeed pursued.
Nonetheless, while correlation methods have been used to uncover the regulatory CNAs of gene expression, they are much less suited for tumor subtype classification. To do so, regression-based techniques capable of extracting feature pairs that account for a large fraction of the observed variability is often preferred. Consequently, these methods will then use the selected features to infer unique alteration patterns that ultimately guide the formation of the disease subgroups. Shen et al. [46], for example, introduced a latent variable regression approach for tumor subtype discovery. By modeling the subtypes as latent variables, inference was conducted by simultaneously capturing genomic patterns that are: (i) consistent across multiple data types, (ii) specific to individual data types, or (iii) weak, yet consistent across datasets that would emerge only as a result of combining levels of evidence [46].
COpy Number and EXpression In Cancer (CONEXIC) [38] is a Bayesian network-based algorithm that identifies driving mutations and the biological processes they influence. CONEXIC is inspired by Module Networks [67], but has been augmented by a number of critical modifications that make it suitable for identifying drivers. CONEXIC uses a score-guided search to identify the combination of modulators that best explains the behavior of a gene expression module across tumor samples and searches for those with the highest score within the amplified or deleted regions.
IMPORTANT BIOLOGICAL FINDINGS RESULTANT OF INTEGRATIVE ANALYSIS
In the following section, we discuss important biological insights that have been uncovered by integrative works over the last decade. This discussion is meant to highlight examples of various biological endpoints. More examples can be found in the Supplementary Table S3. The most important success of the integrative analysis approaches has been identifying genes targets of genomic CNA and altered pathways in primary tumors. Tonon et al. [68] identified WHSC1L1 and TPX2 as two candidates likely targeted for amplification in both pancreatic ductal adenocarcinoma and nonsmall-cell lung cancer. Garraway et al. [57] identified MITF as a potential ‘lineage addiction’ oncogene necessary for tissue-specific cancer development and progression. Deletion of the transcription factor RUNX3 was shown to play an important role in primary breast cancer [69]. Overexpression of VEGFA via 6p21 gain in hepatocellular carcinomas was found to be a novel, noncell-autonomous mechanism of oncogene activation [48]. Adler et al. [36] used the SLAMS algorithm to identify CSN5 and MYC as two genetic regulators of the breast cancer. Taylor et al. identified the nuclear receptor coactivator NCOA2 as an oncogene in ∼11% of prostate tumors [70].
Woo et al. [41] identified NCSTN and SCRIB among others as potential drivers in hepatocellular carcinoma progression. Akavia et al. [38] identified TBC1D16 and RAB27A as drivers of melanoma using their CONEXIC algorithm and suggested that abnormal regulation of protein trafficking contributes to proliferation in melanoma. In T-cell prolymphocytic leukemia, 734 genes including those involved in lymphomagenesis, cell cycle regulation, apoptosis and DNA repair were differentially expressed and significantly enriched in genomic regions affected by recurrent chromosomal imbalances [71]. Lee et al. showed that 7p13 were significantly correlated with epidermal growth factor receptor signaling pathway in glioblastoma multiforme, chr 13q with NF-κB cascades in bladder cancer and chr 11p with Reck pathway in breast cancer with their bi-clustering algorithm.
Integrative analysis has been used to identify tumor subtypes or patient groups that have different characteristics including patient survival, and response or resistance to the therapy. Myllykangas et al. [72] showed statistically significant differences in immunopositivity of ERBB2 and MUC1 genes in the intestinal and diffuse subtypes of gastric cancer. Using both the GE and CNA information simultaneously, Shen et al. [46] clustered breast and lung datasets. In the breast data, three distinct clusters were identified. One cluster was separated based on cell line differences, the second based on HER2/ERBB2 concordant amplification and overexpression and the third based on consistent amplifications at the end of chromosome 17q. Interestingly, the second cluster was associated with poor survival. Similarly, the lung tumors were separated into four clusters. The first was characterized by 8p/underexpression and was also highly correlated with EGFR mutation and DUSP4 deletion. The second was highlighted by 12q amplification, a region with known oncogenes CDK4 and MDM2, and the final two were formed based on the extent of 8p loss and EGFR mutation.
Zhang et al. [45] identified a very poor prognostic group by integrating CNA and GE data on lymph node-negative primary breast tumors that was putatively more resistant to preoperative paclitaxel and 5-fluorouracil-doxorubicin-cyclophosphamide combination chemotherapy, particularly against the doxorubicin compound, while potentially benefiting from etoposide. Based on their analysis, Rinaldi et al. [73] suggested B-cell associated tyrosine kinase Syk as a possible therapeutic target in mantle cell lymphoma. Findings of Olejniczak et al. [74] suggested that 18q21-23 CN could be a clinically relevant predictor for sensitivity of SCLC to Bcl-2 family inhibitors in small-cell lung carcinoma. Etemadmoghadam et al. [44] showed that amplification of 19q12, containing CCNE1 and 20q11.22-q13.12, mapping immediately adjacent to the steroid receptor coactivator NCOA3, was significantly associated with poor response to primary treatment in ovarian carcinomas. They also identified a cell-cycle independent role for CCNE1 in modulating chemoresponse.
GUIDELINES FOR USING EXISTING INTEGRATIVE ANALYSIS METHODS
The choice for the appropriate analysis method(s) depends upon the desired endpoint. However, a central aim of integrative analysis combining GE profiles with CNA is to identify driver CNA that elicit cancer through aberrant gene expression from myriads of passenger CNA. Therefore, given a paired GE and CNA dataset, interested readers can use following guidelines to make most out of their data using existing methods.
For a given dataset, first task would be to determine genes with CNA that are differentially expressed. Moreover, an important task would be to identify presence of tumor subtypes in the data that is influenced by CNA. It would be equally prudent to check whether large CNA (e.g. at the whole chromosome or arm level) is affecting expression of majority of the genes in that region. The iCluster methodology [46] can be used to identify the tumor subtypes characterized by concordant CNA and GE changes. This is important, as the presence of tumor subtypes or chromosome aneuploidy can adversely affect downstream analysis. A good example of importance of identifying tumor subtypes was the identification of lineage-specific master regulators by the methodology of Garraway et al. [57].
Many software packages are available to identify list of genes that may be enriched in oncogenes and tumor suppressor genes using dosage effect (SIGMA2 [75], ACE-it [52]), concordant changes in CNA/GE (DRI [76], SODEGIR [26], CNAmet [77], remap [78]) or Bayesian frameworks (CONNEXIC [38]). The bi-clustering algorithm of Lee et al. [19] can be used to identify cancer-type specific biological pathways. The integrative methodology of Adler et al. [36] can be used to identify genetic regulators when distinguishing gene expression signatures are available. It is important to note that choice of the gene expression signature to divide the tumor samples will play a very important role in successfully identifying the regulators.
DISCUSSION
Recent studies have estimated that >15% of heritable gene expression variation can be directly attributed to CN variants in normal cells [31]. It is natural to ask how much stronger this relationship is in various cancer types. Moreover, it is important to know how much a decade's worth of efforts in generating paired data on gene expression and CNA in large cohorts for multiple cancer types have advanced our understanding of cancer biology and help improve the clinical care. It is equally important to identify weaknesses of current methodologies and previous analysis efforts so that novel algorithms can be developed.
Indeed, integrative methodologies have greatly advanced our understanding of genomic CNA and their downstream implications in cancer. While the estimates vary depending upon the cancer type and the analysis methodology, it is estimated that ∼60% of the genes show differential expression concordant to their CN status. These analyses have suggested that the global correlation between GE and CN is relatively weak but consistent across studies. There is a strong evidence for a cis-dosage effect of CNA on GE, and segmenting the CNA levels and probe filtering helps to improve these observed relationships [59]. The exploratory analysis has also provided deeper insights into transcription regulation. Increased gene expression in response to gene amplifications may suggest that most genes are not subject to specific auto-regulation of dosage compensation, yet it is equally clear that most of these genes are incapable of completely overriding transcription regulatory mechanisms. Analyzing the genomic distribution of expressed genes may permit the inference of DNA CN aberrations, particularly in aneuploidy (where gene expression can be averaged across large chromosomal regions). Although elevated expression of an amplified gene cannot be considered as strong independent evidence of a candidate oncogene's role in tumorigenesis, there exists a possible role for widespread DNA CNA in tumorigenesis beyond the amplification (or deletion) of specific oncogenes (or TSGs) [79, 80]. Widespread DNA CNAs and concomitant gene expression imbalances may disrupt critical stoichiometric relationships in cell metabolism and physiology (i.e. proteosome, mitotic spindle), possibly promoting further chromosomal instability that directly contributes to tumor development and progression. A substantial portion of the phenotypic uniqueness (and by extension, the heterogeneity in clinical behavior) among patients’ tumors may be traced to underlying variations in the DNA CN. Potential cancer therapeutics can exploit specific or global imbalances in gene expression.
Over the past decade, these methods have gradually progressed from exploratory tools to specialized techniques that uncover novel biology. These methods have helped in identifying gene targets of CNA during the process of tumor formation, drivers and subtype-specific genes for multiple cancer types. Integrative analysis has been used to identify tumor subtypes or patient groups that have different characteristics including patient survival, and response or resistance to the therapy. Although, the advantage provided by integrative approaches as oppose to carrying out the same analysis using one data type only has not been quantified in any published work.
Whereas, we have provided guidelines to the interested readers in utilizing existing methods in the previous section, future integration methods will benefit by adopting following general guidelines, individual parts of which has been shown to work efficiently in the literature. (i) The data integration methods should use efficient dimensionality reduction methods, as genomic data are very high dimensional and attempting to integrate paired data only exacerbate the dimensionality problem, (ii) the uncertainty of the discrete CNs should be propagated to test statistics for differential expression between CN groups, or use the call probabilities of CN altered regions instead of the actual calls, (iii) to reduce tumor heterogeneity, tumor subclasses could be identified before integrating the two data types for identifying gene targets of tumorigenesis and driver genes, (iv) the emergence of indirect relationships (interactions not restricted by physical location) point out the need for methodologies to simultaneously model both interaction types. Most integrative methods start analysis without taking advantage of the gene interaction and regulatory network information present in the literature and from other functional genomics dataset. Moreover, functional enrichment analysis, and utilization of clinical information is primarily seen as postanalysis interpretation tool rather than assistance for inference. Methods incorporating network and clinical information during the inference process will be more powerful in achieving desired endpoints and (v) finally, none of the methods described above infer causal associations between gene expression and disease that is governed by CNA. Causal analysis methods followed by experimental validation could help.
There is still a lot to be desired on the analysis side. For example, it is interesting to note that while the impact of CNA on GE is well explored, that of LOH and UPD is not well established. It is not clear as to what is the statistical power of the many available data integration methodologies and how much noise, which is inherent in the functional genomics datasets, they can tolerate. It is not clear as to what is the minimum number of samples that are required to achieve both high sensitivity and specificity for the desired analysis endpoints. There is also a lack of a gold standard, with which we can compare the newly developed methodologies. Only one comparative analysis, exploring the impact of CNA on GE, has been reported for five cancer types in the literature [59]. No comparative analysis of important biological endpoint has been carried out. Such an analysis would be invaluable in terms of understanding the evolutionary pathways of cancer. An interesting, yet completely unexplored question is to understand the role of chromosomal aneuploidy in cancer. The origins of these genomic abnormalities remain a subject of debate even to this day [81]. While some view them as the central initiator of tumor formation [82–84], others believe that they merely exist as side effects of deranged cell division cycles [85, 86]. This question can be answered using presently available paired datasets, but it will require development of new methodologies.
We hope that, with the reducing cost of next generation sequencing, more paired datasets providing information on GE and CNA will be available. This will reduce some of the analysis issues due to probe bias on the microarray platforms, as well as, provide additional information. For example, expression profiles with next generation sequencing can also provide information on alternative splicing and miRNA expression [87]. Genomic DNA profiles with next generation sequencing can provide additional informations like mutations, and chromosomal structural variations like fusion and inversion apart from the CNA [88, 89]. Furthermore, integration methodologies of future will integrate additional paired information such as epigenetic methylation and histone modification. They will not only provide more detailed insights on cancer, but also benefit clinical care.
SUPPLEMENTARY DATA
Supplementary data are available online at http://bib.oxfordjournals.org/.
Key Points.
Oncogenes and tumor suppressor genes can be identified from genome-wide CNA profiles of cancer patients but the critical challenge is in differentiating between alterations that drive the cancer growth and other seemingly random alterations that accumulate through instability induced by tumorigenesis.
There is a strong evidence for a cis-dosage effect of CNA on gene expression and this relationship can help identification of novel genes involved in cancer as well as other aspects of tumor biology and clinical care
Currently available integration methodologies can be grouped based on methodological approach like stepwise integration or joint integration, or integration endpoints like gene/gene-set discovery or subtype clustering.
There is a need for comparative analysis of paired CNA and gene expression data for multiple cancer types toward identification of different biological endpoints.
There is a need for development of novel integration methods toward improving current methodologies and for studying effects of anuploidy in cancer.
FUNDING
The cancer training grant (NIH T32 CA09337 to Yi Li and N.H.); Claudia Adams Barr Award for basic innovative research in cancer;Multiple Myeloma Career Development Award that is a part of DFCI/HCC Multiple Myeloma SPORE (grant NIH 5P50 CA100707-07 to P.K.S.); dChip grant (NIH 1R01GM077122 to C.L.).
Supplementary Material
Biographies
Norman Huang received his Master’s degree from the Department of Biostatistics of the Harvard Graduate School of Arts and Sciences. He is currently a graduate student in the Department of Biostatistics and Computational Biology of Dana-Farber Cancer Institute and Harvard School of Public Health. His research interests include methods for integrating copy number and gene expression.
Parantu K. Shah, PhD, is a research fellow in the Department of Biostatistics and Computational Biology of the Dana-Farber Cancer Institute and Harvard School of Public Health. His research interests include developing tools for managing, analyzing and integrating large-scale data to understand cancer biology and improve patient care.
Cheng Li, PhD, is an associate professor of Biostatistics in the Department of Biostatistics and Computational Biology of the Dana-Farber Cancer Institute and Harvard School of Public Health. His research interests are aneuploidy in cancer and genomics-based prognosis.
References
- 1.Vogelstein B, Kinzler KW. The Genetic Basis of Human Cancer. New York, USA: McGraw-Hill Professional; 2002. [Google Scholar]
- 2.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–76. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 4.Alitalo K, Schwab M, Lin CC, et al. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci USA. 1983;80:1707–11. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinds PW, Dowdy SF, Eaton EN, et al. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–13. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahl GM, Padgett RA, Stark GR. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979;254:8679–89. [PubMed] [Google Scholar]
- 7.Schimke RT, Kaufman RJ, Alt FW, Kellems RF. Gene amplification and drug resistance in cultured murine cells. Science. 1978;202:1051–5. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- 8.Koivisto P, Kononen J, Palmberg C, et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res. 1997;57:314–9. [PubMed] [Google Scholar]
- 9.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 11.Orlow I, Lacombe L, Hannon GJ, et al. Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst. 1995;87:1524–9. doi: 10.1093/jnci/87.20.1524. [DOI] [PubMed] [Google Scholar]
- 12.Nagai MA, Yamamoto L, Salaorni S, et al. Detailed deletion mapping of chromosome segment 17q12-21 in sporadic breast tumours. Genes Chromosomes Cancer. 1994;11:58–62. doi: 10.1002/gcc.2870110109. [DOI] [PubMed] [Google Scholar]
- 13.Cavenee WK, Dryja TP, Phillips RA, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–84. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 14.Baker SJ, Preisinger AC, Jessup JM, et al. P53 Gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–22. [PubMed] [Google Scholar]
- 15.Ruivenkamp CA, van Wezel T, Zanon C, et al. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat Genet. 2002;31:295–300. doi: 10.1038/ng903. [DOI] [PubMed] [Google Scholar]
- 16.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37(Suppl):S11–7. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 18.National Cancer Institute. The Cancer Genome Atlas Homepage. http://cancergenome.nih.gov (11 May 2011, date last accessed) [Google Scholar]
- 19.Lee H, Kong SW, Park PJ. Integrative analysis reveals the direct and indirect interactions between DNA copy number aberrations and gene expression changes. Bioinformatics. 2008;24:889–96. doi: 10.1093/bioinformatics/btn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monni O, Barlund M, Mousses S, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–16. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 22.van de Wiel MA, Picard F, van Wieringen WN, Ylstra B. Preprocessing and downstream analysis of microarray DNA copy number profiles. Brief Bioinform. 2011;12:10–21. doi: 10.1093/bib/bbq004. [DOI] [PubMed] [Google Scholar]
- 23.Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief Funct Genomic Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 24.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staaf J, Lindgren D, Vallon-Christersson J, et al. Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome SNP arrays. Genome Biol. 2008;9:R136. doi: 10.1186/gb-2008-9-9-r136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bicciato S, Spinelli R, Zampieri M, et al. A computational procedure to identify significant overlap of differentially expressed and genomic imbalanced regions in cancer datasets. Nucleic Acids Res. 2009;37:5057–70. doi: 10.1093/nar/gkp520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–5. [PubMed] [Google Scholar]
- 28.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips JL, Hayward SW, Wang Y, et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 2001;61:8143–9. [PubMed] [Google Scholar]
- 30.Jarvinen AK, Autio R, Haapa-Paananen S, et al. Identification of target genes in laryngeal squamous cell carcinoma by high-resolution copy number and gene expression microarray analyses. Oncogene. 2006;25:6997–7008. doi: 10.1038/sj.onc.1209690. [DOI] [PubMed] [Google Scholar]
- 31.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf M, Mousses S, Hautaniemi S, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004;6:240–7. doi: 10.1593/neo.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masayesva BG, Ha P, Garrett-Mayer E, et al. Gene expression alterations over large chromosomal regions in cancers include multiple genes unrelated to malignant progression. Proc Natl Acad Sci USA. 2004;101:8715–20. doi: 10.1073/pnas.0400027101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soroceanu L, Kharbanda S, Chen R, et al. Identification of IGF2 signaling through phosphoinositide-3-kinase regulatory subunit 3 as a growth-promoting axis in glioblastoma. Proc Natl Acad Sci USA. 2007;104:3466–71. doi: 10.1073/pnas.0611271104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Adler AS, Lin M, Horlings H, et al. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–30. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin LX. An integrative analysis of microRNA and mRNA expression–a case study. Cancer Inform. 2008;6:369–79. doi: 10.4137/cin.s633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akavia UD, Litvin O, Kim J, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweet-Cordero A, Tseng GC, You H, et al. Comparison of gene expression and DNA copy number changes in a murine model of lung cancer. Genes Chromosomes Cancer. 2006;45:338–48. doi: 10.1002/gcc.20296. [DOI] [PubMed] [Google Scholar]
- 40.Bergamaschi A, Kim YH, Kwei KA, et al. CAMK1D amplification implicated in epithelial–mesenchymal transition in basal-like breast cancer. Mol Oncol. 2008;2:327–39. doi: 10.1016/j.molonc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo HG, Park ES, Lee JS, et al. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–66. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Broet P, Richardson S. Detection of gene copy number changes in CGH microarrays using a spatially correlated mixture model. Bioinformatics. 2006;22:911–8. doi: 10.1093/bioinformatics/btl035. [DOI] [PubMed] [Google Scholar]
- 44.Etemadmoghadam D, deFazio A, Beroukhim R, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–27. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Martens JW, Yu JX, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795–801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 46.Shen R, Olshen AB, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–12. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kingsley CB, Kuo WL, Polikoff D, et al. Magellan: a web based system for the integrated analysis of heterogeneous biological data and annotations; application to DNA copy number and expression data in ovarian cancer. Cancer Inform. 2007;2:10–21. [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck AH, Lee CH, Witten DM, et al. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene. 2010;29:845–54. doi: 10.1038/onc.2009.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruano Y, Mollejo M, Ribalta T, et al. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao J, Weremowicz S, Feng B, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–78. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 52.van Wieringen WN, Belien JA, Vosse SJ, et al. ACE-it: a tool for genome-wide integration of gene dosage and RNA expression data. Bioinformatics. 2006;22:1919–20. doi: 10.1093/bioinformatics/btl269. [DOI] [PubMed] [Google Scholar]
- 53.Schafer M, Schwender H, Merk S, et al. Integrated analysis of copy number alterations and gene expression: a bivariate assessment of equally directed abnormalities. Bioinformatics. 2009;25:3228–35. doi: 10.1093/bioinformatics/btp592. [DOI] [PubMed] [Google Scholar]
- 54.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 55.van de Wiel MA, Wieringen WN. CGH regions: dimension reduction for array CGH data with minimal information loss. Cancer Inform. 2007;3:55–63. [PMC free article] [PubMed] [Google Scholar]
- 56.Van Wieringen WN, Van De Wiel MA. Nonparametric testing for DNA copy number induced differential mRNA gene expression. Biometrics. 2009;65:19–29. doi: 10.1111/j.1541-0420.2008.01052.x. [DOI] [PubMed] [Google Scholar]
- 57.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 58.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu W, Choi H, Ghosh D. Global associations between copy number and transcript mRNA microarray data: an empirical study. Cancer Inform. 2008;6:17–23. doi: 10.4137/cin.s342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotliarov Y, Kotliarova S, Charong N, et al. Correlation analysis between single-nucleotide polymorphism and expression arrays in gliomas identifies potentially relevant target genes. Cancer Res. 2009;69:1596–603. doi: 10.1158/0008-5472.CAN-08-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menezes RX, Boetzer M, Sieswerda M, et al. Integrated analysis of DNA copy number and gene expression microarray data using gene sets. BMC Bioinformatics. 2009;10:203. doi: 10.1186/1471-2105-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berger JA, Hautaniemi S, Mitra SK, Astola J. Jointly analyzing gene expression and copy number data in breast cancer using data reduction models. IEEE/ACM Trans Comput Biol Bioinform. 2006;3:2–16. doi: 10.1109/TCBB.2006.10. [DOI] [PubMed] [Google Scholar]
- 63.Tsukamoto Y, Uchida T, Karnan S, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol. 2008;216:471–82. doi: 10.1002/path.2424. [DOI] [PubMed] [Google Scholar]
- 64.Lipson D, Ben-Dor A, Dehan E, Yakhini Z. Joint analysis of DNA copy numbers and gene expression levels. Algorithms Bioinformatics. 2004;3240:135–46. [Google Scholar]
- 65.Soneson C, Lilljebjorn H, Fioretos T, Fontes M. Integrative analysis of gene expression and copy number alterations using canonical correlation analysis. BMC Bioinformatics. 2010;11:191. doi: 10.1186/1471-2105-11-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez I, DeJean S, Martin P, et al. Highlighting relationships between heterogeneous biological data through graphical displays based on regularized canonical correlation analysis. J Biol Syst. 2008;17:173–99. [Google Scholar]
- 67.Segal E, Shapira M, Regev A, et al. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 68.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–30. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen W, Salto-Tellez M, Palanisamy N, et al. Targets of genome copy number reduction in primary breast cancers identified by integrative genomics. Genes Chromosomes Cancer. 2007;46:288–301. doi: 10.1002/gcc.20411. [DOI] [PubMed] [Google Scholar]
- 70.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durig J, Bug S, Klein-Hitpass L, et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32) Leukemia. 2007;21:2153–63. doi: 10.1038/sj.leu.2404877. [DOI] [PubMed] [Google Scholar]
- 72.Myllykangas S, Junnila S, Kokkola A, et al. Integrated gene copy number and expression microarray analysis of gastric cancer highlights potential target genes. Int J Cancer. 2008;123:817–25. doi: 10.1002/ijc.23574. [DOI] [PubMed] [Google Scholar]
- 73.Rinaldi A, Kwee I, Taborelli M, et al. Genomic and expression profiling identifies the B-cell associated tyrosine kinase Syk as a possible therapeutic target in mantle cell lymphoma. Br J Haematol. 2006;132:303–16. doi: 10.1111/j.1365-2141.2005.05883.x. [DOI] [PubMed] [Google Scholar]
- 74.Olejniczak ET, Van Sant C, Anderson MG, et al. Integrative genomic analysis of small-cell lung carcinoma reveals correlates of sensitivity to bcl-2 antagonists and uncovers novel chromosomal gains. Mol Cancer Res. 2007;5:331–9. doi: 10.1158/1541-7786.MCR-06-0367. [DOI] [PubMed] [Google Scholar]
- 75.Chari R, Coe BP, Wedseltoft C, et al. SIGMA2: a system for the integrative genomic multi-dimensional analysis of cancer genomes, epigenomes, and transcriptomes. BMC Bioinformatics. 2008;9:422. doi: 10.1186/1471-2105-9-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salari K, Tibshirani R, Pollack JR. DR-Integrator: a new analytic tool for integrating DNA copy number and gene expression data. Bioinformatics. 2010;26:414–6. doi: 10.1093/bioinformatics/btp702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louhimo R, Hautaniemi S. CNAmet: an R package for integrating copy number, methylation and expression data. Bioinformatics. 2011;27:887–8. doi: 10.1093/bioinformatics/btr019. [DOI] [PubMed] [Google Scholar]
- 78.Peng J, Zhu J, Bergamaschi A, et al. Regularized multivariate regression for identifying master predictors with application to integrative genomics study of breast cancer. Ann Appl Stat. 2010;4:53. doi: 10.1214/09-AOAS271SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li R, Yerganian G, Duesberg P, et al. Aneuploidy correlated 100% with chemical transformation of Chinese hamster cells. Proc Natl Acad Sci USA. 1997;94:14506–11. doi: 10.1073/pnas.94.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rasnick D, Duesberg PH. How aneuploidy affects metabolic control and causes cancer. Biochem J. 1999;340(Pt 3):621–30. [PMC free article] [PubMed] [Google Scholar]
- 81.Marx J. Debate surges over the origins of genomic defects in cancer. Science. 2002;297:544–6. doi: 10.1126/science.297.5581.544. [DOI] [PubMed] [Google Scholar]
- 82.Li R, Sonik A, Stindl R, et al. Aneuploidy vs. gene mutation hypothesis of cancer: recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci USA. 2000;97:3236–41. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duesberg PH. Are cancers dependent on oncogenes or on aneuploidy? Cancer Genet Cytogenet. 2003;143:89–91. doi: 10.1016/s0165-4608(02)00938-x. [DOI] [PubMed] [Google Scholar]
- 84.Shih IM, Zhou W, Goodman SN, et al. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61:818–22. [PubMed] [Google Scholar]
- 85.Zimonjic D, Brooks MW, Popescu N, et al. Derivation of human tumor cells in vitro without widespread genomic instability. Cancer Res. 2001;61:8838–44. [PubMed] [Google Scholar]
- 86.Lamlum H, Papadopoulou A, Ilyas M, et al. APC mutations are sufficient for the growth of early colorectal adenomas. Proc Natl Acad Sci USA. 2000;97:2225–8. doi: 10.1073/pnas.040564697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–64. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 88.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.