Abstract
There is an urgent need to develop sustainable solutions to convert solar energy into energy carriers used in the society. In addition to solar cells generating electricity, there are several options to generate solar fuels. This paper outlines and discusses the design and engineering of photosynthetic microbial systems for the generation of renewable solar fuels, with a focus on cyanobacteria. Cyanobacteria are prokaryotic microorganisms with the same type of photosynthesis as higher plants. Native and engineered cyanobacteria have been used by us and others as model systems to examine, demonstrate, and develop photobiological H2 production. More recently, the production of carbon-containing solar fuels like ethanol, butanol, and isoprene have been demonstrated. We are using a synthetic biology approach to develop efficient photosynthetic microbial cell factories for direct generation of biofuels from solar energy. Present progress and advances in the design, engineering, and construction of such cyanobacterial cells for the generation of a portfolio of solar fuels, e.g., hydrogen, alcohols, and isoprene, are presented and discussed. Possibilities and challenges when introducing and using synthetic biology are highlighted.
Keywords: Cyanobacteria, Design and engineering, Renewable energy, Microbial cells, Solar fuels, Synthetic biology
Introduction
Cyanobacteria are prokaryotic microorganisms, unicellular to filamentous, with the same type of photosynthesis as higher plants. They are present in highly diverse and extreme environments with significant fluctuations in, e.g., temperature, salinity, pH, and water availability. Since 1995 when the first cyanobacterial genome became public, a large number of cyanobacterial genomes are now sequenced and genetic tools for traditional as well as synthetic biology-based molecular technologies are available and also further developed. The ability of cyanobacteria to use solar energy and atmospheric CO2 as energy and carbon sources, respectively, together with their faster growth rates (compared to plants) and the relative ease with which they can be genetically engineered (compared to algae), make cyanobacteria stand out from all other organisms used in biotechnological applications (Angermayr et al. 2009; Heidorn et al. 2011).
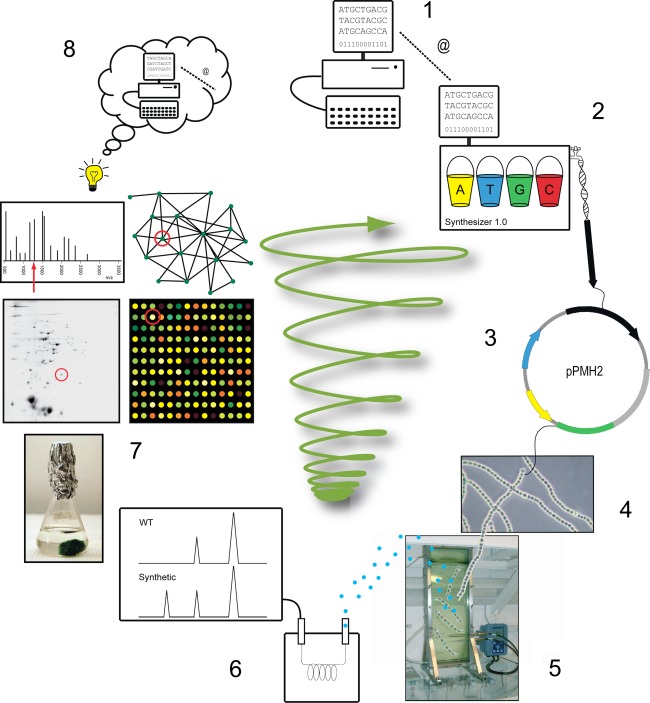
We are using a general approach developed by many laboratories to design, engineer, construct, and analyze cyanobacteria for targeted and improved biofuel production. The experimental design is followed by the construction, characterization, and analyses of the cyanobacterial cells as detailed below (see also Fig. 1):
Hypothesis generation. Bioinformatic investigations to (a) Codon optimization and (b) Experimental design.
Synthesis of DNA parts such as promoters, ribosomal binding sites and genes to fit and be optimized for the specific host.
Assembly of the genetic parts into the genetic vehicle.
Insertion of the novel genetic constructs into the cyanobacterial host.
Controlled cultivation in purpose designed photobioreactors.
Analysis of the culture for biofuel production performance.
High-throughput data acquisition from multiple levels; transcriptome, proteome, metabolome, and bioinformatic network analyses to create gene association, regulatory and metabolic networks. Data interpretation and validation of results.
Identification of processes of importance to further improve production. Identify bottlenecks and suggest modifications, etc. Hypothesis generation.
Based on the new knowledge generated through the above process start a new cycle at 1.
Fig. 1.
Overview of the general approach adopted in our laboratory to design, engineer, and analyze cyanobacteria for improved biofuel production. After establishing an initial hypothesis, bioinformatic investigations and careful literature reviews are carried out to, e.g., design genetic constructs, optimize codon utilization, and to develop the experimental set up (1). Once this is accomplished, follows the synthesis of DNA parts such as promoters, ribosomal binding sites, and genes to fit the specific host (2), which are then assembled in appropriate genetic vehicles (3). These genetic constructs are subsequently transferred to the cyanobacterial host (4) and the new engineered cyanobacterium can now be cultivated in purpose designed bioreactors under specific controlled conditions (5). After optimizing selected parameters, the engineered cyanobacterium is thoroughly tested to examine its novel capacities in biofuel production (6)—in the present figure, the biofuel produced is envisioned to be in the gas phase. In order to identify, e.g., bottlenecks and limiting steps in the metabolic pathways leading to the biofuel production, high-throughput data is acquired from multiple levels, including analyses of the organism’s transcriptome, proteome, and metabolome (7). Integrating these varied aspects into bioinformatic analyses can then elucidate further regulatory and metabolic networks (7). The identification of processes of importance to promote biofuel production and/or efficiency will lead to the generation of new hypotheses (8), which will then guide the process to a new level of engineering. Therefore, this process can cycle iteratively for additional improvements
Native and Engineered Cyanobacteria for Photobiological H2 Production
Cyanobacteria can be used for the production of molecular hydrogen (H2), a possible future energy carrier. As cyanobacteria can use sunlight as energy source, water as an electron source and air as a carbon (CO2) and a nitrogen (N2) source, the use of cyanobacteria for the production of renewable energy carriers like H2 would be sustainable and also give the possibility of a high energy conversion efficiency. In cyanobacteria, two natural pathways for hydrogen production can be used: H2 production as a by-product during nitrogen fixation and as a product of the activity of the bidirectional hydrogenase. Using these pathways, only limited amounts of hydrogen can be evolved with wild-type strains, levels which are not at all sufficient for a commercial production of H2 as a competitive energy carrier. Several bottlenecks have been identified, some of which have been addressed: (i) H2 uptake by the cells (uptake hydrogenase)—genetically engineered strains with a non-functional/deleted uptake hydrogenase have been produced (see Tamagnini et al. 2007), (ii) low energy efficiency and turnover of the nitrogenase and/or the hydrogenase, (iii) limiting amounts of active H2-evolving enzymes, (iv) high oxygen sensitivity of the nitrogenase and/or the hydrogenase, (v) electron flow inhibition by accumulation of ATP in a hydrogenase-driven system, (vi) low quantum efficiency due to too large antennas in both Photosystem II (PSII) and PSI, and (vii) electron-consuming pathways competing with an efficient electron transfer to the H2 enzymes (Tamagnini et al. 2007; Angermayr et al. 2009). Addressing the last point, a significantly increased hydrogen production was recently achieved when redirecting the electron supply toward the hydrogenase via genetic engineering of the electron-competing nitrate assimilation pathway in the unicellular cyanobacterium Synechocystis 6803 (Baebprasert et al. 2011).
Recently, the bacterial [FeFe] hydrogenase HydA from Clostridium acetobutylicum has been expressed in Synechococcus elongatus PCC 7942 together with the necessary maturation enzymes, and, under anoxic conditions, the resulting light-dependent hydrogen evolution could be shown to be increased >500 times, compared to H2 evolution from the endogenous [NiFe] hydrogenase (Ducat et al. 2011a). These results clearly show the high potential of heterologous expression of suitable enzymes and generation of valuable products in cyanobacterial strains. In order to realize this potential, a new approach aims to introduce engineering principles into molecular biology: synthetic biology.
Standardized Genetic Engineering, Synthetic Biology
Synthetic biology is the design and construction of new biological parts, devices, and systems, as well as the redesign of existing biological systems for useful purposes (http://syntheticbiology.org). Basic principles of synthetic biology are the (i) use of standardized and well-characterized building blocks (e.g., BioBricks), (ii) hierarchical design of nature-inspired, artificial genetic circuits and proteins in silico, and (iii) use of chemical DNA synthesis which allows production of DNA sequences that are not found in nature (Huang et al. 2010). For the rational design of artificial genetic circuits, the behavior of each individual part has to be well characterized. Especially promoters are key parts for the heterologous expression of proteins, and in Huang et al. (2010) promoters have been characterized in cyanobacteria in a standardized experimental setup. The vision of cyanobacterial synthetic biology is to build up a repository of standard genetic parts for use in the hosts that are shared by the cyanobacterial community and can speed up successful genetic engineering of cyanobacterial strains for the sustainable production of valuable products.
Native Transcriptional Regulation, Importance of Non-Native Systems
Each time a gene or protein is identified through, e.g., microarray, proteomics, genetic engineering, as being important in a given metabolic pathway, it becomes crucial to proceed with its characterization. The decision to follow up on this gene or protein may emerge from its identification as a bottleneck in the pathway, or simply because it is extremely differentially expressed with unknown effects on that pathway. Regardless of the motivation, further characterization can lead to an optimization of the overall reaction of interest. If little is known about this gene and respective protein product, then transcriptional studies are usually the first step of the characterization. These regularly include analyses of the transcript level in different environmental conditions (Northern blotting, RT-qPCR), determination of its promoter structure (primer extension, 5′ RACE), identification of transcription factors controlling its expression (DNA affinity assays, mobility shift assays), and depending on the genetic context, assessment of transcript units (monocistronic vs. polycistronic—Northern blotting, RT-PCR). Obtained results will shed light into the environmental conditions controlling the gene’s expression and therefore it becomes possible to assign it to particular regulatory network(s). Furthermore, detailed transcriptional studies are also of utmost importance to determine specific gene expressions in different cells, e.g., heterocysts and/or vegetative cells in filamentous, heterocyst-forming cyanobacteria as well on the subcellular level. In the given example, the data may indicate additional roles of this gene in cell differentiation or expose further metabolic interactions. In the end, the combination of all transcriptional analyses with a biochemical characterization may lead to a better understanding of the gene’s functions.
Transcription factors are DNA-binding proteins interacting with the promoter element and influencing positively or negatively the transcription of the downstream gene(s). Recently in our laboratory, we have been able to demonstrate that the transcription factor LexA is under post-transcriptional control in the cyanobacterium Synechocystis (Oliveira and Lindblad 2011). In addition, post-translational modifications were also observed, which seem to affect the DNA-binding capacity of LexA (Oliveira and Lindblad 2011). However, transcriptional studies need to be assessed cautiously, as an increasing number of reports indicate that massive non-coding and antisense RNA is expressed in cyanobacterial cells (Georg et al. 2009; Mitschke et al. 2011), even though their function(s) is yet unclear.
Thorough transcriptional analyses of the gene in study may lead to novel functionalities and utilities. For example, if the gene has a specific cell expression, or if it responds specifically and with an appropriate strength to an environmental condition, its promoter can be used to control the expression of other genes (native or foreign) in order to introduce novel capacities into cyanobacteria. For instance, the promoter of petE, encoding the electron carrier plastocyanin has been widely used to control the expression of various genes based on the amount of copper present in the medium; alternatively, the promoter of patS, encoding a heterocyst-inhibiting signaling peptide has been used to target gene expression specifically to heterocysts (Yoon and Golden 1998, 2001; Oliveira and Lindblad 2008). However, regardless of all the efforts to characterize the promoter and understand its behavior in different conditions, it is clear that it will still be dependent on the cell transcription machinery. Therefore, undesired cross-talk between various transcriptional networks may be too difficult to insulate, resulting in unexpected expression patterns and leading to abnormal metabolic flux.
Consequently, orthogonal promoters are required to control the expression of genes when metabolic pathways are to be introduced or modified. As an example, the hybrid promoters tac and trc have been used to regulate the expression of various genes in cyanobacteria. However, these well-described promoters used in Escherichia coli have been shown to respond differently in a cyanobacterial background (Huang et al. 2010; Heidorn et al. 2011), further supporting the notion that all these parts have to be initially and thoroughly characterized in cyanobacteria.
Gene Optimization
Using cyanobacteria for generating interesting products not naturally made by the system requires expression of heterologous genes. These may originate from any organism in possession of the desired metabolic capabilities. Their expression in the cyanobacterial host necessitates systems for propagation or integration in the host genome, as well as control of transcription and translation (Heidorn et al. 2011). It may not be enough, however, to merely introduce the gene sequences of interest under control of a suitable promoter. Since different organisms have different preferences for codon use, optimizing the heterologous gene sequences to fit the host organism may be necessary to enhance expression of the encoded proteins.
Codon optimization is done by comparing the sequence of the gene with the codon use preferences of the intended host organism, and then changing the sequence of the gene to create a better fit for the host. A basic level of codon optimization can be achieved by simply identifying and changing codons that are very rarely used in the host, in order to prevent delay of translation at the point of those codons. However, with DNA synthesis technology routinely available, the entire gene sequence may be reconstructed starting from the amino acid sequence of the encoded protein. This allows for optimization of all the codons to fit better with the host’s preferences, and can also be used to minimize problems caused by strong secondary structures in the mRNA at the 5′ end of the gene. Furthermore, repetitive sequences can be avoided, and new features such as protein tags and other modifications may also be introduced (Welch et al. 2011). Downstream cloning strategies can be facilitated through removal and introduction of restriction sites. Software is available to help design the synthetic gene with all required features (e.g., “Gene Designer” from the company DNA 2.0, Welch et al. 2011).
In cyanobacteria, there is to our knowledge so far only one example where codon optimization of an entire gene sequence has been demonstrated to enhance expression of a heterologous protein. This was done in a study where the plant enzyme isoprene synthase was expressed in the unicellular cyanobacterium Synechocystis PCC 6803. It was found that when the codon usage of the gene was adapted to the preferences of the host bacterium, expression of the protein was substantially increased, with a factor of about 10, compared to when the original plant gene sequence was expressed in the same cyanobacterium (Lindberg et al. 2010). It should be noted, however, that higher levels of protein expression does not necessarily correlate with higher levels of activity of the enzyme in vivo, or with an increase in formation of a desired product, since other limitations on the reaction may apply in the cell.
Examples of Genetically Engineered Cyanobacteria Producing a Solar Fuel
Cyanobacteria have been genetically engineered and demonstrated to produce H2 (Tamagnini et al. 2007; Baebprasert et al. 2011; Ducat et al. 2011a), ethanol (Deng and Coleman 1999; Dexter and Fu 2009), 1-butanol (Lan and Liao 2011), isobutyraldehyde and isobutanol (Atsumi et al. 2009), isoprene (Lindberg et al. 2010), ethylene (Takahama et al. 2003), sugars, and lactic acid (Niederholtmeyer et al. 2010)—for recent reviews, see Angermayr et al. (2009) and Ducat et al. (2011b).
Analyze Modified Cells with Proteomics, Identify Bottlenecks, and Suggest Further Steps
The complexity of processes involved in photobiological fuel production, requires systems-level strategies to exploit the full capacity of cyanobacteria as efficient future biofuel producers. By using such a strategy, including integration of analysis of growth and enzyme activity with quantitative mass spectrometry-based proteomics, we identified potential targets for future genetic engineering to improve production of H2 from the heterocyst-forming cyanobacterium Nostoc punctiforme. In this study, an engineered H2 producing strain of N. punctiforme was compared to the wild type and bottlenecks, such as amount of substrate carbohydrates, and O2 level, was identified, and could be directly related to specific cellular pathways and regulatory activities of importance for the H2 production. This study (Ekman et al. 2011) highlights the significance of a system-wide analysis, on proteome level, of cyanobacterial cells engineered to produce a biofuel. The limitations on the engineered cyanobacteria, in large-scale cultivations in bioreactors or during different cultivation condition, could be identified at an early stage and therefore we will use systems analyses on proteome level for characterizations of the novel synthetic biology engineered cyanobacteria.
From our work, we have concluded that quantitative large scale proteomics provides a useful method to identify targets for directed genetic/physiological modification to optimize the system for a biotechnological approach such as production of sustainable energy carriers.
The final goal in the systems biology approach is the translation of the quantitative proteomic data into mathematical models, which will aid in demonstrating the metabolic and regulatory network as a dynamic in silico model.
Design, Engineering, and Construction of Cyanobacterial Cells for Biofuel Production
Using the above outlined strategy to design, engineer, construct, and analyze cyanobacteria for improved biofuel production (schematically presented in Fig. 1), there is a solid platform to generate efficient photosynthetic microbial cell factories for future large-scale production of solar fuels.
Acknowledgments
Our research on photosynthetic microbial cell factories for direct solar fuel production is supported by the Swedish Energy Agency, the Knut, and Alice Wallenberg Foundation (project MoSES), the Nordic Energy Agency (project AquaFEED), and the EU/Energy FP7 project SOLAR-H2 (contract # 212508).
Biographies
Peter Lindblad
is a Professor at Uppsala University. His research interests include microbial chemistry, cyanobacterial biotechnology, microbial solar fuels, transcriptional regulation, and transcription factors.
Pia Lindberg
is a Researcher at Uppsala University. Her research interests include microbial biotechnology, especially metabolic engineering of cyanobacteria for production of biofuels.
Paulo Oliveira
was a Researcher at Uppsala University until autumn 2011 when he moved to Porto University. His research interests include regulation of cyanobacterial hydrogenases, different transcription networks, cell division in cyanobacteria, and subcellular localization of proteins.
Karin Stensjö
is an Associate Professor at Uppsala University. Her research interests include microbial systems biology, proteome dynamics in cyanobacteria, oxidative stress, and redox regulations.
Thorsten Heidorn
was a Researcher at Uppsala University until the end of 2011 when he moved to BioForsk. His research interests include the development of photosynthetic microbial cell factories, synthetic biology, and bioprocess development in photobioreactors.
References
- Angermayr SA, Hellingwerf K, Lindblad P, Teixeira de Mattos MJ. Energy biotechnology with cyanobacteria. Current Opinion in Biotechnology. 2009;20:257–263. doi: 10.1016/j.copbio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nature Biotechnology. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- Baebprasert W, Jantaro S, Khetkorn K, Lindblad P, Incharoensakdi A. Increased H2 production in the cyanobacterium Synechocystis sp. strain PCC 6803 by redirecting the electron supply via genetic engineering of the nitrate assimilation pathway. Metabolic Engineering. 2011;13:610–616. doi: 10.1016/j.ymben.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Deng MD, Coleman JR. Ethanol synthesis by genetic engineering in cyanobacteria. Applied and Environmental Microbiology. 1999;65:523–528. doi: 10.1128/aem.65.2.523-528.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J, Fu PC. Metabolic engineering of cyanobacteria for ethanol production. Energy & Environmental Science. 2009;2:857–864. doi: 10.1039/b811937f. [DOI] [Google Scholar]
- Ducat DC, Sachdeva G, Silver PA. Rewiring hydrogenase-dependent redox circuits in cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3941–3946. doi: 10.1073/pnas.1016026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat DC, Way JC, Silver PA. Engineering cyanobacteria to generate high-value products. Trends in Biotechnology. 2011;29:95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Ekman M, Ow SY, Holmqvist M, Zhang X, Wagenen J, Wright PC, Stensjö K. Metabolic adaptations in a H2 producing heterocyst-forming cyanobacterium: potentials and implications for biological engineering. Journal of Proteome Research. 2011;10:1772–1784. doi: 10.1021/pr101055v. [DOI] [PubMed] [Google Scholar]
- Georg J, Vosz B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Molecular Systems Biology. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn T, Camsund D, Huang H–H, Lindberg P, Oliveira P, Stensjö K, Lindblad P. Synthetic biology in cyanobacteria: engineering and analyzing novel functions. Methods in Enzymology. 2011;497:540–579. doi: 10.1016/B978-0-12-385075-1.00024-X. [DOI] [PubMed] [Google Scholar]
- Huang H–H, Camsund D, Lindblad P, Heidorn T. Design and characterisation of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Research. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbondioxide. Metabolic Engineering. 2011;13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metabolic Engineering. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voß B, Steglich C, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederholtmeyer H, Wolfstadter BT, Savage DF, Silver PA, Way JC. Engineering cyanobacteria to synthesize and export hydrophilic products. Applied and Environmental Microbiology. 2010;76:3462–3466. doi: 10.1128/AEM.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira P, Lindblad P. An AbrB-like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803. Journal of Bacteriology. 2008;190:1011–1019. doi: 10.1128/JB.01605-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira P, Lindblad P. Novel insights into the regulation of LexA in the cyanobacterium Synechocystis sp. strain PCC 6803. Journal of Bacteriology. 2011;193:3804–3814. doi: 10.1128/JB.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama K, Matsuoka M, Nagahama K, Ogawa T. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene forming enzyme at the psbA1 locus. Journal of Bioscience and Bioengineering. 2003;95:302–305. doi: 10.1016/s1389-1723(03)80034-8. [DOI] [PubMed] [Google Scholar]
- Tamagnini P, Leitão E, Oliveira P, Ferreira D, Pinto F, Harris D, Heidorn T, Lindblad P. Cyanobacterial hydrogenases: Diversity, regulation and applications. FEMS Microbiology Reviews. 2007;31:692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- Welch M, Villalobos A, Gustafsson C, Minshull J. Designing genes for successful protein expression. Methods of Enzymology. 2011;498:43–66. doi: 10.1016/B978-0-12-385120-8.00003-6. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Golden JW. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Golden JW. PatS and products of nitrogen fixation control heterocyst pattern. Journal of Bacteriology. 2001;183:2605–2613. doi: 10.1128/JB.183.8.2605-2613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]