During the last decade, the active usage of Public Access to the World Wide Web increased in such a huge way, that an ongoing data paucity would be hard to believe. Already in 1993, Cohen et al. (1993) correctly urged other ecologists to combine themselves for a concrete and necessary improvement of food-web knowledge. However, in a recent lecture, Joel E. Cohen still reported one—according to him—“shocking” paucity of directly observed, appropriately organized, and publicly availablewebs (Cohen et al. 2009). The basis for this claim was a brilliant analysis of three datasets on the abundance and distribution of organisms in aquatic ecosystems. Did, thus, the ecological community react in a different way to the World Wide Web, possibly due to missing ways to capture the semantics of complex ecological data (Madin et al.2007) in comparison to the flexible ontology of bio-medical research?
Ecological datasets with comparable complexity are not very frequent, but have been documented on previous occasions. Indeed, real sharing of raw ecological data remains uncommon, but online dataset collections and diffuse scientific networks are known to occur widely. For information security, relational databases stored in scientific journals are often not standardized (too many native forms) but in most cases more reliable. The main advantage of project-driven network collections is their user-intuitive interface, developed according to protocols for the taxonomic concept authorities, the quantification of species properties (traits) and the environmental metadata.
In all cases, core hypotheses and key questions might change in time, and according to the query, one database can become less useful. Well-organized food webs, therefore, do not (and can never) exist, in contrast to comprehensive food webs. To make a food web comprehensive, the abundances of different organisms (i.e., population densities) should be coupled to traits like metabolism, size, or body-mass average. The recent focus on these functional traits raised in popularity, possibly through studies on body-mass development and long-term field monitoring. Life-history experiments in laboratory, and extensive studies in space and time, even as investigations at geological scales, are now widespread (e.g., Ernest et al. 2009; Jones et al.2009). In this way, valuable metabolism—mass and biovolume equations became available in digital internet-accessible forms, together with an increasing amount of (cor)relationships investigated at broad scales of complexity (Hayward et al.2009; Jones et al.2009; Mulder et al.2009).
To improve understanding of any comprehensive food web, environmental data are necessary. With data mining, complete food webs can be developed from publicly accessible data. Impressive collective food webs were analyzed (Rossberg et al.2006; Stouffer et al.2007). Aquatic food webs were constructed with all necessary data from a set of 52 lakes (http://www.adirondacklakessurvey.org/ltmloc.html), and geotagged food webs were realized for the aquatic macrofauna from Massachusetts to Virginia from prominent publicly accessible US-EPA sets (Hale et al.2002). Besides accessible datasets on plant traits, there are also free-downloadable food webs for terrestrial microflora, microfauna, and mesofauna, with plenty of environmental data (Mulder and Elser 2009). Containing more than 16000 multitrophic interactions as Open Data, the latter research article shows an evident response of most soil organisms to the elementary (chemical) factors of the environment.
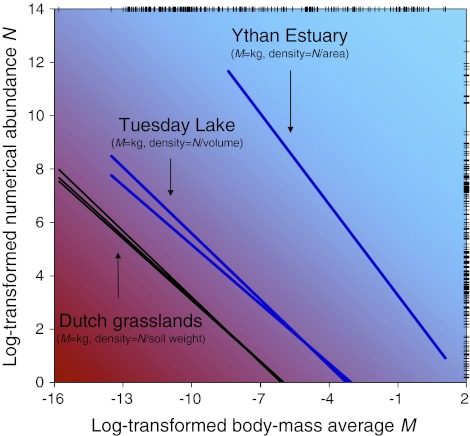
Elementary factors, in fact, determine the ability of specific populations to become established or even competitive. Farming and fishing have been historically constrained by climate, technology, and human pressures. Dramatic changes have accelerated extinctions, broken trophic links, and exacerbated the fragmentation of ecosystems. Abiotic conditions are compulsory because they influence the biotic community as a whole, whereas chemical properties influence individual species fitness and govern how single species interact with their environment (Sterner and Elser 2002; Mulder and Elser 2009). Although there is meanwhile a general agreement that, at population level, the body-mass average remains a statistically reliable predictor of the numerical abundance in food webs (Fig. 1), it is not the identification of any universal structure, but the full mechanistic explanation of structural variations beyond which is challenging (Mulder et al.2005, 2008, 2009). Such results become even more interesting if seen in the dynamic framework of interspecific competition: different sensitivity to the elemental resources allows to understand better population dynamics and to increase the realism of extrapolations.
Fig. 1.
Abundance and distribution of populations occurring in six publicly available food webs: three terrestrial habitats (brown webs corner, bottom left), one freshwater habitat (sampled in 1984 and 1986), and one interface environment (blue webs corner, upper right); units kept as in the original studies (Mulder et al.2005; Cohen et al.2009). Logarithmic transformation is necessary to reduce the influence of outliers, as it places all living organisms into the same geometric domain in which allometric scaling is represented consistently
Functional trait responses to environmental constraints remain invaluable for modeling the biological populations and their complex interactions. To investigate further these trends and the many other comparable patterns, the existing possibilities offered online cannot be ignored. As a matter of fact, metadata collections (http://esapubs.org/archive) ensuring long-term access sensu Science Commons (e-Depots), nonprofit Open Access journals (like http://www.plos.org) and online articles, peer-reviewed alerts (F1000–Biology), open repositories (http://www.arXiv.org) and showcases (http://precedings.nature.com), E-books (http://books.google.com), and advanced search engines are available for research.
One of the most serious problems remains the ignorance of these possibilities. This is rather surprising, according to Bollen et al. (2009), seen that the scientific literature is mostly accessed online, as shown by many web portals that have attempted to measure scientific impact from usage log data (article metrics). The Web not only helps several researchers in a way comparable to interactive media, but also avoids to copy something (or to make wrong claims) simply because “it’s already there, and it’s indexed to Google” (Butler 2005; Brumfiel 2009). Thus, the World Wide Web requires a high responsibility online. Notwithstanding the great potential, up to now, too many scientists seem fully unaware of the new interactive benefits (Bourne et al. 2008). Recently, the algorithm that Google uses for ranking web pages solved mathematically the prediction of the consequences of species’ extinction in complex food webs (Allesina and Pascual 2009). Ecological networks (i.e., multitrophic food webs) and electronic networks (digital internet) are, therefore, much more close together than commonly thought. The Web provides the opportunity to investigate mechanisms underlying patterns in all scientific disciplines. Suggesting data paucities in food-web science, for instance, is not helpful, and nor is it credible on the Web. Maybe, we have them without knowing of it (Hemingway 1952).
Footnotes
This synopsis was not peer reviewed.
References
- Allesina S, Pascual M. Googling food webs: Can an Eigenvector measure species’ importance for coextinctions? PLoS Computational Biology. 2009;5:E1000494. doi: 10.1371/journal.pcbi.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen J, Sompel H, Hagberg A, Chute R. A principal component analysis of 39 scientific impact measures. PLoS ONE. 2009;4:E6022. doi: 10.1371/journal.pone.0006022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne PE, Fink JL, Gerstein M. Open access: Taking full advantage of the content. PLoS Computational Biology. 2008;4:E1000037. doi: 10.1371/journal.pcbi.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumfiel G. Breaking the convention? Nature. 2009;459:1050–1051. doi: 10.1038/4591050a. [DOI] [PubMed] [Google Scholar]
- Butler D. Join efforts. Nature. 2005;438:548–549. doi: 10.1038/438548a. [DOI] [PubMed] [Google Scholar]
- Cohen JE, Beaver RA, Cousins SH, DeAngelis DL, Goldwasser L, Heong KL, Holt RD, Kohn AJ, et al. Improving food webs. Ecology. 1993;74:252–258. doi: 10.2307/1939520. [DOI] [Google Scholar]
- Cohen JE, Schittler DN, Raffaelli DG, Reuman DC. Food webs are more than the sum of their tritrophic parts. Proceedings of the National Academy of Science of the USA. 2009;106:22335–22340. doi: 10.1073/pnas.0910582106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest, S.K.M., T.J. Valone, and J.H. Brown. 2009. Long-term monitoring and experimental manipulation of a Chihuahuan Desert ecosystem near Portal, Arizona, USA. Ecological Archives E090–118. [DOI] [PubMed]
- Hale, S.S., M.M. Hughes, C.J. Strobel, H.W. Buffum, J.L. Copeland, and J.F. Paul. 2002. Coastal ecological data from the Virginian Biogeographic Province, 1990–1993. Ecological Archives E083–057.
- Hayward A, Khalid M, Kolasa J. Population energy use scales positively with body size in natural aquatic microcosms. Global Ecology and Biogeography. 2009;18:553–562. doi: 10.1111/j.1466-8238.2009.00459.x. [DOI] [Google Scholar]
- Hemingway Ernest. The old man and the sea. 1. New York: Charles Scribner’s Sons; 1952. [Google Scholar]
- Jones, K.E., J. Bielby, M. Cardillo, S.A. Fritz, J. O’Dell, C.D.L. Orme, K. Safi, W. Sechrest, et al. 2009. PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecological Archives E090–184.
- Madin J, Bowers S, Schildhauer M, Krivov S, Pennington D, Villa F. An ontology for describing and synthesizing ecological observation data. Ecological Informatics. 2007;2:279–296. doi: 10.1016/j.ecoinf.2007.05.004. [DOI] [Google Scholar]
- Mulder C, Elser JJ. Soil acidity, ecological stoichiometry and allometric scaling in grassland food webs. Global Change Biology. 2009;15:2730–2738. doi: 10.1111/j.1365-2486.2009.01899.x. [DOI] [Google Scholar]
- Mulder C, Cohen JE, Setälä H, Bloem J, Breure AM. Bacterial traits, organism mass, and numerical abundance in the detrital soil food web of Dutch agricultural grasslands. Ecology Letters. 2005;8:80–90. doi: 10.1111/j.1461-0248.2004.00704.x. [DOI] [Google Scholar]
- Mulder C, Hollander HA, Hendriks AJ. Aboveground herbivory shapes the biomass distribution and flux of soil invertebrates. PLoS ONE. 2008;3:E3573. doi: 10.1371/journal.pone.0003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder C, Hollander HA, Vonk JA, Rossberg AG, Jagers op Akkerhuis GAJM, Yeates GW. Soil resource supply influences faunal size-specific distributions in natural food webs. Naturwissenschaften. 2009;96:813–826. doi: 10.1007/s00114-009-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossberg AG, Matsuda H, Amemiya T, Itoh K. Food webs: Experts consuming families of experts. Journal of Theoretical Biology. 2006;241:552–563. doi: 10.1016/j.jtbi.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Sterner, Robert W., and James J. Elser. 2002. Ecological stoichiometry: The biology of elements from molecules to the biosphere. Princeton: Princeton University Press.
- Stouffer DB, Camacho J, Jiang W, Amaral LA. Evidence for the existence of a robust pattern of prey selection in food webs. Proceedings of the Royal Society of London. 2007;274 B:1931–1940. doi: 10.1098/rspb.2007.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]