Abstract
Numerous genetic variants have been successfully identified for complex traits, yet these genetic factors only account for a modest portion of the predicted variance due to genetic factors. This has led to increased interest in other approaches to account for the “missing” genetic contributions to phenotype, including joint gene-gene or gene-environment analysis.
A variety of methods for such analysis have been advocated. However, they have seldom been compared systematically. To facilitate such comparisons, the developers of the Multifactor Dimensionality Reduction (MDR) simulated 100 data replicates for each of 96 two-locus models displaying negligible marginal effects from either locus (16 variations on each of 6 basic genetic models). The genetic models, based on a dichotomous phenotype, had varying minor allele frequencies and from 2 to 8 distinct risk levels associated with genotype. The basic models were modified to include “noise” from combinations of missing data, genotyping error, genetic heterogeneity, and phenocopies. This study compares the performance of three methods designed to be sensitive to joint effects (MDR, Support Vector Machines (SVM), and the Restricted Partition Method (RPM)) on these simulated data.
In these tests, the RPM consistently outperformed the other two methods for each of the 6 classes of genetic models. In contrast, the comparison between other two methods had mixed results. The MDR outperformed the SVM when the true model had only a few, well-separated risk classes; while the SVM outperformed the MDR on more complicated models. Of these methods, only MDR has a well-developed user interface.
Keywords: epistasis, missing heritability, simulated data, Multifactor Dimensionality Reduction (MDR), Support Vector Machine (SVM), Restricted Partition Method (RPM)
INTRODUCTION
A key challenge for genetic analysis today is to account for the bulk of the phenotypic variance in complex traits attributable to genetic factors. Traditional univariate statistical genetic analysis methods have been highly successful: by early 2011, genome-wide association studies (GWAS) alone have identified over 4400 genetic variants contributing to disease (Hindorff et al. 2009). However, for many complex traits (e.g. obesity, smoking, diabetes), the variants identified by studies with large samples and dense genome-wide genotyping account for only a modest fraction of the phenotypic variance estimated to be attributable to genetic contributions [Goldstein 2009; Hirschhorn 2009; Kraft and Hunter 2009]. This has led to increased interest in other approaches to account for the “missing” genetic contributions to phenotype.
Several mechanisms have been suggested that could account for genetic effects that are not identified by current GWAS strategies. These include rare variants or other variants not surveyed by current GWAS chips, structural variants (e.g. copy number variants such as insertion/deletions or copy neutral variation such as inversions and translocations), genetic heterogeneity, parent-of-origin effects, and joint effects of multiple factors (such as gene-gene interactions, and gene-environment interactions) [Galvan, et al. 2010; Manolio, et al. 2009].
Though it is likely that all these mechanisms play some role in the “missing” genetic heritability for complex diseases, the possibility of joint effects presents a particularly appealing target for research. It is known that such mechanisms play an important part in biology, with well-documented examples in model organisms of epistasis having substantial impact on phenotypes ranging from gross morphology to longevity to efficiency of reproduction [Anholt, et al. 2003; Gerke, et al. 2009; Mackay 2010; Vieira, et al. 2000; Wolf, et al. 2005]. Further, many important traits of medical interest (such as heart disease, hypertension, diabetes, cancer, and infection) arise from biological systems controlled by interacting genetic factors [Churchill, et al. 2004; Lander and Schork 1994; Phillips 2008; Routman and Cheverud 1995; Schork 1997; Szathmary, et al. 2001]. The term “interaction” is used with multiple meanings in biological research [Wang, et al.]. In this manuscript, “interaction” will be used in the broad sense of “joint effects”, including, but not limited to, the statistical definition of interaction.
Numerous approaches have been suggested for examining joint effects. Several of these are particularly designed to be sensitive to joint effects even if one or more of the contributing factors displays little to no marginal effect in univariate analysis. Of these, one of the most popular is the Multifactor-dimensionality reduction (MDR) approach [Ritchie, et al. 2001]. Others include the Restricted Partition Method (RPM) [Culverhouse, et al. 2004], and a machine learning approach based on Support Vector Machines (SVM) [Chen, et al. 2008].
Although each of these methods has intellectual appeal and their own strengths and weaknesses, a comparison of results on identical data can be useful for researchers who are choosing a method for data analysis. The Ritchie group provided a convenient setting for such a comparison by simulating data for 96 two-locus disease risk models (100 data replicates each), publishing the results of the MDR on these data, and making the datasets publicly available [Ritchie, et al. 2003]. A subsequent publication [Ritchie, et al. 2007] revised the power reported for the MDR upward in the case of many of the models involving genetic heterogeneity to correct an overly stringent definition of “success” used in the 2003 publication. The revised definition calls the method a success in cases of genetic heterogeneity if the top result was either of the two causal pairs of SNPs included in the generating model.
In 2008, Chen et al. took up the challenge to test their Support Vector Machine (SVM) approach on these same data [Chen, et al. 2008]. They used the updated definition for power from the 2007 Ritchie et al. paper for their method, but compared their results to the MDR results published in the 2003 Ritchie et al. paper (success for models containing genetic heterogeneity only if the first of two causal pairs in the data was the top result of the analysis). As a consequence, instead of the split decision resulting from using the updated definition consistently, it appeared as if the SVM was superior to the MDR for each of the 6 broad model classes from which the 96 tested models were derived.
In this paper, the results of the three methods (MDR, SVM, RPM) on these simulated data are compared using the updated definition of success (i.e. the top pair of loci are causative) for all three methods.
METHODS
Data
The 96 data models are based 6 basic genetic interaction models, each modified in 16 ways to include all combinations 4 challenges for genetic analysis: genotyping error, missing data, phenocopies, and genetic heterogeneity. For each of these 96 combination models, 100 replicate datasets were generated and made available for researchers who wished to test alternative methods. (Data available upon request from the authors of [Ritchie, et al. 2003].)
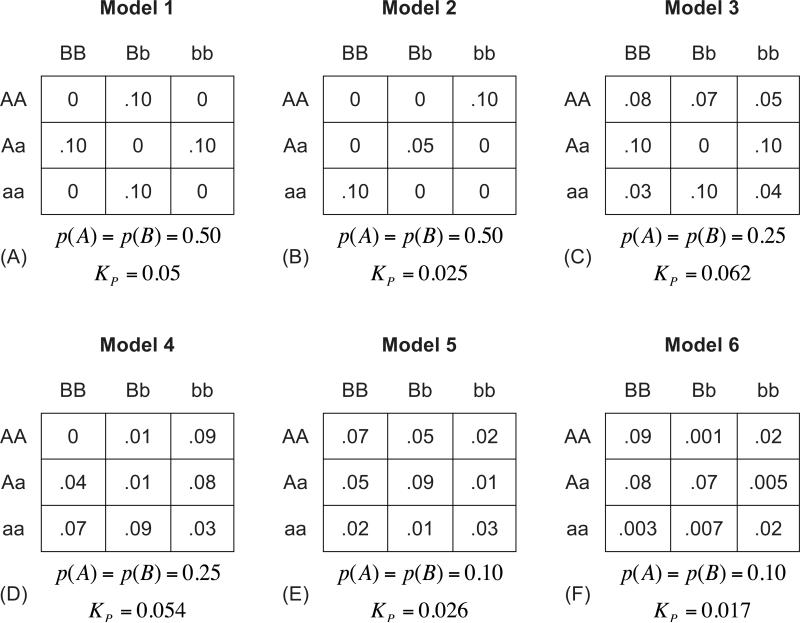
For each genetic model, the functional loci are single-nucleotide polymorphisms (SNPs). The six epistatic models (Figure 1) were chosen to represent a range of allele frequencies and patterns of risk-genotype associations. The polymorphisms associated with risk (the causative SNPs) in the first two models have minor allele frequency (MAF) = 50%; in Models 3 and 4, both SNPs have MAF = 25%; and in the final two models, the MAF for both of the causative SNPs is 10%. A second key difference between the models is the number of levels of risk. The first model displays only two levels of risk, the second model contains an additional intermediate risk level, while the other models have even more distinct risk levels. A third difference between the models is the level of risk for “low risk” individuals. In the first 3 models, genotypes carried by a substantial portion of the population (37.5%, 62.5%, and 25% respectively) carry no risk of disease. Thus, in a case/control setting, no cases would be sampled from these “low risk” joint genotypes. In contrast, Model 4 contains a single cell that is absolutely protective (representing only 0.4% of the population) while in Models 5 and 6 every joint genotype carries some risk.
Figure 1.
Multilocus penetrance functions and allele frequencies (p, q) used to simulate case-control data exhibiting gene-gene interactions in absence of main effects. For each model, the marginal penetrances all equal the population disease frequency, KP.
All of the models were selected to display interaction effects but little to no main effects when genotypes were generated according to Hardy-Weinberg proportions. The models were chosen to represent a wide range of two-locus models with this property (varying minor allele frequencies, varying population prevalence, and varying numbers of risk levels). In addition, some of them have been previously been discussed in the literature. For instance, the second model (Fig 1B) was initially described by [Frankel and Schork 1996]. In this model, high risk of disease is dependent on inheriting exactly two high-risk alleles (A and/or B) from two different loci. The high-risk genotype combinations in this model are AAbb, AaBb, and aaBB, with penetrances of 0.1, 0.05, and 0.1, respectively, corresponding to a population disease prevalence of 2.5%. This model has been shown to achieve the maximum heritability possible for a two locus purely epistatic model with the given disease prevalence (KP = 2.5%) and allele frequencies (p(A) = p(B) = 0.5) [Culverhouse, et al. 2002]. In fact, this model is part of a class of models that provide maximum genetic contribution to phenotypic variance whenever the prevalence of the phenotype is less than 25%. A discussion of each of the other models can be found in [Ritchie, et al. 2003].
Each dataset consists of 200 cases and 200 controls, each with genotypes for 10 unlinked SNPs, 2 of which are associated with the phenotype. Each of the non-associated SNPs had MAF equal to that of the associated SNPs in the generating model (i.e. MAF = 0.5 for Models 1 and 2, MAF=0.25 for Models 3 and 4, and MAF = 0.1 for Models 5 and 6). Genotypes were generated under Hardy-Weinberg equilibrium.
Sources of noise
For each of the six epistasis models, 16 submodels were produced based on the presence or absence of combinations of the following types of noise typical for genetic association data: genetic heterogeneity (GH), phenocopies (PH), genotyping error (GE), and missing genotypes (MS). For each submodel, 100 datasets with were simulated. This resulted in a total of 6 × 16 = 96 different testable models, each with 100 replicates available for the evaluation of power. Because of improvements in genotyping technology since the original publication of these simulated data, the genotyping error rates and missing data rates simulated by Ritchie et al. are higher than typical for current genetic studies.
Genetic heterogeneity was simulated having two different two-locus combinations associated with the risk of disease. In each case, both pairs of causative variants were included in the simulated genotype data and both had the same generating model. Half of the affected individuals were due to one pair of the causative loci, while the other half were associated with the other pair of causative loci.
Phenocopies were simulated such that 50% of the individuals labeled as affected were chosen at random, independent of the genetic model. These individuals were assumed to be affected due to random environmental factors.
Genotyping error was simulated using a directed-error model [Akey, et al. 2001]. This model simulates systematic genotyping errors that result in overrepresentation of one allele. For each locus, a bias towards the a or the A allele was prescribed. Five percent of the genotypes were selected and, unless it was already homozygous in the biased direction, the genotype was changed so that it had one more of the overrepresented alleles. Although this rate was common at the time the data was generated, it is considerably higher than is common today.
Missing data was simulated by randomly selecting 5% of the individuals in a dataset. These individuals were deemed to have failed genotyping and were excluded from the analysis.
Analytic Methods
Restricted Partition Method (RPM)
The RPM is an exploratory tool to investigate, in a model agnostic manner, joint effects of genetic and environmental factors contributing to quantitative or dichotomous phenotypes. The method partitions multilocus genotypes (or genotype-environmental exposure classes) into statistically distinct “risk” groups, then evaluates the resulting model for phenotypic variance explained. It is sensitive to factors whose effects are apparent only in a joint analysis, and which would therefore be missed by many other methods.
The RPM algorithm is an iterative search procedure for finding an optimized partition of the genotypes. Genotypes are sequentially merged based on the similarity of the mean values of their phenotypic trait. Selection of which genotypes to merge at each step is based on statistical criteria from a multiple comparisons test. Initially, each multi-locus genotype forms its own group. The algorithm proceeds as follows:
A multiple comparisons test is performed to identify which (if any) genotype groups have different mean quantitative trait values. The procedure halts if all groups have different means.
Pairs of genotype groups with means that are not significantly different from each other are ranked according to the difference in means between the two groups.
The pair from step 2 with the smallest difference (i.e., most similar mean values) is merged to form a new group.
The algorithm returns to step 1.
To provide a measure of the importance of the final results, the variance attributable to the joint genotypes in the final model [i.e., R2 = (between group variance)/(total phenotypic variance)] is computed. A natural consequence of this definition is that, if the genotypes are merged into a single group at the end of the algorithm, then R2 = 0, reflecting the lack of evidence for quantitative trait differences between the genotypes. This does not indicate that the mean values of the genotypes are identical, only that there is not sufficient evidence to reject the null.
The Games–Howell variant of Tukey's Honestly Significant Difference (HSD) multiple comparison method with α = 0.05 is the default and was used for these analyses. (The Games–Howell version allows for the variance and sample sizes to vary between groups [Games and Howell 1976].)
Because the R2 estimated by this procedure has a distribution that is difficult to parameterize, the RPM software uses a permutation-based strategy to estimate p-values. Phenotype values are permuted among the individuals. For each permutation of the data, the RPM model R2 is computed, cumulatively producing an empirical null distribution for the model in question. The test statistic from the unpermuted data is compared to this empirical null to approximate the p-value for the model.
Although the RPM algorithm was designed for quantitative phenotypes, subsequent empirical evaluation demonstrated its utility for dichotomous traits [Culverhouse 2007]. The violation of distributional assumptions for the merging rule affects the statistical interpretation of the final groups, but does not affect the validity of the permutation-based p-values of the final model.
Multifactor Dimensionality Reduction (MDR)
The MDR method is designed to analyze the association of dichotomous traits and combinations of discrete predictors (genetic or environmental exposures). The MDR method can analyze an arbitrary number of simultaneous predictors. For clarity, this description will focus on how the method works for pairwise analyses of SNPs. The approach is easily generalized to more predictors that need not be restricted to SNPs, but can include any predictor that can be discretized into distinct strata.
A key part of the MDR is cross-validation. The process begins by dividing the data into equal sized subsets (e.g. 10). One subset is set aside as testing data and the rest of the data is combined to be a training data set.
The first pair of SNPs is selected and the corresponding joint genotypes are represented in a table, each cell representing one of the 9 genotypes aabb, aabB, aaBB, aAbb, aAbB, aABB, AAbb, AAbB, AABB. Each of these 9 cells are labeled as either high-risk or low risk: high-risk if the ratio of affected individuals to unaffected individuals exceeds some threshold T, and low-risk otherwise. The threshold would typically be equal to the ratio of cases to controls in the data. Since cells labeled “low risk” will typically contain some cases, and cells labeled “high risk” will contain some controls, model will have an associated misclassification rate. After going through every possible pair of SNPs, the pair with the lowest misclassification rate in the training set will be chosen. The prediction error based on how well this model classifies the testing data is then recorded.
Next, a new subset of the data is chosen to be the testing set, and the rest of the data (including the old testing set) becomes the new training set. The process is repeated for every pair of predictors, and again a best model from the training data is selected and its accuracy in the testing data is recorded.
After each of the subsets has been used as the test data, one will have a collection of best pairs and their associated testing errors. In an ideal situation, the same pair would be chosen every time. In general, the pair chosen most frequently (i.e. the pair with the greatest cross-validation consistency) is selected as the top pair. If there is a tie, it can be broken by choosing the pair with the lowest average misclassification rate in the test data.
For a more complete description of the MDR, see [Ritchie, et al. 2001].
Support Vector Machine (SVM)
The SVM approach also focuses on dichotomous phenotypes, with the aim of finding a hyperplane, H, in the space of genotypes that maximizes prediction accuracy (i.e. does the “best” job of separating the cases from the controls). Under the ideal conditions (i.e. there is a hyperplane that perfectly separates the two classes), the hyperplane is defined by H : wTx + b = 0 such that for any control, i, its corresponding genotype vector xi satisfies the equation wTxi + b ≤ −1, and for any case, j, its corresponding genotype vector x j satisfies the equation wTx j + b ≥ 1 and the minimum distance of any of the data points to the plane, , is as large as possible. Vectors representing the genotypes of the cases and controls that lie on the boundaries (i.e. wTxi + b ≤ − 1 and wTx j + b ≥ 1, respectively) are called support vectors.
In general, it will not be possible to find a hyperplane that separates the cases from the controls perfectly. In this case, the optimization function will include a penalty for misclassified points proportional to their distance from the boundary. The resulting set of support vectors includes the vectors for misclassified subjects as well as the boundary subjects.
If, as would typically be the case for epistatic interactions, the decision boundary is inherently non-linear, the SVM approach can be modified by use of a non-linear transformation to project the data non-linearly into a higher dimensional space, where they are more likely to be linearly separable [Cover 1965]. Technical details of the suggested approach, including the selection of additional parameters, are provided in [Chen, et al. 2008].
RESULTS
The statistic used for comparison in the previous analyses of these data ([Chen, et al. 2008; Ritchie, et al. 2007; Ritchie, et al. 2003]) is a simple count of how often the top ranked two-locus SNP-pair from the analysis was a causative pair. To make the comparisons as straightforward as possible, the same statistic will be used in this study. Table I lists how often MDR, SVM, and RPM found a causative pair of SNPs (a true signal) as the top signal in these data. The numbers listed for the MDR and the SVM were results from the developers of the methods performing the analyses ([Chen, et al. 2008; Ritchie, et al. 2007]).
Table I. Comparison of MDR, SVM, and RPM two-locus analyses in the Ritchie test data.
Cells list the number of times the top result was a causative pair from 45 sampled pairs of SNPs If Genetic Heterogeneity (GH) is not present, there is only one causative pair. When GH is present, there are two. Results are based on 100 data replicates for each model-noise combination
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | Model 6 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of noise | MDR | SVM | RPM | MDR | SVM | RPM | MDR | SVM | RPM | MDR | SVM | RPM | MDR | SVM | RPM | MDR | SVM | RPM |
| None | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | 99 | 99 | 100 | 82 | 90 | 97 | 84 | 92 | 99 |
| GH | 70 | 42 | 100 | 34 | 53 | 100 | 42 | 35 | 75 | 41 | 46 | 76 | 20 | 29 | 64 | 19 | 32 | 78 |
| PC | 90 | 90 | 98 | 99 | 98 | 100 | 45 | 41 | 52 | 32 | 42 | 41 | 30 | 44 | 46 | 32 | 46 | 53 |
| GE | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 97 | 100 | 100 | 80 | 91 | 97 | 92 | 93 | 100 |
| MS | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 99 | 100 | 97 | 100 | 100 | 82 | 90 | 93 | 87 | 96 | 97 |
| GH+PC | 24 | 21 | 43 | 35 | 30 | 72 | 9 | 9 | 21 | 8 | 7 | 21 | 7 | 10 | 12 | 5 | 9 | 19 |
| GH+GE | 69 | 54 | 100 | 34 | 55 | 100 | 42 | 36 | 74 | 41 | 42 | 76 | 20 | 31 | 64 | 19 | 37 | 72 |
| GH+MS | 65 | 47 | 100 | 40 | 55 | 100 | 42 | 35 | 72 | 31 | 37 | 73 | 18 | 28 | 57 | 22 | 34 | 76 |
| PC+GE | 94 | 87 | 98 | 99 | 99 | 100 | 41 | 47 | 51 | 48 | 43 | 49 | 28 | 48 | 61 | 33 | 51 | 67 |
| PC+MS | 96 | 91 | 96 | 99 | 99 | 100 | 42 | 54 | 61 | 43 | 49 | 54 | 14 | 27 | 35 | 16 | 27 | 46 |
| GE+MS | 100 | 100 | 100 | 100 | 100 | 100 | 98 | 100 | 100 | 98 | 99 | 99 | 74 | 88 | 96 | 84 | 92 | 99 |
| GH+PC+GE | 27 | 18 | 52 | 35 | 41 | 69 | 10 | 13 | 19 | 8 | 6 | 14 | 7 | 10 | 19 | 6 | 10 | 19 |
| GH+PC+MS | 23 | 18 | 52 | 38 | 36 | 77 | 9 | 9 | 19 | 10 | 10 | 18 | 4 | 10 | 22 | 6 | 11 | 25 |
| GH+GE+MS | 64 | 49 | 100 | 44 | 58 | 100 | 42 | 36 | 72 | 41 | 44 | 73 | 16 | 24 | 65 | 11 | 25 | 73 |
| PC+GE+MS | 94 | 86 | 97 | 100 | 100 | 100 | 48 | 40 | 59 | 42 | 50 | 43 | 18 | 31 | 36 | 16 | 32 | 43 |
| GH+PC+GE+MS | 31 | 18 | 48 | 36 | 29 | 72 | 9 | 6 | 20 | 7 | 8 | 20 | 4 | 6 | 16 | 3 | 10 | 16 |
GH = genetic heterogeneity (50% of cases are associated with each of 2 pairs of SNPs in the data)
PC = phenocopies (50% of cases are not associated with the measured genetic variants: perhaps due to environmental exposures)
GE = genotyping error (5% of genotypes selected at random. If not homozygous for the biased allele, the allele count was increased by one)
MS = missing data (5% of the samples were assumed to be of poor quality (failed genotyping) and were deleted
The highest estimated power for each model/error combination is in boldface. When the power of one method surpassed the other two, it is underlined.
The overall results can be summarized as follows: for 78/96 (81%) of the model-noise combinations examined the RPM model outperformed both of the other methods. For 16/96 (17%) of the model-noise combinations, the RPM tied for highest power with at least one of the other two methods. For the final 2/96 (2%) of the model-noise combinations, the RPM performed better than the MDR, but worse than the SVM. For none of the model-noise combinations did the RPM perform more poorly than both of the other methods.
In addition to this global summary of results, it can be informative to examine individual results in more detail. One can observe that although differences in power to identify the causative SNP pair were sometimes dramatic, there were numerous instances, even when there are no ties, where the differences were modest. The combination of genetic Model 6 (which had 8 different risk levels) together with noise from genetic heterogeneity and missing data is a model that displayed a dramatic difference: the RPM identified the a causative pair of interacting SNPs 76% of the time, compared to 22% for MDR and 34% for SVM. In contrast, all three methods performed well for the combination of genetic Model 2 (displaying 3 risk levels) and noise from phenocopies: the RPM correctly identified the causative pair in all 100 of the datasets, the MDR identified the correct pair in 99% of the replicates, and the SVM in 97%.
Even though some of the more modest differences between the methods could simply be the result of sampling variability, the differences between the methods are clearly not random, even within each of the six genetic models. Under the null hypothesis that each method is equally good (so when the RPM and another method do not tie for the best, which one “wins” is random), the RPM performed significantly better than the other two methods for each of the 6 model classes. For models 5 and 6 (with at least 6 different risk levels, all > 0, and MAF=0.1), there are no ties and the RPM outperforms both of the other methods in each of the 16 model-noise combinations (p = (1/3)16 = 2.3 × 10−8 that this could be due to chance). These two models also contain many examples where there were dramatic differences in power.
For model 3 (7 risk levels, 14% of the population has 0 genetic risk, and MAF=0.25), the RPM out performs the other two methods for 13 of the model-noise combinations and ties the best other method for the other 3 combinations (p = (1/3)13 = 6.3 × 10−7). For models 1 and 2 (no more than 3 risk levels, at least half of the population with 0 genetic risk, and MAF=0.5), the RPM outperforms the other methods in each of the 11 settings where it does not tie for the best (p = 5.6 × 10−6). Finally, for model 4 (7 risk levels, <1% of the population having no genetic risk, MAF=0.25), of the 13 settings where the RPM does not tie for the best power, the point estimate for the power of the RPM is greater than those for the other two methods in 11, and is less than only one of the other methods in the other 2 settings (p = 1.1 × 10−4).
If the RPM is removed from consideration and only the MDR and SVM are compared, the situation is more complicated. Model 1 fits the basic framework of the MDR ideally, with only two, well-separated levels of risk. For this model, the MDR is clearly superior to the SVM and never ranks lower than the SVM for any of the error/noise combinations (p = 1.1 × 10−4). In contrast, for Models 2, 3, and 4 (where number of risk levels increase, the MAF decreases to 0.25, and the proportion of the population with no genetic risk decreases), the two methods perform similarly, but begin to trend to the SVM as the models become more complicated (p = 0.50, p = 0.27, p=0.11, respectively). Finally, for models 5 and 6, (at least 6 risk levels, no genotype is risk-free, MAF=0.1) there are no ties between MDR and SVM, with SVM demonstrating more power in each setting (p = 1.5 × 10−5). Thus, which of the MDR or SVM has superior power is highly model dependent.
DISCUSSION
Each of the three methods (RPM, MDR, SVM) has strengths and weaknesses as analysis methods for detecting joint or interaction effects of multiple loci. The MDR is the easiest to use given its polished user interface. It is also the most commonly used of the three approaches and, as a consequence, is something of a standard for comparison. Given this, it seemed appropriate to test the methods using data generated by the developers of the MDR. Although these data represent only a limited range of dichotomous phenotype models for the comparison of these methods, the results provide insight into the strengths and weaknesses of the three approaches, some of which could have been predicted from the basic features of the methods.
For these data sets and the designated metric (the number of times the top pair from the analysis was a “causal” pair), the RPM was the most successful of the three, demonstrating significantly superior power for each of the 6 classes of models tested. In a secondary comparison between the MDR and SVM, each demonstrated superiority over the other in certain settings. Though the deviations in performance were sometimes small, and may in part be due to sampling variation, the distributions of these deviations revealed highly significant differences between the methods.
An examination of the analytic underpinnings of these methods can help us understand the differences in performance seen for these data. First, the MDR and RPM have more in common with each other than either has with the SVM approach. Both the MDR and RPM are based on evaluating a single partition of the multi-locus genotypes for association to phenotype. The most important differences being (i) the RPM was designed to deal with quantitative phenotypes and is also appropriate for dichotomous traits [Culverhouse 2007]) while the basic MDR is inherently a tool for dichotomous traits; (ii) the RPM separates the multi-factor cells into statistically distinct risk strata (the number determined by the data), while the MDR, dichotomizes the cells into low and high risk; (iii) the MDR utilizes a cross-validation procedure as part of its evaluation of the models; (iv) the MDR focuses on finding the single best 2-SNP (or n-SNP) model, while the RPM assumes that there may be more than one multi-SNP combination contributing to phenotypic variability; and (v) (as a consequence of (iv)) the permutation tests to evaluate statistical significance are different. For example, for a two-locus analysis, permutations are performed in the RPM only for pairs of SNPs demonstrating statistically distinct risk strata, and a multiple test correction must be used. In contrast, the MDR extreme value approach requires every pair of SNPs in the data to be evaluated for each permutation, but the result does not require further correction for multiple tests.
The key similarity between the MDR and the SVM that separates both from the RPM is the fact that both MDR and SVM dichotomize the multi-locus genotypes (the MDR based on a fixed threshold, typically the sample disease prevalence; the SVM adaptively chosen to fit the data), while the RPM allows for more (or fewer) distinct risk levels. These basic differences may provide an essential part of the explanation for which situations are optimal for the use of each methods. First, when comparing the MDR to the SVM, one sees that Model 1 fits the basic framework of the MDR ideally, with only two, well-separated levels of risk. For this model, the MDR is clearly superior to the SVM and never ranks lower than the SVM, no matter which of the error factors have been added. Model 2 also suits the MDR approach well, with genotypes containing 62.5% of the population having no chance of containing a case (i.e. again, the high and low risk cells are well separated). In contrast, for models 5 and 6, each displaying several distinct risk levels, the SVM is clearly more powerful than the MDR (even though they both dichotomize). One possible explanation for this is the greater flexibility provided to the SVM through the use of a data-derived threshold for dichotomization, rather than the a priori threshold used by the MDR. Another possible contributing factor is the use of cross-validation by the MDR. These particular models, with gradations in risk, contain larger fractions of the populations with genotype-associated risk near the population prevalence. In data sampled from these models, particularly those with noise, specific genotypes may be less consistently classified as high or as low risk in the cross-validation portion of the MDR algorithm.
The strategy of the RPM to allow its model to contain intermediate risk groups may be the reason it outperforms the two dichotomizing methods for these models. In addition, because the RPM explicitly makes use of the degree of separation between risk groups when defining its models, it is also strengthened when there are well-separated risk groups. This may help explain why the RPM maintains higher power in genetic models 1 and 2 for many of the noise combinations that considerably weaken the other two approaches.
There are substantial computational benefits to choosing an a priori threshold to dichotomize the cells into low and high risk (as the MDR does): the method greatly simplifies the evaluation space (all the 21,146 ways to partition the 9 cells of a two-SNP model) to a single partition. The RPM may take up to 8 iterations of the algorithm to choose the partition. This simplification can provide a substantial computational saving when multiplied by all pairs of SNPs. The computational difference becomes even more significant when models involving more than two factors are examined.
However, as we have seen, the simplification has associated costs: First, by making distinctions between cells that are have essentially the same risk (e.g. a cell with 50.0% affected individuals would be called “low risk” while a cell with 50.1% affected would be called “high risk”), the algorithm (particularly the cross-validation) may be unstable and produce sub-optimal results. Second, by collapsing multiple distinct risk levels to only 2, some of the information about association to the trait may be lost.
The SVM also dichotomizes the data but uses a transformation of a hyperplane determined by the data to choose the best “cut point”. Whether it is due to the flexibility of the choice of cut point, or simply the lack of the cross-validation feature, the SVM performed better than the MDR when the data contained genotypes with associated risks near the overall population risk.
Another key contrast between the RPM and MDR is that the RPM takes an open attitude to multiple signals while the MDR is clearly focused on identifying the single best set of predictors is the data for any level of analysis (e.g. two-locus, three-locus, etc.). The approach of the RPM requires much more post-analysis interpretation from the user. This difference in focus has a secondary impact on the permutation approaches used by the MDR and RPM for statistical evaluation. The MDR, focused on the single top signal, uses an extreme value comparison to evaluate significance. After identifying the top signal in the original data, it permutes the data multiple times, each time running the full MDR and selecting the single top result from each permutation. By comparing the original top signal to the distribution of top signals from the permutations, the multiple comparisons have already been taken into account. This is another way that the interpretation of MDR results is straightforward. In contrast, the RPM does not assume that all the SNP pairs have the same null distribution and so generates individual null distributions for each SNP pair of interest. As a result, a post-analysis correction for multiple tests must be applied. However, this approach can result in considerable computational savings as permutations need only be performed on the top signals rather than on every pair in the data. The SVM, as a well-known approach derived from standard machine learning algorithms, is primarily a source of hypothesis generation and has no specific approach to statistical significance. However, in general, the SVM approach requires a complete optimization of the parameters that would be computationally much more expensive than the MDR fixed threshold. To ameliorate this problem, the authors suggest several ways in which the parameter space could be sampled in a computationally efficient manner to obtain approximately optimal penalization parameters. The requirement for data specific parameter optimization, combined with the lack of software provided by the authors, will make this approach somewhat daunting for many researchers.
Other points of comparison between the methods include their applicability to quantitative as well as dichotomous phenotypes, incorporation of covariate information, and utilization of family data. The RPM is appropriate for either quantitative or dichotomous phenotypes without modification. Covariates can be addressed by analyzing the residual after a regression including the covariates for a quantitative phenotype or converted to a categorical (or ordinal) variable to be included directly in the analysis for either quantitative or dichotomous phenotypes. Similarly, though family structure cannot be directly modeled in an RPM analysis, at least two plausible options are available. For a quantitative trait, the residuals from a mixed model taking pedigree relationships into account could be analyzed (as described in [Aulchenko, et al. 2007]). In addition, it had been demonstrated that the crude approach of increasing sample size by naively including related individuals from modestly sized families can increase power for either quantitative or dichotomous traits without substantially increasing false positive rates [Culverhouse, et al. 2009]. The SVM is inherently limited to the analysis of dichotomous phenotypes and has not been extended to incorporate covariates or related individuals. Although the basic MDR is limited to dichotomous phenotypes and categorical covariates for unrelated individuals, several variants of the method have been developed. Not all of these focus on identifying the single “best model”. Among these are the MDR-PDT [Martin, et al. 2006], which is appropriate for nuclear family data, and the pedigree-based GMDR [Lou, et al. 2008], which is appropriate for both quantitative and dichotomous traits, can adjust for covariates, and can use data from arbitrary pedigree structures in a statistically appropriate way.
This study has numerous limitations. The most obvious is that the six genetic models cannot possibly represent the full space of possible two-locus models, much less the space of higher order interactions that would be required to fully model true biology. In fact, they only represent 3 different sets of minor allele frequencies. Similarly, both the sample size and the specific noise models included represent only isolated points in a high dimensional space. The relatively small sample size for an examination for interactions (200 cases, 200 controls) may have been particularly problematic for the cross-validation methodology used by the MDR, which exacerbates the problem of sparse cells in these data. Other limitations include the fact that test data included only a handful of markers instead of the thousands or millions and that the only estimate of power or the false positive rate is how often the “top” signal was causative. In spite of these limitations and more, these data do provide a range of both genetic models and error that can be used as a common testing ground for multiple analytic methods and which highlight several differences between the three methods examined here.
In summary, for these data, the RPM was consistently the most powerful for each of the 6 basic genetic models, while the MDR and SVM each surpassed the other for some models. The genetic models in these analyses each consisted of two interacting dichotomous loci, with MAF ranging from 0.1 to 0.5, and with between 2 and 8 distinct risk levels associated with the 9 two-locus genotypes in each model. The data included in these comparisons consisted of the 6 genetic models, each modified by the 16 combinations of the presence or absence of 4 types of data noise: genetic heterogeneity, phenocopies, missing data, and genotyping error. Key factors that appear to distinguish the performance of the MDR from that of the SVM are that MDR performed at its best when there were few, well-separated genetic risk levels, while the SVM outperformed the MDR for more complicated two-locus genetic models. The MDR software is available at http://www.multifactordimensionalityreduction.org. The SVM results reported by [Chen, et al. 2008] used software from a library for SVM (LIBSVM) developed by Chang and Lin [2005]. This software is available at http://www.csie.ntu.edu.tw/~cjlin/libsvm. Software implementing the RPM is available from the author.
ACKNOWLEDGEMENTS
This work was supported by NIDA grants R03 DA023166 and R21 DA033827 and NCI grant P01 CA089392. The author would like to thank Dr. Brian Suarez and Dr. Laura Bierut for their careful reading and thoughtful comments on the manuscript.
REFERENCES
- Akey JM, Zhang K, Xiong M, Doris P, Jin L. The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures. Am J Hum Genet. 2001;68(6):1447–56. doi: 10.1086/320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RR, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, Rollmann SM, Kamdar KP, Mackay TF. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35(2):180–4. doi: 10.1038/ng1240. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177(1):577–85. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: A library for support vector machines. 2005 Software is available at http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- Chen SH, Sun J, Dimitrov L, Turner AR, Adams TS, Meyers DA, Chang BL, Zheng SL, Gronberg H, Xu J, Hsu FC. A support vector machine approach for detecting gene-gene interaction. Genet Epidemiol. 2008;32(2):152–67. doi: 10.1002/gepi.20272. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O'Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–7. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Cover TC. Geometrical and statistical properties of systems of linear inequalities with applications in pattern recognition. IEEE Transactions on Electronic Computers. 1965;EC-14:326–334. [Google Scholar]
- Culverhouse R. The use of the restricted partition method with case-control data. Hum Hered. 2007;63(2):93–100. doi: 10.1159/000099181. [DOI] [PubMed] [Google Scholar]
- Culverhouse R, Jin W, Jin CH, Hinrichs A, Suarez BK. Power and false positive rates for the Restricted Partition Method (RPM) in a large candidate gene dataset. BMC Proceedings. 2009;3(Suppl 7):S74. doi: 10.1186/1753-6561-3-s7-s74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse R, Klein T, Shannon W. Detecting epistatic interactions contributing to quantitative traits. Genet Epidemiol. 2004;27(2):141–52. doi: 10.1002/gepi.20006. [DOI] [PubMed] [Google Scholar]
- Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002;70(2):461–71. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel WN, Schork NJ. Who's afraid of epistasis? Nat Genet. 1996;14(4):371–3. doi: 10.1038/ng1296-371. [DOI] [PubMed] [Google Scholar]
- Galvan A, Ioannidis JP, Dragani TA. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet. 2010;26(3):132–41. doi: 10.1016/j.tig.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Games PA, Howell JF. Pairwise multiple comparison prodedures with unequal N's and/or variances: A monte carlo study. Journal of Educational Statistics. 1976;1:113–125. [Google Scholar]
- Gerke J, Lorenz K, Cohen B. Genetic interactions between transcription factors cause natural variation in yeast. Science. 2009;323(5913):498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–8. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360(17):1701–3. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Mangold JE, Zhu J, Elston RC, Li MD. A combinatorial approach to detecting gene-gene and gene-environment interactions in family studies. Am J Hum Genet. 2008;83(4):457–67. doi: 10.1016/j.ajhg.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. Mutations and quantitative genetic variation: lessons from Drosophila. Philos Trans R Soc Lond B Biol Sci. 2010;365(1544):1229–39. doi: 10.1098/rstb.2009.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet Epidemiol. 2006;30(2):111–23. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9(11):855–67. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Edwards TL, Fanelli TJ, Motsinger AA. Genetic heterogeneity is not as threatening as you might think. Genet Epidemiol. 2007;31(7):797–800. doi: 10.1002/gepi.20256. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24(2):150–7. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routman EJ, Cheverud JM. Gene effects on a quantitative trait: two-locus epistatic effects measured at microsatellite markers and at estimated. QTL Evolution. 1995;51:1654–1662. doi: 10.1111/j.1558-5646.1997.tb01488.x. [DOI] [PubMed] [Google Scholar]
- Schork NJ. Genetics of complex disease: approaches, problems, and solutions. Am J Respir Crit Care Med. 1997;156(4 Pt 2):S103–9. doi: 10.1164/ajrccm.156.4.12-tac-5. [DOI] [PubMed] [Google Scholar]
- Szathmary E, Jordan F, Pal C. Molecular biology and evolution. Can genes explain biological complexity? Science. 2001;292(5520):1315–6. doi: 10.1126/science.1060852. [DOI] [PubMed] [Google Scholar]
- Vieira C, Pasyukova EG, Zeng ZB, Hackett JB, Lyman RF, Mackay TF. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics. 2000;154(1):213–27. doi: 10.1093/genetics/154.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elston RC, Zhu X. The meaning of interaction. Hum Hered. 2010;70(4):269–77. doi: 10.1159/000321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Leamy LJ, Routman EJ, Cheverud JM. Epistatic pleiotropy and the genetic architecture of covariation within early and late-developing skull trait complexes in mice. Genetics. 2005;171(2):683–94. doi: 10.1534/genetics.104.038885. [DOI] [PMC free article] [PubMed] [Google Scholar]