Abstract
Structural genomic variations represent a major driving force of evolution and a burst of large segmental gene duplications occurred in the human lineage during its separation from non-human primates. SRGAP2, a gene recently implicated in neocortical development, has undergone two human-specific duplications. Here we find that both duplications (SRGAP2B and SRGAP2C) are partial and encode a truncated F-BAR domain. SRGAP2C is expressed in the developing and adult human brain and dimerizes with ancestral SRGAP2 to inhibit its function. In the mouse neocortex, SRGAP2 promotes spine maturation and limits spine density. Expression of SRGAP2C phenocopies SRGAP2 deficiency. It underlies sustained radial migration and leads to the emergence of human-specific features, including neoteny during spine maturation and increased density of longer spines. These results suggest that inhibition of SRGAP2 function by its human-specific paralogs has contributed to the evolution of the human neocortex and plays an important role during human brain development.
Keywords: evolution, cortex, dendritic spine, F-BAR, genomic, gene duplication, human, human-specific paralogs, synapse
INTRODUCTION
In recent years, many potential genetic mechanisms have been proposed to participate in human brain speciation. These include changes in transcriptional regulation (Enard et al., 2002; Konopka et al., 2009), accelerated evolution of small non-coding RNAs (Pollard et al., 2006), changes in the activity and/or region-specificity of enhancer elements (McLean et al., 2011; Prabhakar et al., 2006; Prabhakar et al., 2008), or changes in patterns of alternative splicing (Calarco et al., 2007). So far, very few studies have assessed the functional consequences of these changes. Another important mechanism is genomic duplication, which generates copies of genetic material that serve as substrates for molecular evolution (Hurles, 2004; Ohno, 1970). In fact, recent lines of evidence have revealed that a burst of large segmental gene duplications occurred in the human lineage during its separation from non-human primates approximately 6 millions years ago (Bailey et al., 2002; Fortna et al., 2004). Several of the genes contained in these duplications are expressed in the developing brain. This has led to the hypothesis that these evolutionarily recent gene duplications might have participated in the emergence of human-specific features of brain development and function (Bailey and Eichler, 2006; Sikela, 2006; Stankiewicz and Lupski, 2010; Varki et al., 2008). However, this hypothesis has never been tested experimentally. The principal obstacles have been that the function of the ancestral genes is often unknown and the duplications are poorly assembled in the current human genome because of their high degree of conservation with the ancestral gene.
In this study, we focused on SLIT-ROBO Rho-GTPase activating protein 2 (SRGAP2), a gene highly expressed during brain development (Bacon et al., 2009; Guerrier et al., 2009). The protein SRGAP2 is composed of three functional domains (Guerrier et al., 2009): a N-terminal F-BAR (Bin, Amphiphysin, Rvs) domain (Itoh et al., 2005) involved in membrane deformation, a central Rho-GAP domain which specifically stimulates the GTPase activity of Rac1 and a C-terminal tail containing a SH3 domain. We recently demonstrated that SRGAP2 controls cortical neuron migration (Guerrier et al., 2009). Recent analysis of large segmental duplications has revealed that SRGAP2 has two main duplicates in the Homo neanderthalensis and Homo sapiens genomes but not in the genome of our closest living relatives, the Chimpanzee, Orangutan and Gorilla (Sudmant et al., 2010). In this study, we mapped the location and genomic structure of the human-specific SRGAP2 paralogs (see also accompanying article by Dennis et al.). We provide evidence that they are expressed in human neurons and encode a truncated F-BAR domain that interacts with ancestral SRGAP2 to inhibit its function. We used in vitro and in vivo approaches to determine the function of SRGAP2 and its human paralogs in the neocortex, the region of the brain, the evolution of which is thought to underlie the emergence of human cognitive abilities. Our results uncover a new function for ancestral SRGAP2 in promoting dendritic spine maturation and indicate that expression of a human-specific paralog of SRGAP2 in mouse pyramidal neurons extends the phase of spine development and leads to an increased density of longer spines in vivo, a feature characterizing pyramidal neurons in the human neocortex (Benavides-Piccione et al., 2002; Elston et al., 2001).
RESULTS
Genomic organization of SRGAP2 paralogs
The gene SRGAP2 was identified using array comparative genomic hybridization (aCGH) to have undergone human-specific duplication resulting in extra copies in the H. sapiens genome, but not in the genomes of four non-human primates (Fortna et al., 2004). However, the number and locations of these human-specific copies were unknown. Using the BLAT search tool, we searched GRCh36/hg18 reference genome (available human genome assembly in 2006 at the start of the project) with the cDNA of human SRGAP2 (GenBank: BC132874.1). We located the ancestral SRGAP2 gene at chromosome 1q32.1 (SRGAP2A) and two duplicates at 1q21.1 and 1p12 (hereon referred to as SRGAP2B and SRGAP2C, respectively) that showed more than 99% similarity to the query sequence (Figure S1A,B). The observation that there are two duplicates of SRGAP2 in the human genome was recently confirmed and validated by analysis of copy number variation in whole-genome shot-gun sequencing data (Sudmant et al., 2010). Even in the most recent reference assembly (GRCh37/hg19), the ancestral SRGAP2A gene is classified as misassembled and SRGAP2B and SRGAP2C lie in recently duplicated regions of the genome that are incompletely assembled and present large contig gaps. In order to determine the approximate genomic organization of these three paralogs, we retrieved human Bacterial Artificial Chromosome (BAC) clones mapping to these three loci. To fill in the gaps in the assembled sequence, we performed BLAST searches against the end sequences database using genome sequences flanking the gaps as inputs. The exon organization of the human BACs was determined by dot blot hybridization with several probes covering different portions of the entire extent of SRGAP2 cDNA (data not shown). In addition, exons in each BAC clone were sequenced from the human BAC DNA either directly or following amplification to reveal the organization of the three SRGAP2 paralogs (data not shown, Figure S1C). The full sequencing and assembly of the three genomic loci and their recent evolution is the focus of the accompanying paper by Dennis et al.. Both analyses revealed that SRGAP2A contains 22 exons and encodes a protein with 1071 amino acids (aa), which is 98% identical to its mouse ortholog SRGAP2, whereas SRGAP2B and SRGAP2C have a 3’-breakpoint located in intron 9 of the ancestral copy and are predicted to express a protein containing 459 aa. The first 452 aa correspond to the first 452 aa of the F-BAR domain of SRGAP2A (aa1–501, 8). Because intron 9 of SRGAP2A becomes 3’ untranslated region (3’ UTR) in SRGAP2B and SRGAP2C, both use an alternate stop codon located in intron 9, which adds 7 aa (VRECYGF; Figure 1A). We identified two potential poly-adenylation sites in the 3’-UTRs of SRGAP2B and SRGAP2C based on the clustering of Expressed Sequence Tags (EST) from the dbEST database. The two 3’-UTRs are approximately 1.4 kb or 5 kb long. Taken together, our results indicate that both paralogs encode a truncated F-BAR domain that lacks the last C-terminal 49 aa.
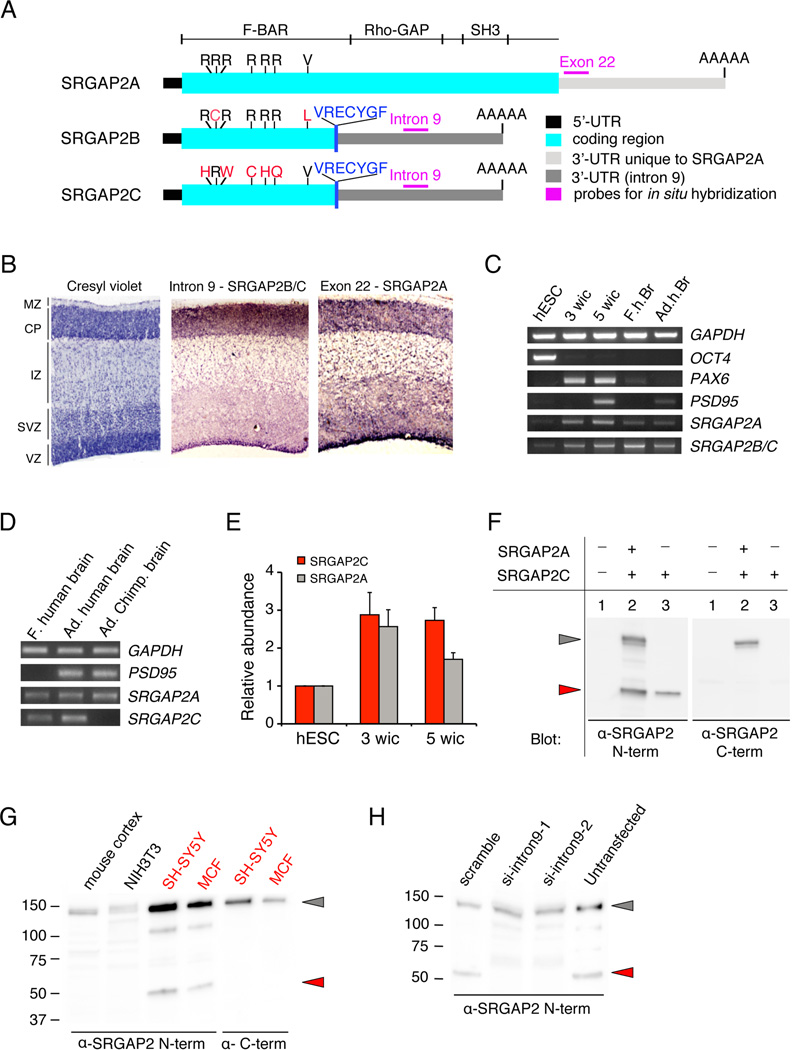
Figure 1. SRGAP2 and its human-specific paralogs are expressed in human neurons.
(A) Schematic representation of transcripts of the three paralogs of SRGAP2. SRGAP2B and SRGAP2C encode a truncated F-BAR domain with non-synonymous mutations marked in red. VRECYGF denotes C-terminal amino-acid residues translated from intron 9 unique to SRGAP2B and SRGAP2C.
(B) RNA in situ hybridization on developing human cortex at Gestational Week (GW) 11 using probes specific for SRGAP2B and SRGAP2C (intron 9) and SRGAP2A (exon 22). Cresyl violet marks the cortical layers: VZ (ventricular zone), ISVZ & OSVZ (Inner & Outer Sub-Ventricular Zone, respectively), IZ (Intermediate Zone), CP (Cortical Plate), MZ (Marginal Zone).
(C) RT-PCR showing that SRGAP2A and SRGAP2B and SRGAP2C are expressed in OCT4+ human ES cells (hESC), neurons derived from hESCs following 3 weeks (PAX6+, PSD95−) or 5 weeks (PSD95+) in culture (wic) as well as fetal human brain, (F.h.br) and Adult human brain (Ad.h.Br).
(D) RT-PCR performed on RNA from fetal (f.) human brain, adult (Ad.) human brain and adult chimpanzee (Ad. Chimp) brain samples. SRGAP2A primers detect the ancestral copy in human and chimp samples. SRGAP2C primers show an amplification band specifically in the human samples indicating stringency of primer hybridization only to SRGAP2C variant.
(E) Quantitative-RT-PCR showing relative abundance of SRGAP2C and SRGAP2A in samples corresponding to lanes 1, 2 and 3 in panel C normalized to levels of GAPDH (mean ± s.d.; standard deviation from 2 technical replicates).
(F) Full length SRGAP2A detected by Western blotting with anti (α)-SRGAP2 N-terminal or anti-SRGAP2 C-terminal antibodies when transfected in HEK293T cells (grey arrowhead). The anti-SRGAP2 N-term, but not the anti-SRGAP2 C-terminal antibody can detect SRGAP2C (red arrowhead).
(G) A translation product corresponding to human-specific paralogs SRGAP2B or SRGAP2C in size (50kDa; red arrowhead) is seen in human cell lines SH-SY5Y and MCF7 but not in mouse developing cortex or cell line NIH3T3, only upon Western blotting with anti-SRGAP2 N-terminal antibody, not with anti-SRGAP2 C-terminal antibody.
(H) Western blotting with anti-SRGAP2 N-terminal antibody showing knock-down of endogenous SRGAP2B or SRGAP2C but not ancestral SRGAP2A by siRNA against intron 9 (si-intron9-1 and -2; lane 2,3) in SH-SY5Y cells.
Expression of SRGAP2 paralogs
We isolated one cDNA clone from a human library (GenBank: BC112927.1) that originates from SRGAP2C which is almost identical in sequence to the first 9 exons of the ancestral SRGAP2A, except for five non-synonymous base-pair mutations (out of 1356 base pairs) mutating 5 arginine (5R) residues (Figure 1A). As shown in the accompanying paper from Dennis et al., SRGAP2B and SRGAP2C are identical apart from a few base pair mutations and both lack the last 49 aa of the F-BAR domain. In order to determine the expression of these human-specific paralogs in the brain, we took advantage of the fact that intron 9 becomes 3’UTR in SRGAP2B and SRGAP2C to perform in situ hybridization (ISH) on cryostat sections from human fetal cortex (gestational week (GW) 11). SRGAP2B and SRGAP2C were therefore detected using an intron 9 probe which does not recognize processed mRNA encoding ancestral SRGAP2A, whereas SRGAP2A was detected using an exon 22 probe which conversely does not recognize SRGAP2B or SRGAP2C (Figure 1A,B and data not shown). This analysis revealed a striking similarity in the pattern of expression of the paralogs in the germinal layers (ventricular zone (VZ), and subventricular zone (SVZ)) where neural progenitors divide to produce post-mitotic neurons. We also detected significant expression in the cortical plate (Figure 1B), where neurons end their migration and undergo terminal differentiation and synapse formation. To refine this gene expression analysis, we took advantage of the possibility to differentiate human Embryonic Stem Cells (hESC) into relatively pure populations of forebrain excitatory pyramidal neurons (Kim et al., 2011). Total RNA was harvested from hESC at different time points during their directed differentiation in vitro (Kim et al., 2011) (Figure 1C), allowing us to perform a temporal analysis using RT-PCR primers (Table S1) amplifying SRGAP2A (exon 10–11 junction) or SRGAP2B and SRGAP2C (intron 9) transcripts. Both ancestral and human-specific paralogs of SRGAP2 were detected at low levels in undifferentiated hESC which express OCT4 (lane 1 in Figure 1C). Their expression increased in a mixture of neural progenitor cells (PAX6+) and immature neurons (TUJ1+, neurons) (3 weeks in culture (wic), Kim et al., 2011; lane 2 in Figure 1C) as well as in more mature, synaptically active, PSD95-expressing hESC-derived neurons (5 wic (Kim et al., 2011), lane 3 in Figure 1C). Both transcripts were also detected using RT-PCR from unpooled mRNA isolated from an individual human fetal brain (GW16; lane 4 in Figure 1C) and from the brain of a 44 years-old adult human (lane 5 in Figure 1C). In order to compare the relative level of expression of transcripts encoded by the paralogs and to tease apart transcripts derived from SRGAP2B and SRGAP2C, we took advantage of a fixed single base pair variation in exon 6 of SRGAP2C to design a set of primers specifically amplifying SRGAP2C transcripts (but not chimpanzee or human SRGAP2A transcripts or human SRGAP2B transcripts; Table S1; Figure 1D). Quantitative-RT-PCR (qRT-PCR) revealed that both SRGAP2C and SRGAP2A transcripts were abundant in hESC-derived neurons cultured for 3 and 5 weeks compared to non-differenciated human-ES cells (Figure 1E). However, SRGAP2C transcript maintained a higher level of expression than SRGAP2A after 5 weeks in culture (5 wic, Figure 1E). This suggests that SRGAP2C and SRGAP2A transcripts do not systematically co-vary and might be under different regulatory mechanisms leading to different transcript abundance in human neurons.
We next wanted to determine if a protein product corresponding to the translation of SRGAP2B or SRGAP2C transcripts could be detected in human cells. In transfected HEK293T cells, which do not express endogenous SRGAP2, SRGAP2A could be detected in a Western blot using an antibody directed against either the C-terminal (aa873–890) or the N-terminal (aa193–205) domain of the protein (Figure 1F, grey arrow). As predicted, SRGAP2C was only detected using the antibody directed against the N-terminal of SRGAP2, as a single band of a lower molecular weight (Figure 1F, red arrow). In human SH-SY5Y neuroblastoma cell line and human MCF7 breast cancer cell line, a 120kDa protein corresponding to SRGAP2A (Figure 1G, grey arrow) and a 50kDa protein (red arrow) were detected with the N-terminal SRGAP2 antibody. The 50kDa product was not detected in the same lysates using the C-terminal SRGAP2 antibody or in lysates isolated from postnatal day (P) 7 mouse cortex or from the mouse fibroblast NIH3T3 cell line, suggesting that it originates from human-specific SRGAP2B or SRGAP2C (Figure 1G). We confirmed that the 50kDa protein product detected with the N-terminal SRGAP2 antibody in neuroblastoma SH-SY5Y cells was specifically translated from SRGAP2B or SRGAP2C using two independent small interfering (si)RNAs (called siRNA intron9-1 and intron9-2) targeting intron 9, i.e. the 3’-UTR of the endogenous SRGAP2B and SRGAP2C mRNA that is absent in endogenous SRGAP2A mRNA (lane 2–3 in Figure 1H). The 50kDa protein product was detected in human SH-SY5Y cells transfected with control scramble siRNA but not in cells transfected with siRNA intron9-1 or intron9-2 (Figure 1H). Overall our results demonstrate that SRGAP2A and its human-specific paralog SRGAP2C are expressed in fetal and adult human brain and neurons.
SRGAP2C interacts with and inhibits ancestral SRGAP2 in vitro
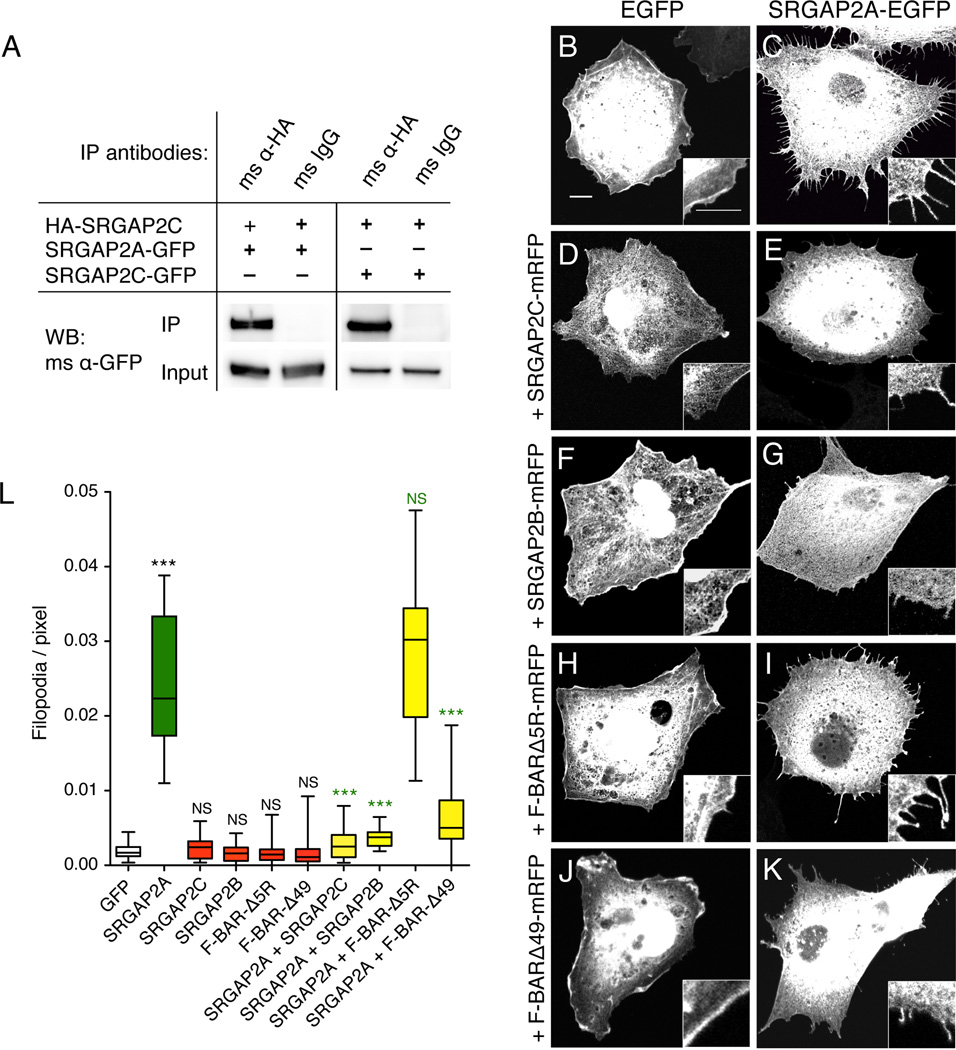
Next we explored the potential function of the human-specific paralogs of SRGAP2. Since the F-BAR domain is a homodimerization domain involved in membrane deformation (Frost et al., 2009) and since SRGAP2C encodes most of the F-BAR domain of the ancestral SRGAP2A protein, we tested if it retained the ability to dimerize with full length SRGAP2. In HEK293T cells, HA-tagged SRGAP2C was co-expressed with either SRGAP2A-GFP or SRGAP2C-GFP followed by HA-immunoprecipitation. Our results showed that SRGAP2C protein can dimerize with both SRGAP2A and SRGAP2C (Figure 2A). This suggests that the few amino acid substitutions in the sequence of SRGAP2C as well as the deletion of the last 49 aa do not alter its ability to dimerize with full length SRGAP2A. In order to explore the biological function of SRGAP2C protein, we employed a robust membrane deformation assay in COS7 cells (Guerrier et al., 2009). As previously shown for mouse SRGAP2 (Guerrier et al., 2009), expression of human SRGAP2A-EGFP induced filopodia protrusions in COS7 cells (Figure 2B–C and 2L). In contrast, expression of SRGAP2C-mRFP failed to induce filopodia (Figure 2D and 2L). Furthermore, when co-expressed with ancestral SRGAP2A-EGFP, SRGAP2C-mRFP efficiently blocked its ability to induce filopodia (Figure 2E and 2L). Expressing a cDNA encoding SRGAP2B (clone provided by Megan Dennis from Dr Eichler’s laboratory) gave similar results (Figure 2F–G, and 2L). Two features of SRGAP2C might account for this ability to block SRGAP2A function: (1) the mutation of several arginine residues that could mediate membrane binding/deformation properties (Shimada et al., 2007) and (2) the truncation of the F-BAR domain deleting the last 49 aa (see Figure 1A). To test the relative contribution of these two duplicate-specific differences, we introduced these mutations individually into the native F-BAR domain of ancestral SRGAP2 (F-BAR-Δ5R and F-BAR-Δ49 respectively). While both mutations abolished the filopodia induction properties of the F-BAR domain (Figure 2H, 2J and 2L), only FBAR-Δ49 mimicked SRGAP2C with regard to its ability to antagonize SRGAP2-mediated membrane protrusions (Figure 2K–L). Combined with the observation that the deletion of the most C-terminal 49 amino acids, but not the 5R mutations, is a common feature of SRGAP2C and SRGAP2B, our data suggest that both human-specific paralogs can interact with ancestral SRGAP2A and inhibit its function.
Figure 2. SRGAP2C dimerizes with SRGAP2A and inhibits its membrane deformation properties in COS7 cells.
(A) Co-immunoprecipitation (Co-IP) of SRGAP2A-GFP or SRGAP2C-GFP along with HA-SRGAP2C in HEK293T cells transfected with the indicated constructs. Co-IP was performed using mouse anti-HA antibody and mouse IgG as negative control. Western blotting (WB) was performed with a mouse (ms) anti (α)-GFP antibody.
(B–K) Representative confocal images of COS7 cells transfected with indicated constructs and visualized by EGFP signal. Scale bar: 5 µm. Inset shows 2-fold magnification.
(L) Box plot showing quantification of the number of filopodia per pixel along cell periphery, for cells represented in B–K. n = 20 cells per condition. *** p<0.001, Mann-Whitney test.
SRGAP2C inhibits SRGAP2 function during cortical neuron migration
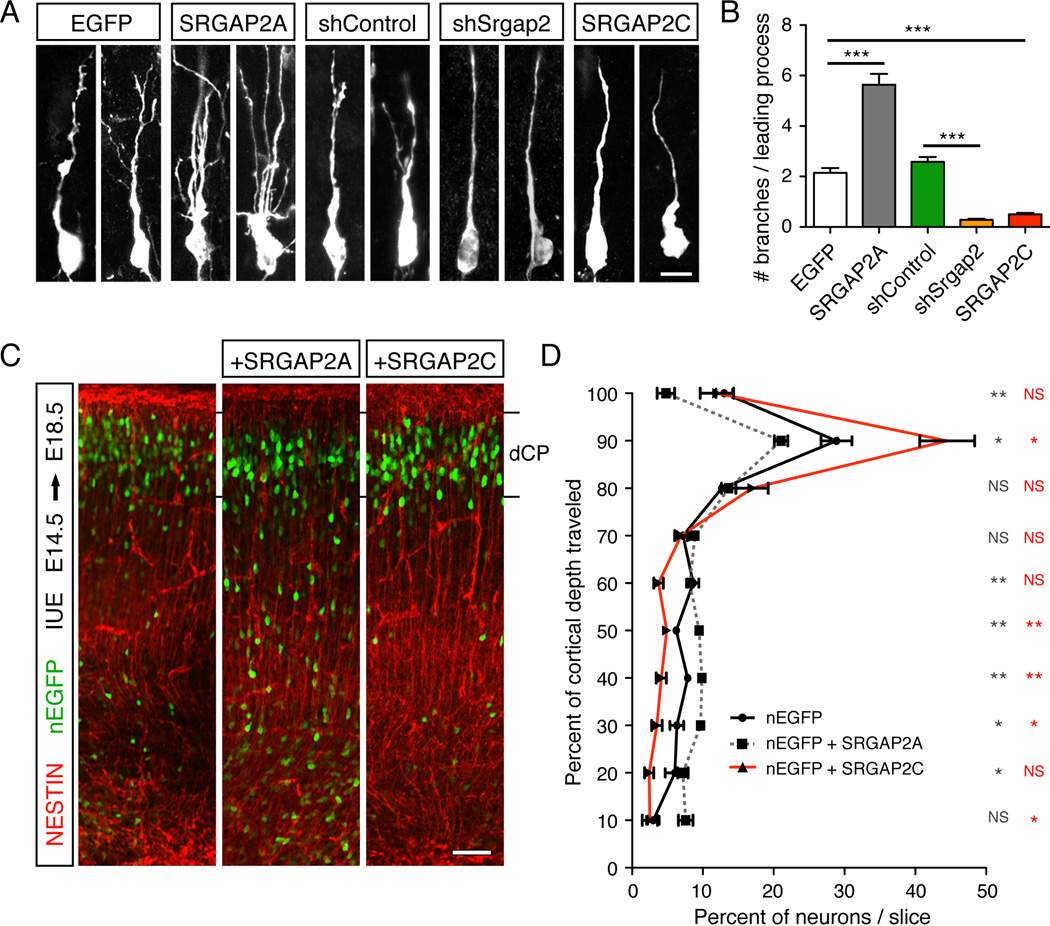
We then decided to test if the human-specific duplicates of SRGAP2 can also inhibit the documented function of SRGAP2 during cortical neuron migration (Guerrier et al., 2009). We focused on SRGAP2C for further experiments because in the human brain, SRGAP2C transcripts are substantially more abundant than SRGAP2B transcripts (see accompanying paper by Dennis et al.). We used in utero electroporation to compare the in vivo function of SRGAP2 and its human-specific paralog SRGAP2C. Mouse cortical progenitors generating layer 2/3 pyramidal neurons were electroporated at E14.5 and analyzed at E18.5, when most control neurons have exited the cell cycle, engaged radial migration and a significant portion have reached their final position in the cortical plate (CP). As previously demonstrated for mouse SRGAP2 in ex vivo electroporated slices of embryonic cortices (Guerrier et al., 2009), over-expression of human ancestral SRGAP2A induced excessive branching of the leading process (LP) of radially migrating neurons compared to control (Figure 3A–B), resulting in slower migration with less neurons accumulating in the CP after 4 days in utero (Fig. 3C–D). Conversely, knock-down of SRGAP2 using a short hairpin RNA (shRNA) (Figure S2) resulted in a reduction of LP branches on migrating neurons compared to neurons expressing control shRNA (Figure 3A–B), which has been shown to increase the rate of migration (Guerrier et al., 2009). Interestingly, neurons expressing SRGAP2C showed a reduction in LP branching very similar to that seen after SRGAP2 knockdown (Figure 3A–B), resulting in an increased rate of radial migration as quantified by the proportion of neurons successfully reaching the CP after 4 days in utero (Figure 3C–D). These results support the notion that the function of ancestral SRGAP2 is conserved between mouse and human and is inhibited by SRGAP2C expression in vivo.
Figure 3. SRGAP2C expression in radially migrating mouse cortical neurons phenocopies Srgap2 knock-down.
(A) Confocal images of optically isolated neurons showing representative morphologies of radially migrating cortical neurons in E18.5 embryos following in utero electroporation (IUE) at E14.5 of the indicated constructs. sh stands for short hairpin. Scale bar: 10 µm.
(B) Mean number of branches (± standard error to the mean; s.e.m) of the leading process of neurons as represented in panel A. n=3 animals/condition, 100–150 neurons/condition.
(C) Low magnification confocal images of E18.5 cortical slices showing migration of in utero electroporated neurons expressing nuclear-EGFP (nEGFP) alone or together with SRGAP2A or SRGAP2C. Staining with anti-GFP antibody shows the position of the electroporated neurons, and anti-NESTIN antibody marks the radial glial scaffold. dCP is dense Cortical Plate.
(D) Quantification of neuron distribution in cortical slices as illustrated in panel C (mean ± s.e.m). n=3 animals/condition, 9–10 slices/condition.
In panels B and D, * p < 0.05; ** p < 0.005; *** < 0.0001; NS (Not significant, p>0.05); Mann-Whitney test.
SRGAP2: a postsynaptic protein that regulates spine morphology in vitro
The expression of SRGAP2C (up to P21) in neurons generated at E15.5 did not alter the final position of neurons in layer 2/3 compared to control (Figure S3). Indeed, although SRGAP2C expression was found to accelerate neuronal migration, this is not expected to alter cell positioning in the context of inside-out cortical neuron migration. The absence of an overt effect on neuronal placement allowed us to address the potential function of SRGAP2 at later stages of brain development. We and others have recently shown that SRGAP2 is highly expressed in the mouse neocortex during the first weeks after birth (Bacon et al., 2009; Guerrier et al., 2009), a period critical for spinogenesis and synaptogenesis in rodents (Miller, 1986; Romand et al., 2011). Therefore, we decided to investigate the currently unknown function of SRGAP2 in dendritic spines. In the neocortex, spines receive most of the excitatory connections and are a critical site of structural and functional synaptic plasticity (Holtmaat and Svoboda, 2009; Yuste and Bonhoeffer, 2001). In mouse cortical neurons cultured for 20 days in vitro (DIV), endogenous SRGAP2 showed a punctate staining that was apposed to the presynaptic marker SYNAPSIN1 and extensively co-localized with the postsynaptic marker HOMER1 (Figure 4A), indicating that SRGAP2 is enriched at excitatory synapses. Over-expression of RFP-tagged SRGAP2 with GFP-tagged HOMER1c showed that SRGAP2 strongly accumulates in the head of dendritic spines with HOMER1 clusters (Figure 4B). Subcellular fractionation further confirmed that SRGAP2 is associated with synaptic membranes and is detected in the postsynaptic density (Figure S4).
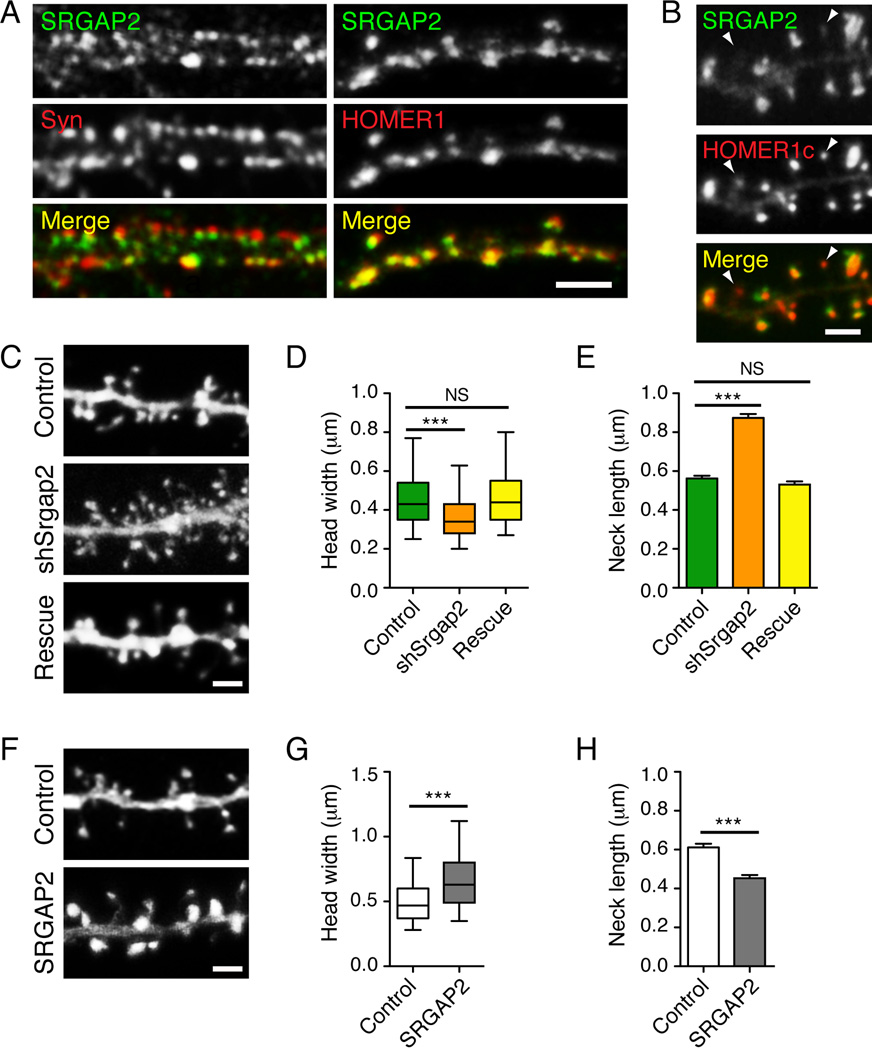
Figure 4. SRGAP2 is accumulated at excitatory synapses and promotes spine head growth in cultured cortical neurons.
(A) Segments of dendrites from cortical neurons (20 days in vitro) stained for SRGAP2 and the presynaptic marker SYNAPSIN1 (Syn) (left) or SRGAP2 and the excitatory postsynaptic marker HOMER1 (right). Scale bar: 5 µm.
(B) When over-expressed, SRGAP2-RFP localized to the head of dendritic spines and largely co-localized with the excitatory postsynaptic marker HOMER1c-GFP. Note that SRGAP2 was barely detectable in spines with small HOMER1c clusters (arrowheads).
(C) Segments of dendrites from cortical neurons expressing a control shRNA (Control), a shRNA targeting mouse Srgap2 (shSrgap2) and shSrgap2 co-expressed with SRGAP2A, which is resistant to this shRNA (rescue). Neurons were transfected 11 days after plating and imaged 9 days after transfection.
(D) Box plot showing the distribution of the width of spine heads in knock-down experiments. nControl = 1537, nshSrgap2 = 1261, nrescue = 910.
(E) Mean length of spine necks in knock-down experiments (± s.e.m).
(F) Segments of dendrites from cortical neurons expressing GFP alone (control), or GFP with ancestral SRGAP2 (SRGAP2). Neurons were transfected 17–18 days after plating and imaged 2 days after transfection.
(G) Distribution of the width of spine heads in gain-of-function experiments. nControl = 907, nSRGAP2 = 1020.
(H) Mean length of spine necks in gain-of-function experiments (± s.e.m).
In panels B, C and F, scale bars represent 2 µm. ***p < 0.001, NS (not significant): p > 0.05, Mann-Whitney test. Data are from a minimum of three independent experiments.
We next performed several loss- and gain-of-function experiments and measured key parameters of spine morphology: (i) the width of the spine head, which correlates with the size of the postsynaptic density, the number of synaptic AMPA glutamate receptors, the number of presynaptic vesicles (Arellano et al., 2007; Bourne and Harris, 2007; Harris and Stevens, 1989; Matsuzaki et al., 2004) and can be used as an indicator of spine maturation (Harris and Stevens, 1989) and (ii) the length of the spine neck, which impacts the biochemical and potentially the electrical isolation of the spine head from the dendritic shaft (Araya et al., 2006; Noguchi et al., 2005; Yuste, 2011). We first studied SRGAP2 function in cultured cortical neurons using RNA interference (see Figure S2). Plasmids expressing a control shRNA or shRNA against mouse Srgap2 (shSrgap2) were introduced into neurons using magnetofection after 11DIV and spine morphology was quantified 9–10 days later. Neurons expressing a control shRNA (Figure 4C) exhibited spines with an average spine head width of 0.458 ± 4 µm (n=1377) (Figure 4D) and an average neck length of 0.56 ± .01 µm (Figure 4E). shSrgap2-expressing neurons displayed immature-looking spines (Figure 4C) with smaller head width (0.367 ± 0.004 µm, n=1139, Figure 4D) and longer spine neck (87 ± 0.02 µm, Figure 4E) than control shRNA-expressing neurons. We also observed an increase in spine density from 1.25 ± 0.04 spine.µm−1 in control neurons (n=15) to 2.14 ± 0.13 spine.µm−1 in shSrgap2-expressing neurons (n=15). The effects of SRGAP2 knockdown on spine morphology and density could be rescued by co-expressing the shRNA against Srgap2 with a shRNA-resistant cDNA (Figure S2 and Figure 4C–E, density after rescue: 1.20± 0.06 spine.µm−1, n=15). Conversely, in gain-of-function experiments, over-expression of SRGAP2 for only 48h induced a striking enlargement of dendritic spines in 20DIV neurons, as visualized by co-transfection with EGFP (Figure 4F). Upon SRGAP2 over-expression, most spines were mushroom shaped, with larger spine heads (Figure 4G) and shorter spine necks (Figure 4H) than in control neurons. Interestingly, SRGAP2 was barely detected in long, thin spines but was strongly accumulated in spines with large heads (see arrowheads in Figure 4B). Altogether, these results suggest that SRGAP2 is a new postsynaptic protein that promotes spine head enlargement.
SRGAP2 promotes spine maturation and limits spine density in the neocortex
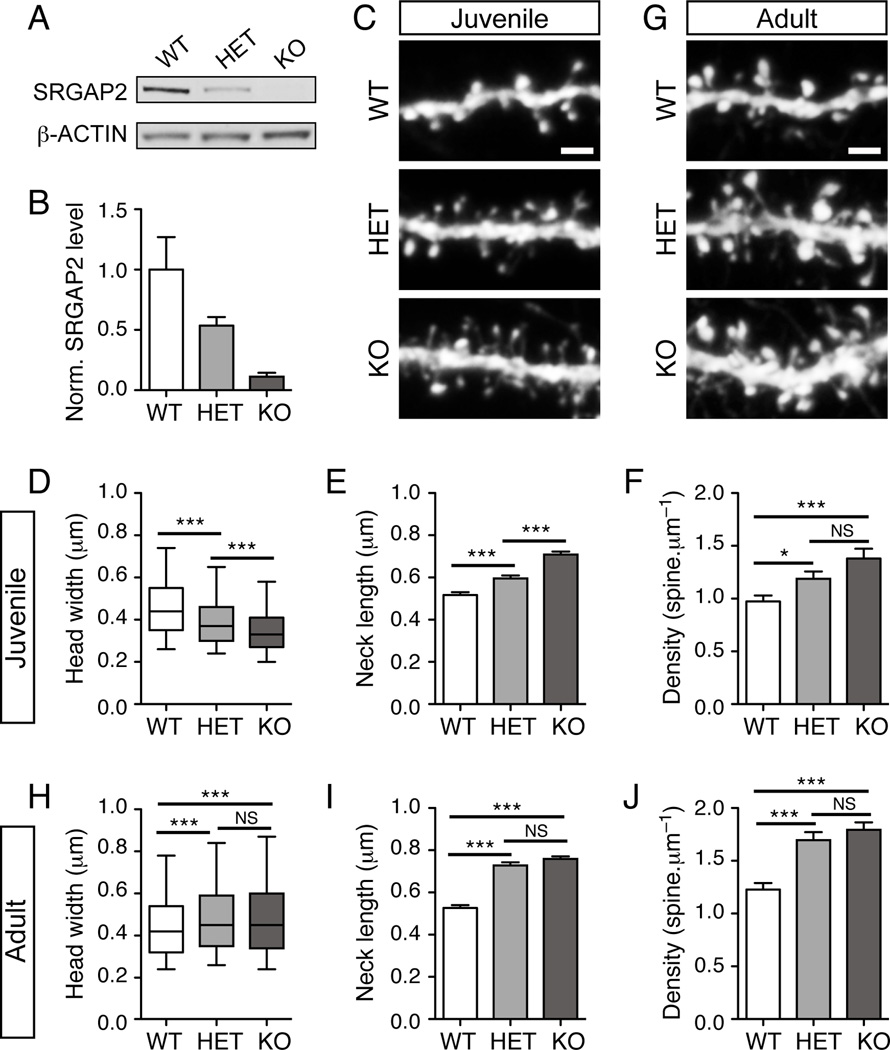
To obtain further insights into the function of SRGAP2 in vivo, we took advantage of a gene trap allele of Srgap2, hereon called Srgap2 knock-out (KO) (SRGAP2Gt(XH102)Byg/Mmcd, available at MMRRC-UC Davis, Figure S5A–C). Mapping of the insertion site in intron 2 using an inverse PCR strategy allowed us to design a PCR genotyping protocol to distinguish the heterozygous from the homozygous mice (Figure S5A–C). The homozygous Srgap2 KO mice showed a ≈90% reduction in the level of Srgap2 transcript (Figure S5D) as well as protein (Figure 5A–B) in the cortex, while heterozygous mice expressed intermediate levels of Srgap2 mRNA and protein. Srgap2 KO mice were viable (although born significantly below the expected Mendelian ratio; see Table S2) and showed no abnormality in cortical lamination (Figure S6A–B). Srgap2 KO mice were crossed with reporter mice expressing YFP in layer 5 pyramidal neurons (Thy1-YFP line H, (Feng et al., 2000), allowing quantitative assessment of dendritic spine morphology (Figure S6C). In juvenile mice (P18–P21), analysis of spines in apical oblique dendrites (see arrows in Figure S6C) of layer 5 pyramidal neurons from wild-type, heterozygous and homozygous KO mice (Figure 5C) revealed that SRGAP2 deficiency decreased the width of spine heads (Figure 5D) and increased the length of spine necks (Figure 5E), which is consistent with the results obtained with cultured neurons. These morphological defects were associated with a marked increase in the density of dendritic spines (Figure 5F). Interestingly, these changes were dose-dependent. Indeed, in juvenile mice heterozygous for Srgap2, spines were thinner, longer and more numerous than in wild-type mice but to a lesser extent than in KO mice (Figure 5C–F).
Figure 5. SRGAP2 deficiency delays spine maturation and increases spine density in vivo.
(A) Representative Western blot showing the relative amount of SRGAP2 in cortical lysates of wild-type (WT), heterozygous (HET) and knock-out (KO) mice.
(B) Quantification of SRGAP2 level in cortical lysates from 3 different animals per genotype normalized (norm.) to β-ACTIN and to the average SRGAP2 level in WT.
(C–F) Dose-dependent effect of SRGAP2 deficiency in juvenile mice (P18–P21). (C) Segments of oblique dendrites from WT, HET and KO mice expressing YFP in layer V pyramidal neurons (Thy1-YFP H line). (D) Distribution of spine head widths, nWT = 1278, nHET = 1307, nKO = 1602. (E) Mean spine neck length (± s.e.m). (F) Mean spine density (± s.e.m). nWT = 26, nHET = 20, nKO = 28.
(G–J) Long-term effect of SRGAP2 deficiency on dendritic spines. (G) Segments of oblique dendrites from adult (P65–P77) WT, HET and KO mice expressing YFP in layer V pyramidal neurons (Thy1-YFP H line). (H) Distribution of spine head widths in adult neurons, nWT = 1394, nHET = 1441, nKO = 2010. (I) Mean spine neck length in adult neurons (± s.e.m). (J) Mean spine density (± s.e.m). nWT = 23, nHET = 23, nKO = 35. Scale bars: 2 µm. ***p < 0.001, NS: p > 0.05, Mann-Whitney test.
We then wondered whether these defects were transient or maintained in adults. In adults (>P65 mice) (Figure 5G), the size of spine heads in wild-type mice was in the same range as in wild-type juveniles (Figure 5D and 5H), indicating that spines were already mature at P21, which is consistent with a previous report on the development of layer 5 pyramidal neurons (Romand et al., 2011). In contrast, heterozygous and KO neurons showed a substantial growth of their spine heads between juvenile and adult stages (Figure 5D and 5H). We observed a 22% and a 40% increase in the mean head width in heterozygous and KO neurons respectively, so that the size of spine heads in these neurons was close to the value measured in wild-type neurons (Figure 5H), even slightly larger (8% and 10% larger in heterozygous and KO neurons respectively and see Figure S6D). These data strongly suggest that SRGAP2-deficient neurons were still immature at P21 but eventually reached maturation in adults. Importantly, adult SRGAP2-deficient neurons maintained spines with longer necks and a spine density ≈1.4-fold higher than wild-type neurons (Figure 5I–J). Taken together, these results indicate that: (1) SRGAP2 promotes spine maturation and limits spine density in vivo and (2) SRGAP2 co-regulates spine density with the length of spine neck. Thus, at low SRGAP2 levels, the phase of spinogenesis is extended and adult neurons display more numerous spines with longer neck, which may have a profound impact on neuronal connectivity and synaptic input integration.
Expression of human-specific SRGAP2C in mouse neurons delays spine maturation and increases spine number
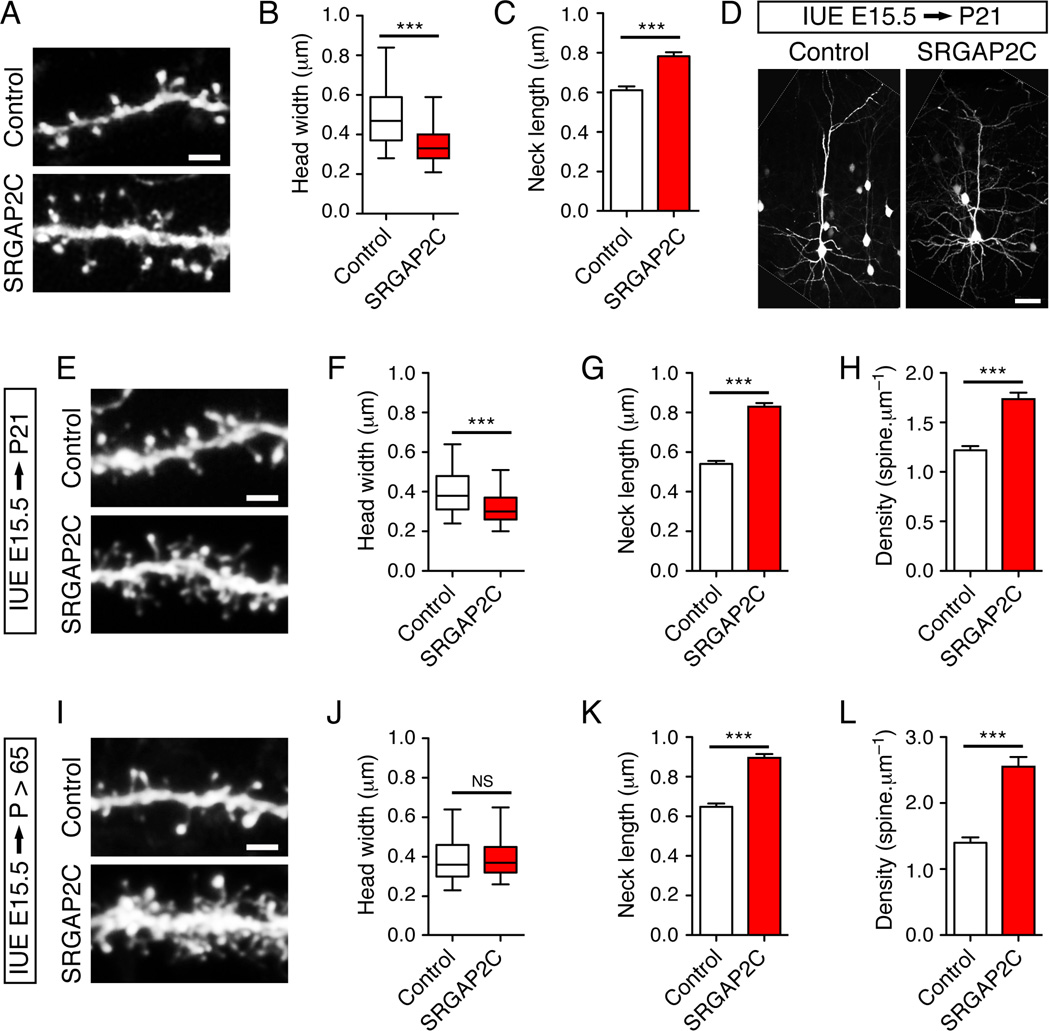
Based on all the results shown so far, we hypothesized that long-term expression of the human protein SRGAP2C would delay spine maturation. In vitro, mouse cortical neurons maintained for 20DIV and analyzed 2 days post-transfection of SRGAP2C displayed significant changes in spine morphology (Figure 6A–C), which were opposite to the over-expression of SRGAP2 (see Figure 4F–H) and similar to the shRNA-mediated knock-down of SRGAP2 (see Figure 4C–E). We next analyzed in juvenile mice the consequences of SRGAP2C expression in layer 2/3 pyramidal neurons following in utero electroporation at E15.5. The co-expression of myristoylated Venus allowed the visualization of neuronal morphology (Figure 6D). As expected, in juvenile mice, SRGAP2C-expressing neurons exhibited numerous immature-looking spines compared to control neurons (Figure 6E). Spines in SRGAP2C expressing neurons had smaller head (Figure 6F), longer neck (Figure 6G) and were present at a higher density (Figure 6H) than in control neurons. In adult SRGAP2C-expressing neurons (Figure 6I), spine head widths were similar to control neurons (Figure 6J) but both spine neck length (Figure 6K) and spine density (Figure 6L) remained significantly higher than in control neurons. Again, these effects were highly similar to what we observed in SRGAP2-deficient layer 5 pyramidal neurons in vivo (Figure 5C–J). Overall these results show that expression of SRGAP2C in the mouse neocortex leads to changes in spine development compatible with an inhibition of endogenous SRGAP2 and resulting in neoteny during dendritic spine maturation.
Figure 6. SRGAP2C expression in mouse cortical neurons phenocopies SRGAP2 deficiency in spines.
(A–C) SRGAP2C expression induces long thin spines in cultured cortical neurons. (A) Segment of dendrites from cortical neurons (20DIV) expressing EGFP alone (Control), or EGFP and SRGAP2C (SRGAP2C). Neurons were imaged 2 days after transfection. (B) Box plot representing the distribution of the width of spine heads. nControl = 907 (same as Figure 4F–H), nSRGAP2C = 1029. (C) Mean length of spine necks (± s.e.m).
(D–H) Effect of SRGAP2C expression in juvenile mice in layer 2/3 pyramidal neurons following in utero electroporation (IUE) at E15.5. (D) Representative layer 2/3 pyramidal neurons expressing a control cDNA (Control, left) or SRGAP2C (right) with a vector encoding mVenus. The white lines delineate the border of the original images. (E) Segments of oblique dendrites from neurons in the conditions described above. (F) Distribution of spine head widths, nControl = 1068, nSRGAP2C = 1167. (G) Mean spine neck length (± s.e.m). (H) Mean spine density (± s.e.m). nControl = 16, nSRGAP2C = 16. (I–L) Long-term effect of SRGAP2C expression on spines in adult mice. (I) Segments of oblique dendrites from control and SRGAP2C-expressing neurons after in utero electroporation at E15.5. (J) Distribution of spine head widths, nControl = 800, nSRGAP2C = 941. (K) Mean spine neck length (± s.e.m). (L) Mean spine density (± s.e.m). nControl = 14, nSRGAP2C = 12. Scale bars represent 2 µm in panels A, E and I and 50 µm in panel D. *** p < 0.001, NS : p > 0.05, Mann-Whitney test.
DISCUSSION
Inhibition of SRGAP2 function leads to sustained radial migration
Our study is among the first to address the functional consequence of human-specific gene duplication during brain development. We show that the human-specific paralogs of SRGAP2 are expressed in developing and adult neurons at the RNA and the protein levels along with the ancestral copy. The SRGAP2 duplicates encode a truncated F-BAR domain that binds to and antagonizes the function of ancestral SRGAP2 during neuronal migration and morphogenesis. We previously showed that in the mouse fetal brain, SRGAP2 is up-regulated at the end of cortical migration where it promotes branching in the leading process and decreases the rate of migration (Guerrier et al., 2009). Here, we found that introduction of human-specific SRGAP2C in utero in mouse pyramidal neurons mimics SRGAP2 deficiency during neuronal migration, leading to a deficit in branching in the leading process of migrating neurons and allowing neurons to reach their final position in the cortical plate faster than control neurons. We propose that these effects of human-specific SRGAP2C expression may underlie a more persistent form of radial migration and delay its termination. This could support the journey of neurons over the total thickness of the cortical wall, which is substantially increased in humans compared to non-human primates or rodents (Dehay and Kennedy, 2007; Rakic, 2009; Sidman and Rakic, 1973).
Temporal control of spine development
Numerous proteins have been implicated in the formation, maturation and maintenance of spines (Bourne and Harris, 2008; Dillon and Goda, 2005; Elias et al., 2008; Hayashi et al., 2009) but the mechanisms regulating the timing of spine development in vivo remain poorly understood. In this study, we demonstrate that SRGAP2 associates with the postsynaptic density to promote spine maturation and limits spine density in vivo. In contrast, SRGAP2-deficiency or expression of SRGAP2C delays spine maturation and leads to an increased density of spines with longer neck in the neocortex. These results suggest that SRGAP2C-induced neoteny extends the period of spine production with long-term consequences on their morphology. This altered developmental trajectory of spine maturation may reflect changes in excitatory synaptic development. Other genes have been shown to produce heterochrony during organ development with critical consequences for limb morphogenesis (Dolle et al., 1993) or for the expansion of the neocortical surface (Lui et al., 2011; Rakic, 2009), supporting the notion that slower developmental processes might underlie increasingly complex morphogenesis.
Studies that have quantitatively compared spine morphology, spine density and the developmental time course of spinogenesis between human and monkeys or mouse have revealed that (1) spinogenesis in prefrontal areas presents neoteny in humans (Huttenlocher and Dabholkar, 1997; Petanjek et al., 2011); (2) spine density in comparable neocortical areas and layers is higher in humans (Benavides-Piccione et al., 2002; Elston et al., 2001) and (3) there are substantial differences in spine morphology between species, human spines having longer neck and larger heads than in the other species analyzed (Benavides-Piccione et al., 2002). Our results suggest that the human-specific duplication of SRGAP2 is involved in the emergence of these differences. In the human brain, the rate of spine development differs between cortical areas (Huttenlocher and Dabholkar, 1997). Further investigations will test if expression of SRGAP2C shows spatial heterogeneity.
Functional implications of human-specific SRGAP2 duplication
Spines are known to enhance synaptic connectivity, enable linear integration of synaptic inputs and implement synapse-specific plasticity (Yuste, 2011). The spine neck represents a physical barrier between the synaptic contact in the spine head and the dendritic shaft. The length of the neck impacts the compartmentalization of synaptic signaling and the filtering of synaptic inputs while the volume of the spine head is an indicator of the size of the post-synaptic density and the strength of the synapse (Bourne and Harris, 2008; Yuste, 2011). Both the neck and the head of spines undergo morphological and functional plasticity (Bloodgood and Sabatini, 2005; Matsuzaki et al., 2004; Yuste and Bonhoeffer, 2001) and spines with larger head and wider, shorter neck have been observed after induction of synaptic potentiation in rodents (Yuste and Bonhoeffer, 2001). We found that SRGAP2 co-regulates the density of spines and the length of the spine neck. Accordingly, SRGAP2-deficient neurons or SRGAP2C-expressing neurons harbored spines with longer necks and higher spine density. In our experiments, spine heads in adult SRGAP2-deficient neurons were comparable in size to wild-type neurons. In this context, we speculate that expression of SRGAP2C might allow human cortical pyramidal neurons to receive and integrate a significantly higher number of synaptic inputs without saturation. Such morphological features have been proposed to introduce more opportunities and more flexibility for input integration and information processing in human cortical circuits (Benavides-Piccione et al., 2002). On the other hand, it might also contribute to the higher susceptibility to neurodegenerative or psychiatric disorders of the human brain (Konopka et al., 2009).
Taken together, our results suggest that the expression of SRGAP2C in the human brain inhibits the function of ancestral SRGAP2 and thereby reduces the rate of spine maturation, leading to changes in spine morphology and density that could have important implications for cognition, learning and memory. SRGAP2 is both a membrane-binding protein and a Rho-GAP for the small GTPase Rac1, which has been associated with neurodevelopmental disorders (Govek et al., 2005). It will be important to address the molecular mechanisms by which SRGAP2 promotes spine maturation and how SRGAP2C interferes with this process. A large genomic alteration affecting the human ancestral SRGAP2A gene has recently been reported in a patient with early-infantile encephalopathy with associated epilepsy (Saitsu et al., 2011). However, this study was based on a single patient presenting a large chromosomal translocation that affected the expression of SRGAP2A as well as other genes. Future investigations will test if alterations in copy number or sequence variation of ancestral SRGAP2A or its human-specific paralogs underlie neurodevelopmental or cognitive defects such as autism spectrum disorders or schizophrenia.
EXPERIMENTAL PROCEDURES
Constructs
Human SRGAP2A (GenBank: BC132874) and SRGAP2C (GenBank: BC112927) cDNA were obtained from the IMAGE consortium. cDNA encoding SRGAP2B was a gift from the Eichler laboratory. See Supplemental Experimental Procedures for details of constructs used in this paper.
RNA in situ hybridization
The human fetal material (GW11) used to perform in situ hybridization was collected and used according to the guidelines of the three relevant local Ethics Committees (Erasme Academic Hospital, University of Brussels, and Belgian National Fund for Scientific Research) on Research Involving Human Subjects. Written informed consent was given by the parents in each case. Exon 22 of SRGAP2A (3316–4271 bp of BC132874) and intron 9 (1992–3128 bp of BC112927) were amplified and cloned into pGEM-T-easy vector system 1 (Promega) and served as templates for generating RNA in situ probes. In situ hybridization on human embryonic brain samples was performed as previously described (Lambert et al., 2011).
Animals
All animals were handled according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at University of North Carolina, Chapel Hill and The Scripps Research Institute, La Jolla. For timed pregnant matings, noon after mating is considered embryonic day 0.5 (E0.5). Juveniles correspond to mice between postnatal day (P) 18 and P21, adults correspond to mice between P65 and P77. For information regarding genetic strains, refer to Supplementary Experimental Procedures.
In utero electroporation
We performed in utero electroporation as per a previously described protocol (Yi et al., 2010). Endotoxin free plasmid DNA was injected at a concentration of 1 µg/µl where single plasmids were electroporated. In case of co-electroporation, we injected equimolar quantities of the two plasmids to ensure co-expression. Please see Supplemental Experimental Procedures for electroporation settings, tissue preparation, slicing and immunostaining of electroporated brains.
Primary neuronal culture, magnetofection and immunocytochemistry
Primary cultures of cortical neurons were prepared from embryonic BALB/c mice at day 17.5–18.5 and transfection performed by magnetofection using NeuroMag (OZ Bioscience) according to the manufacturer’s protocol at the indicated times. See details in Supplementary Experimental Procedures.
Morphometric analysis of spines
Morphometric analyses of dendritic spines were performed on z-projections for cultured neurons and in the depth of the z-stack for slices using NIS-Elements. Head width was defined as the largest length of the head that was perpendicular to the neck. Neck length was measured from the point of attachment of the dendrite to the beginning of the spine head, as estimated by the investigator. When the neck could not be distinguished from the head, the neck length was considered to be 0. When the neck was not clearly visible, it was considered to be the shortest distance between the base of the head and the closest surface of the dendrite. Only spines arising form the lateral surfaces of the dendrites were taken into account. For analyses in slices, spines were quantified in the proximal part of oblique dendrites directly originating from the apical trunk. Spines were quantified over an average of 60 µm of dendrite in juvenile mice and 40 µm in adult mice. Spine density was defined as the number of quantified spines divided by the length over which the spines were quantified. See Supplementary Experimental Procedures for more details.
Subcellular fractionation
Subcellular fractionation was performed from P15 mouse brains as described in (Perez-Otano et al., 2006) with minor modifications. See Supplementary Experimental Procedures for details of antibodies used.
Research Highlights.
Article highlights can have up to 4 bullet points and upto 85 characters with spaces. This is what we came up with:
SRGAP2 has undergone two partial duplications specifically in the human genome.
One copy (SRGAP2C) is expressed in the human brain and antagonizes ancestral SRGAP2.
Ancestral SRGAP2 promotes dendritic spine maturation and limits spine density in vivo.
Human SRGAP2C induces neoteny and leads to higher density of spines with longer necks.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Marie Rougié and Virginie Courchet for excellent technical support and management of the mouse colonies and members of the Polleux laboratory for useful discussions. We thank Jason Yi and Michael Ehlers for their initial help with the long-term neuronal cultures, Maxime Camo for help with the initial characterization of the SRGAP2 knockout, Daniel Choquet for providing HOMER1c expression plasmid and Evan Eichler’s lab for reagents and useful discussions. N.L. is a clinical-scientist of the FNRS and P.V. a Research Director of the FNRS. This work was partially supported by grants of the FNRS, Belgian Queen Elizabeth Foundation, Welbio and Excellence Programs of the Walloon Region (to P.V.), Fonds Erasme and the Belgian Kid's Fund (to NL), NIH RO1NS067557 (FP) and ADI-Novartis funds (FP), NIH F31NS061610 (J.dM.-P.), NIH F31NS068038 (J.C-B.), by NSFC (No.31171033) and "973" Project (No.2011CB933101) (W-L.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONTRIBUTIONS
C.C., K.J., T.S. and F.P. conceived the experiments and C.C., K.J., J.C-B., J.dM, T.S., N.L., P.V. performed the experiments. W-L.J. provided the anti-SRGAP2 antibodies. J-E.K. and A.G. provided RNA from cultured human ES cells. C.C., K.J. and F.P. prepared the manuscript.
REFERENCES
- Araya R, Jiang J, Eisenthal KB, Yuste R. The spine neck filters membrane potentials. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17961–17966. doi: 10.1073/pnas.0608755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano JI, Benavides-Piccione R, Defelipe J, Yuste R. Ultrastructure of dendritic spines: correlation between synaptic and spine morphologies. Front Neurosci. 2007;1:131–143. doi: 10.3389/neuro.01.1.1.010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C, Endris V, Rappold G. Dynamic expression of the Slit-Robo GTPase activating protein genes during development of the murine nervous system. The Journal of comparative neurology. 2009;513:224–236. doi: 10.1002/cne.21955. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nature reviews Genetics. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Benavides-Piccione R, Ballesteros-Yanez I, DeFelipe J, Yuste R. Cortical area and species differences in dendritic spine morphology. Journal of neurocytology. 2002;31:337–346. doi: 10.1023/a:1024134312173. [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310:866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annual review of neuroscience. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Caceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. Global analysis of alternative splicing differences between humans and chimpanzees. Genes & development. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nature reviews Neuroscience. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Dillon C, Goda Y. The actin cytoskeleton: integrating form and function at the synapse. Annual review of neuroscience. 2005;28:25–55. doi: 10.1146/annurev.neuro.28.061604.135757. [DOI] [PubMed] [Google Scholar]
- Dolle P, Dierich A, LeMeur M, Schimmang T, Schuhbaur B, Chambon P, Duboule D. Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell. 1993;75:431–441. doi: 10.1016/0092-8674(93)90378-4. [DOI] [PubMed] [Google Scholar]
- Elias GM, Elias LA, Apostolides PF, Kriegstein AR, Nicoll RA. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20953–20958. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, DeFelipe J. The pyramidal cell in cognition: a comparative study in human and monkey. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:RC163. doi: 10.1523/JNEUROSCI.21-17-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS biology. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes & development. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nature reviews Neuroscience. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Hurles M. Gene duplication: the genomic trade in spare parts. PLoS biology. 2004;2:E206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. The Journal of comparative neurology. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Developmental cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kim JE, O'Sullivan ML, Sanchez CA, Hwang M, Israel MA, Brennand K, Deerinck TJ, Goldstein LS, Gage FH, Ellisman MH, et al. Investigating synapse formation and function using human pluripotent stem cell-derived neurons. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3005–3010. doi: 10.1073/pnas.1007753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Lambot MA, Bilheu A, Albert V, Englert Y, Libert F, Noel JC, Sotiriou C, Holloway AK, Pollard KS, et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PloS one. 2011;6:e17753. doi: 10.1371/journal.pone.0017753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW. Maturation of rat visual cortex. III. Postnatal morphogenesis and synaptogenesis of local circuit neurons. Brain research. 1986;390:271–285. doi: 10.1016/s0006-8993(86)80236-0. [DOI] [PubMed] [Google Scholar]
- Noguchi J, Matsuzaki M, Ellis-Davies GC, Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron. 2005;46:609–622. doi: 10.1016/j.neuron.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution of Gene Duplication. Springer-Verlag; 1970. [Google Scholar]
- Perez-Otano I, Lujan R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nature neuroscience. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nature reviews Neuroscience. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romand S, Wang Y, Toledo-Rodriguez M, Markram H. Morphological development of thick-tufted layer v pyramidal cells in the rat somatosensory cortex. Frontiers in neuroanatomy. 2011;5:5. doi: 10.3389/fnana.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Osaka H, Sugiyama S, Kurosawa K, Mizuguchi T, Nishiyama K, Nishimura A, Tsurusaki Y, Doi H, Miyake N, et al. Early infantile epileptic encephalopathy associated with the disrupted gene encoding Slit-Robo Rho GTPase activating protein 2 (SRGAP2) Am J Med Genet A. 2011 doi: 10.1002/ajmg.a.34363. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain research. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Sikela JM. The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS genetics. 2006;2:e80. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annual review of medicine. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nature reviews Genetics. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-beta signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annual review of neuroscience. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.