Abstract
Wig-1 is a transcriptional target of the p53 tumor suppressor and encodes an mRNA stability-regulating protein. We show here that Wig-1 knockdown causes a dramatic inhibition of N-Myc expression and triggers differentiation in neuroblastoma cells carrying amplified N-Myc. Transient Wig-1 knockdown significantly delays development of N-Myc-driven tumors in mice. We also show that N-Myc expression is induced upon moderate p53-activating stress, suggesting a role of the p53-Wig-1-N-Myc axis in promoting cell cycle re-entry upon p53-induced cell cycle arrest and DNA repair. Moreover, our findings raise possibilities for the improved treatment of poor prognosis neuroblastomas that carry amplified N-Myc.
Keywords: Wig-1, N-Myc, p53, mRNA regulation, AU-rich elements
The p53 tumor suppressor is a transcription factor that responds to cellular stress including DNA damage and oncogene activation (reviewed in Vousden and Prives1). p53 activation can result in a wide range of responses depending on the stress agent and cellular context. The classical outcomes of p53 activation are cell cycle arrest, senescence, and apoptosis. Low levels of p53 caused by mild stress may induce arrest and damage repair and maintain cell survival, while high p53 levels in response to severe stress triggers cell death by apoptosis.1 Basal p53 levels also regulate antioxidant activities and metabolism, promoting survival.2 The importance of p53-mediated tumor suppression is demonstrated by the high frequency of p53 mutations in human tumors (reviewed in Soussi and Wiman 3 see also www-p53.iarc.fr and free.p53.fr). Excessive p53 activity is implicated in a number of neurological disorders characterized by untimely neuronal cell death.1
Wig-1 (for wild-type p53-induced gene 1) is a direct p53 target gene identified in mouse,4 rat,5 and human cells.5, 6, 7 It is most strongly expressed in the nervous system, but is present in all cell types with enrichment in stem cells as compared with differentiated tissues.8, 9 Furthermore, Wig-1 is induced after cellular stress in a p53-dependent manner.8 Wig-1 is a zinc finger protein that binds to double-stranded RNA,10, 11 and interacts with the RNA-binding proteins hnRNPA2/B1 and RNA Helicase A through RNA.12 Wig-1 localizes mainly to the nucleus but can shuttle between the nucleus and the cytoplasm.6, 11, 13, 14 Wig-1 is highly conserved during evolution from amoeba to man.8 We have recently shown that Wig-1 can bind to a U-rich element in the 3′UTR of p53 mRNA, thereby stabilizing p53 mRNA by preventing its de-adenylation.13 The U-rich region in the p53 3′UTR is a subclass of the AU-rich elements (AREs) well known for their role in regulating mRNA stability and/or translation efficiency. AREs are generally found in mRNAs of genes whose expression must be tightly controlled, such as genes encoding proteins that regulate cell growth or response to external factors.15, 16 ARE-interacting proteins are important nodes of regulation as they can potentially control the levels of many different mRNAs in the cell, and have been shown to have important roles in cancer as well as development.15, 17 Thus we hypothesized that Wig-1 regulates other ARE-containing mRNAs in addition to that of p53. The mRNA of the N-Myc oncogene contains both U-rich and AU-rich regions that are bound by the nervous system-specific HuR family member HuD.18, 19 N-Myc belongs to the Myc network of transcription factors that can affect a wide variety of cellular processes including cell growth, cell cycle entry, differentiation, and apoptosis.20 N-Myc is normally expressed mainly in nervous tissues early during embryonic development, and is absent in most adult tissues. However, amplification of N-Myc occurs in 20–30% of neuroblastoma, a childhood tumor derived from precursors or immature cells of the sympathetic nervous system.21 N-Myc amplification is the most consistent marker of poor prognosis and aggressive disease in neuroblastoma.20, 22 N-Myc knockdown by siRNA in neuroblastoma cells carrying N-Myc amplification can induce differentiation,23 and antisense strategies that target N-Myc inhibit mouse neuroblastoma tumorigenesis in vivo.24
Results
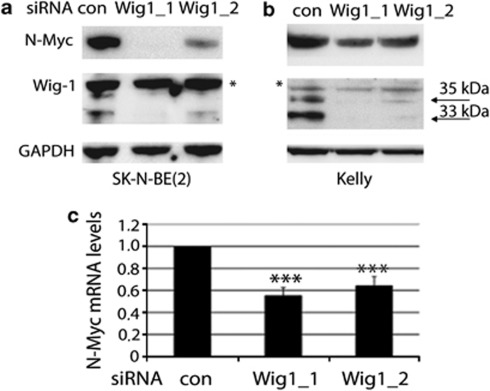
As the N-Myc mRNA is reported to be regulated through AREs18, 19 we hypothesized that Wig-1 could affect N-Myc protein and mRNA levels. We knocked down Wig-1 using two different siRNA oligos in two neuroblastoma cell lines carrying N-Myc amplification, SK-N-BE(2) and Kelly (Figures 1a and b), and detected a significant reduction in N-Myc protein levels in both cell lines with both siRNA oligos. The effect was more pronounced using Wig-1 siRNA 1, which was also more efficient than Wig-1 siRNA 2 in knocking down Wig-1. We next tested if the effect of Wig-1 on N-Myc could be detected also at the mRNA level, using the two different Wig-1 siRNA and assessing N-Myc mRNA levels with the Quantigene assay. Figure 1c demonstrates a reduction in N-Myc mRNA levels compared with GAPDH mRNA levels by 45% and 36% using siRNA oligo 1 and 2, respectively.
Figure 1.
N-Myc protein and mRNA levels are decreased upon Wig-1 knockdown. Wig-1 knockdown using two different siRNA oligos leads to decreased N-Myc protein levels in SK-N-BE(2) (a) and Kelly (b) cells carrying N-Myc amplification. Arrows indicate the two major Wig-1 protein species, and * indicates a non-specific band. (c) Similarly, Wig-1 knockdown leads to decreased N-Myc mRNA levels in SK-N-BE(2) cells, by 45% (P=1.9 × 10−5) with Wig-1 siRNA oligo 1 and by 36% (P=0.0001) by Wig-1 siRNA oligo 2 (n=4), as assessed by the Quantigene assay. The figure shows N-Myc mRNA levels relative to GAPDH mRNA levels. Error bars show S.D.
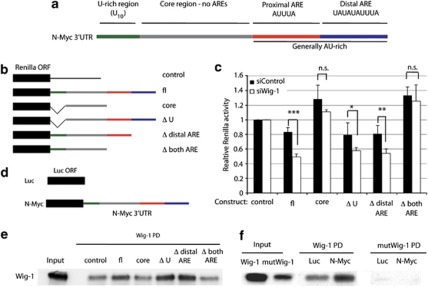
Next, we investigated if the AREs in the N-Myc 3′UTR are required for Wig-1-mediated regulation. The N-Myc 3′UTR contains a U-rich stretch and a general AU-rich region containing two canonical AREs (Figure 2a, Supplementary Figure S1). To examine the effect of Wig-1 on the N-Myc 3′UTR, we used the psiCheck2 luciferase vector in which the 3′UTR of interest is inserted downstream of renilla luciferase. Using this system, we generated constructs containing either full-length or partially deleted N-Myc 3′UTRs, as well as a negative control containing part of the p53 3′UTR lacking the U-rich region (and therefore not regulated by Wig-113) (Figure 2b, Supplementary Figure S1). Wig-1 was knocked down in HCT116 cells that were subsequently transfected with the luciferase constructs. Renilla luciferase activity was assessed and normalized to firefly luciferase expressed from the same vector. HCT116 rather than SK-N-BE(2) cells were used in this experiment because of the low plasmid transfection efficiency in the SK-N-BE(2) cells. Wig-1 expression and efficient knockdown in HCT116 cells are shown in Supplementary Figure S2. We observed a decrease in renilla luciferase activity relative to firefly activity of the construct containing the full-length N-Myc 3′UTR after Wig-1 knockdown (Figure 2c), demonstrating that Wig-1 exerts its effect on N-Myc through the N-Myc 3′UTR. Similar effects on relative renilla luciferase activity were observed for the constructs containing the proximal ARE, but not for the constructs lacking the proximal ARE, indicating that Wig-1 regulates N-Myc mRNA through this region. It is noteworthy that constructs lacking the proximal ARE generated higher relative renilla luciferase activity, suggesting that this region acts as a negative regulatory element in the N-Myc 3′UTR (Figure 2c).
Figure 2.
Wig-1 regulates N-Myc mRNA by binding to the 3′UTR. The N-Myc 3′UTR contains a U-rich stretch and a general AU-rich region containing two canonical AREs (a). To determine if Wig-1 regulates N-Myc through any of these regions, we generated the constructs in (b) linked to renilla luciferase and assessed renilla activity compared with control luciferase activity. (c) Statistically significant reduction after Wig-1 knockdown compared with control siRNA of relative renilla activities was observed with the constructs containing the proximal ARE, that is, full-length N-Myc 3′UTR (fl) (P=7.3 × 10−5); Δ distal ARE (P=0.006); and ΔU (P=0.044), but not with the constructs lacking the proximal ARE (core and Δ both ARE, n.s. stands for not significant) (n=4). To determine if Wig-1 binds to the N-Myc 3′UTR – and in that case, identify the region required for binding – we used the constructs in panel (b) as templates for making biotinylated RNA probes that were mixed with cell lysates from HCT116 cells ectopically expressing wild-type Wig-1. Biotinylated probes were captured using streptavidin beads, washed, and bound proteins were analyzed by western blotting. (e) Wig-1 binds to the probes generated from constructs in panel (b) that contain the proximal ARE but not to those lacking it. PD=pulldown. To investigate if the RNA binding capacity of Wig-1 is required for the binding to N-Myc mRNA, we used luciferase constructs either without a 3′UTR (‘control') or with the 3′UTR of N-Myc (‘N-Myc') (d) as templates for making biotinylated RNA probes that were mixed with cell lysates from HCT116 cells ectopically expressing either wild-type Wig-1 (‘Wig-1') or a zinc finger point mutant form of Wig-1 that cannot bind to RNA10 (‘mutWig-1') and performed the pulldown as above. (f) Binding of wild-type but not mutant Wig-1 to the 3′UTRs of N-Myc, but not to the control RNA. Densitometry analysis using ImageJ showed 1.4 times more wt Wig-1 than mutant Wig-1 protein in the input, most likely reflecting decreased protein stability of the mutant protein. However, 64.5 times more wt than mutant Wig-1 was bound to the N-Myc probe (f)
As we previously showed that Wig-1 is an RNA-binding protein that regulates p53 mRNA through direct binding to the 3′UTR, we tested if Wig-1 can bind to the N-Myc 3′UTR. We generated probes by in vitro transcription of the constructs in Figure 2b in the presence of biotinylated UTP. The biotinylated RNA probes were used in pulldown assays with ectopically expressed FLAG-tagged Wig-1 from HCT116 cell lysates. Figure 2e shows that Wig-1 binds to the probes containing the proximal AU-rich element of the N-Myc 3′UTR, but not to the regions lacking it. Thus, Wig-1 binds to the same region that is required for the Wig-1-mediated regulation of the N-Myc 3′UTR. To investigate if the RNA-binding properties of Wig-1 are required for this interaction, we performed a biotin pulldown assay using probes generated by in vitro transcription of either the luciferase open reading frame (ORF) alone (control) or the luciferase upstream of the N-Myc 3′UTR (Figure 2d). Figure 2f shows that Wig-1 binds to the N-Myc 3′UTR probe but not to the control probe, while a zinc finger point mutant form of Wig-1 which cannot bind to RNA10, 11 is unable to bind to any of the biotinylated probes. Thus, we conclude that Wig-1 binds to and exerts its effect through the proximal AU-rich element in the N-Myc 3′UTR.
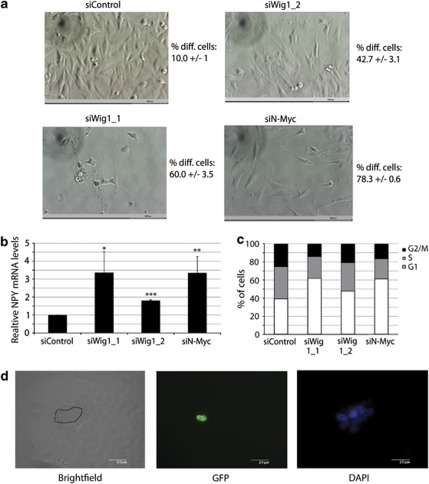
N-Myc knockdown causes differentiation in SK-N-BE(2) cells carrying amplified N-Myc.23 As Wig-1 knockdown significantly decreases N-Myc levels in these cells, we asked if Wig-1 knockdown has a corresponding effect on differentiation. We transfected cells with control siRNA or siRNA targeting Wig-1 or N-Myc for 4 days and recorded morphological changes by phase-contrast imaging (Figure 3a). At this time point, control transfected cells had grown to confluence but remained largely as solitary cells, while cells transfected with N-Myc or Wig-1 siRNA ceased to grow and underwent morphological changes with neurite protrusions that formed network-like patterns between the cells. Moreover, we observed upregulation of the differentiation marker Neuropeptide Y (NPY) at the mRNA level after Wig-1 and N-Myc knockdown, confirming that inhibition of N-Myc expression resulted in differentiation (Figure 3b). We also found an increased fraction of cells in the G1 phase of the cell cycle, in agreement with the observed growth arrest and differentiation (Figure 3c). Wig-1 siRNA 1 gave a more dramatic phenotype than Wig-1 siRNA 2 in all experiments, consistent with its higher efficiency in silencing Wig-1 and downregulating N-Myc expression as shown by western blotting (Figure 1a). To investigate if Wig-1 knockdown induces differentiation in other neuroblastoma cell lines with amplified N-Myc, we knocked down Wig-1 or N-Myc in the cell lines Kelly and LAN-5, and recorded morphological changes as for SK-N-BE(2). We noted differentiation-associated morphology in both Kelly and LAN-5 (Supplementary Figure S3). In Kelly, we also noted significant cell death upon long-term N-Myc or Wig-1 knockdown.
Figure 3.
Wig-1 knockdown causes differentiation in SK-N-BE(2) cells carrying N-Myc amplification. (a) Phase-contrast images of SK-N-BE(2) cells 4 days after siRNA transfection at × 20 magnification. Scale bars indicate 1000 μm. Control transfected cells are confluent and cells are growing as solitary cells with square-like morphology. Wig-1 knockdown using two different siRNA oligos or N-Myc knockdown causes morphological changes associated with differentiation to varying degree. Cells become less confluent and neurite outgrowths appear. Numbers next to images indicate the average percentage of differentiated cells ± standard deviation of three independent experiments, of at least 50 counted cells per experiment. To examine the effect of Wig-1 knockdown on the differentiation-associated marker NPY we transfected cells with siRNA oligos against Wig-1 or N-Myc, harvested cells after 4 days, isolated RNA and performed quantitative real-time PCR. (b) Increased levels of NPY (P=0.025 with siWig-1_1 and 4.7 × 10−6 for siWig-1_2) after Wig-1 knockdown (n=3) compared with control PGK1 levels. Error bars show S.D. (b). To examine the effect or Wig-1 and N-Myc knockdown on cell cycle distribution in SK-N-BE(2) cells, we performed FACS-PI cell cycle analysis 4 days after siRNA transfection. Wig-1 knockdown resulted in a 58% (P=0.0002; siWig-1_1) or 22% (P=0.032; siWig-1_2) increase in the G1 fraction of cells, whereas N-Myc knockdown resulted in a 56% (P=0.0001; siN-Myc) increase (c) (n=5). To confirm that the effect of Wig-1 on differentiation is caused by downregulation of N-Myc, we cotransfected SK-N-BE(2) cells with Wig-1 siRNA and GFP-tagged N-Myc ORF construct lacking the 3′UTR (and therefore not regulated by Wig-1). Cells were fixed and mounted for immunofluorescence 5 days after transfection. (d) shows a GFP-N-Myc ORF-expressing cell (cell borders marked in black) surrounded by non-transfected cells. The GFP-N-Myc-expressing cell remains undifferentiated whereas the surrounding cells that do not express GFP-N-Myc show a differentiation-associated morphology. All images are at × 63 magnification. Scale bars indicate 20 μm. For more examples, see Supplementary Figure 2a
To confirm that the observed effect of Wig-1 knockdown on differentiation was indeed mediated through reduced N-Myc levels, we overexpressed GFP-tagged N-Myc ORF (lacking the 3′UTR regulated by Wig-1), while Wig-1 was knocked down by siRNA. We found that most GFP-positive cells had an undifferentiated phenotype as opposed to their non-transfected, differentiated neighboring cells (Figure 3d; Supplementary Figure S4a). Furthermore, we knocked down Wig-1 in SH-SY5Y neuroblastoma cells that do not carry amplified N-Myc. We did not observe any signs of differentiation upon Wig-1 knockdown in these cells, while differentiation was readily induced by treatment with all-trans retinoic acid (ATRA) as described25 (Supplementary Figures S4b and c). Thus, Wig-1 knockdown triggers differentiation in SK-N-BE(2) cells carrying amplified N-Myc, and this effect is most likely due to attenuated N-Myc expression.
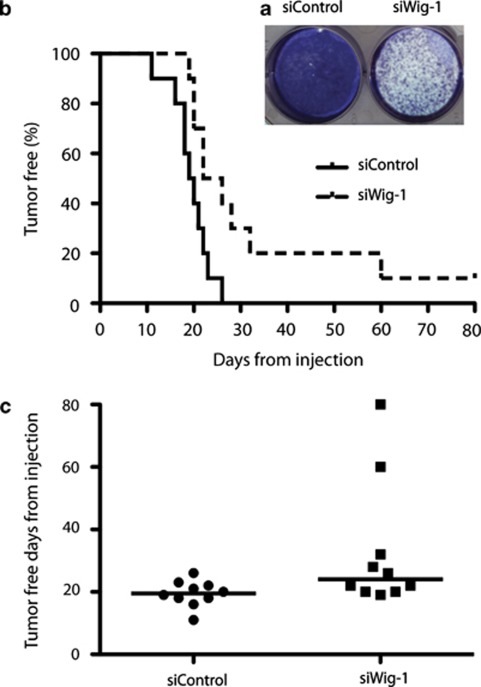
We next investigated if Wig-1 knockdown had any impact on tumor formation in vivo. We first performed a colony formation assay to determine the long-term effect of Wig-1 knockdown by transient siRNA transfection. SK-N-BE(2) cells were transfected with siRNA against Wig-1 or control siRNA, cultured for 8 days post transfection, and stained with Giemsa staining. We noted a clear reduction in the number of colonies after Wig-1 siRNA knockdown as compared with control (Figure 4a). Having established this effect, we transfected SK-N-BE(2) cells with either Wig-1 siRNA or control siRNA and inoculated them subcutaneously in nude mice, using 10 mice for inoculation with siWig-1-transfected cells and 10 mice for inoculation with siControl-transfected cells. Efficient Wig-1 knockdown was confirmed by western blotting in an aliquot of the cells before inoculation (Supplementary Figure S5a). We observed a significantly longer time to tumor take (as defined by a tumor volume of 0.1 ml or more) using Wig-1 siRNA-transfected cells as compared with control siRNA-transfected cells. Average time to tumor take was 24 days in the mice inoculated with Wig-1 siRNA-treated cells, as compared with 19.5 days in the siControl group (Figure 4b and c). We noted a similar difference using tumor volume of 0.2 ml or more as definition of tumor take (Supplementary Figure S5b). One out of ten mice in the siWig-1 xenograft group remained tumor-free for the entire experiment, indicating that even transient loss of Wig-1 may be sufficient to eliminate the tumor-forming potential of the SK-N-BE(2) cells. Once tumors were well established (tumor volume ≥0.2 ml), we noted no further difference in growth rates between the groups (Supplementary Figure S5c), which was expected considering the transient Wig-1 knockdown. In general, transient siRNA transfection does not cause a significant decrease in target protein expression beyond 96 h post transfection, meaning that once tumors were formed they were expected to have reverted to baseline Wig-1 and N-Myc expression levels. Indeed, analysis of several of the tumors confirmed that Wig-1 and N-Myc expression levels were similar in established tumors from both siWig-1 and siControl groups (Supplementary Figure S5d).
Figure 4.
Wig-1 knockdown delays tumor take in nude mice. Transient Wig-1 knockdown is sufficient to cause long-term effects on SK-N-BE(2) cell growth. (a) A colony formation assay with SK-N-BE(2) cells that were transfected either with siRNA targeting Wig-1 or with control siRNA, incubated 8 days after transfection, and fixed and stained with Giemsa staining. The figure shows representative results from one out of four independent experiments (a). Tumor take in the xenograft model is delayed in SK-N-BE(2) cells pre-transfected with siRNA targeting Wig-1 compared with control siRNA. (b) Kaplan–Meier plots in which the median time to tumor take defined by a tumor volume of 0.1 ml is 19.5 days in cells pre-transfected with siControl and 24 days in cells pre-transfected with siWig-1 (P=0.012). (c) shows the same data represented in a scatter plot (P=0.025)
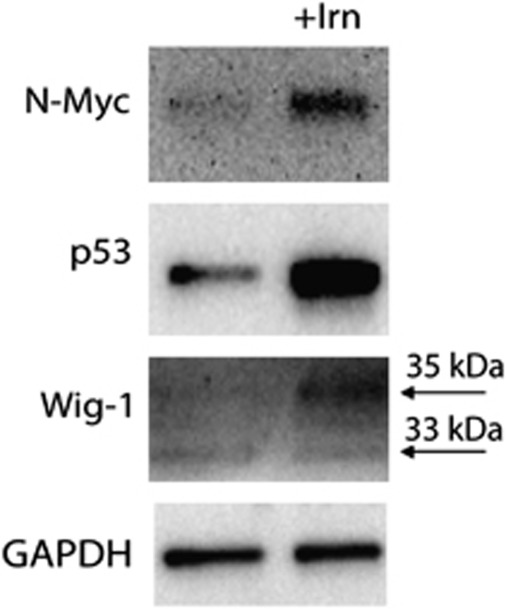
To further explore the biological significance of a p53-Wig-1-N-Myc axis, we hypothesized that N-Myc may be induced in stress situations that will ultimately be resolved, in order to facilitate cell cycle re-entry. To investigate if N-Myc is induced after p53-activating stress, we treated wild-type p53-carrying IMR-32 neuroblastoma cells with the chemotherapeutic drug Irinotecan (Irn). We detected increased p53 and Wig-1 protein levels, as expected, and also observed a marked induction of N-Myc expression (Figure 5).
Figure 5.
Upregulation of N-Myc upon p53 and Wig-1-inducing stress. Moderate stress induced by the topoisomerase 2 inhibitor Irn induces p53 and Wig-1 at the protein level, which leads to induction of N-Myc protein in the wild-type p53-carrying neuroblastoma line IMR-32
Discussion
In this study we have identified Wig-1 as a novel and crucial regulator of N-Myc mRNA. We show that Wig-1 binds and regulates N-Myc mRNA through an ARE-containing region in the N-Myc 3′UTR, and that Wig-1 knockdown leads to reduced N-Myc levels followed by differentiation in SK-N-BE(2) cells with amplified N-Myc. We chose SK-N-BE(2) cells for the differentiation experiments because they are known to differentiate upon downregulation of N-Myc.23 As ARE-binding proteins such as Wig-1 generally have many target mRNAs,16 we cannot formally exclude that some other putative Wig-1-regulated mRNA besides N-Myc may influence differentiation after Wig-1 knockdown. However, N-Myc is likely to be a critical Wig-1 target for induction of differentiation as the SH-SY5Y neuroblastoma cells that do not express N-Myc fail to differentiate upon Wig-1 knockdown (Supplementary Figure S3b and c). In addition, the induction of differentiation-associated morphology of Wig-1-depleted SK-N-BE(2) cells can be prevented by ectopic expression of N-Myc ORF (Figure 3d, Supplementary Figure S2a), indicating that N-Myc alone, in the absence of other putative Wig-1 targets, is sufficient to maintain an undifferentiated phenotype.
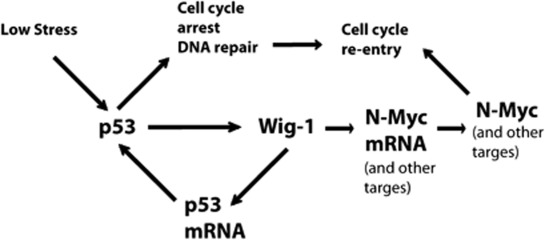
The fact that Wig-1 – target of the tumor suppressor p53 and stabilizer of p53 mRNA13 – sustains expression of the oncogene N-Myc is a striking example of the intricate regulatory network that governs cellular life-and-death decisions. While high levels of cellular stress in general are associated with a p53-dependent apoptotic response, low or moderate stress may induce p53-dependent cell cycle arrest and DNA repair followed by cell cycle re-entry.1 The marked induction of N-Myc expression along with p53 and Wig-1 expression after treatment of wild-type p53-carrying IMR-32 neuroblastoma cells with the chemotherapeutic drug Irn (Figure 5) suggests that upregulation of N-Myc via Wig-1 may have an important role in promoting the recovery and cell survival after p53-induced cell cycle arrest in response to stress. At the same time, Wig-1-mediated stabilization of p53 mRNA will lower the threshold for subsequent p53 activation and thus increase cellular alertness to DNA damage and other types of stress (Figure 6). This notion is supported by the fact that the Wig-1 promoter has a perfect p53 response element,4, 6 which is typically found in promoters of cell cycle arrest-associated p53 target genes like p21.26 Under physiological conditions, it is likely that the regulation of N-Myc by Wig-1 is most important in actively cycling cells relying on N-Myc for their survival, such as neuronal and embryonic stem cells. This hypothesis is consistent with the observed enrichment of Wig-1 in stem cells and indications of a role for Wig-1 in stem cell maintenance.9, 27 Indeed, we have noted that Wig-1 expression is required for stem cell survival, as demonstrated by the early embryonic lethality of Wig-1 knockout mice (Vilborg et al., unpublished results). Thus we propose that Wig-1 has a pro-survival role in cells by sustaining N-Myc expression, and that the simultaneous enhancement of p53 mRNA levels provides a safe-guard mechanism for rapid induction of p53 protein upon stress. The details of such an intricate regulation remain to be elucidated. It is interesting to note that the region of N-Myc 3′UTR that is required for Wig-1-mediated regulation consists of the pentamer AUUUA with an AU-rich region surrounding it (this study), while the ARE bound by Wig-1 in the p53 3′UTR is a stretch of continuous U's.13 Although there are no known preferences for ARE-binding proteins with regard to different AREs,16 one could speculate that some modification of the Wig-1 protein might change its relative affinities for the N-Myc and the p53 AREs under different cellular conditions.
Figure 6.
The p53-Wig-1-N-Myc axis. Model for p53-dependent cell cycle arrest and DNA repair induced by low to moderate cellular stress, followed by cell cycle re-entry and resumed cell growth. Wig-1 may facilitate cell cycle progression and survival post stress by sustaining levels of growth-promoting mRNAs such as N-Myc, while simultaneously stabilizing p53 mRNA to facilitate subsequent p53 activation in case of remaining damage. It is also conceivable that low levels of p53 in unstressed cells can promote survival via induction of Wig-1. In case of severe stress, cells will die from p53-mediated apoptosis
Finally, the observation that transient Wig-1 knockdown in the aggressive human neuroblastoma SK-N-BE(2) cells is sufficient to impair tumor development in nude mice, and, in 1 case out of 10, completely prevent tumor formation, raises interesting therapeutic possibilities. It will be important to investigate the effect of Wig-1 knockdown in vivo on already established tumors. A Wig-1-based therapeutic strategy could either aim at specifically inhibiting Wig-1 binding to N-Myc mRNA, or more broadly targeting Wig-1 expression or RNA binding. The latter approach is most likely to be useful in neuroblastoma with mutant p53. p53 mutations occur in a subset of recurrent and therapy-resistant neuroblastoma.28 Mutant p53 has gain-of-function activities that promote tumor growth.29 We have found that Wig-1 knockdown decreases levels of mutant p53 in the SK-N-BE(2) cells (Supplementary Figure S6). Therefore, therapeutic targeting of Wig-1 expression or RNA binding in mutant p53-carrying tumors may have the added benefit of inhibiting mutant p53 gain-of-function activities. Thus, our findings strongly suggest that Wig-1 knockdown, or interference with Wig-1 RNA binding in general or N-Myc mRNA binding in particular, would be fruitful strategies for treatment of aggressive and recurrent neuroblastoma, which is a significant cause of death among young children.
Materials and Methods
Cell culture, transfection and colony formation assay
For details on p53 and N-Myc status of neuroblastoma cell lines used in this study, see Supplementary Table S1. Cells were grown in IMDM (HyClone, Logan, UT, USA) with 1% glutamine (Sigma Aldrich, St. Louis, MO, USA), 10% FBS (HyClone) and 50 μg/ml Plasmocin (InVivoGen, San Diego, CA, USA). siRNA transfections were performed in media without antibiotics using HiPerFect (Qiagen, Hilden, Germany) according to the manufacturer's protocol using the following siRNA: wig1_1 wig1_2 N-Myc_5 and all stars negative control (all from Qiagen), at a concentration of 10 nM (for Quantigene assays, luciferase assays, mouse xenograft experiments and three out of six of the differentiation experiments in Kelly cells) or 20 nM (for western blotting, differentiation assays, and colony formation assays). For differentiation assays in SH-SY5Y cells harvested 5 days after transfection, and LAN-5 and Kelly cells harvested 6 days after transfection, cells were transfected with siRNA twice, the day following plating and 2 days after the first transfection. For differentiation rescue assays, SK-N-BE(2) cells were plated in 6-well plates and transfected the following day with pEGFP-C1-N-Myc, which was generated by subcloning the N-Myc ORF from pLenti-III-Tet-h-MYCN-2A-dEGFP (Capital Biosciences, Rockville, MD, USA) into the BglII site of the pEGFP-C1 vector by TOPO cloning using the pCR4 vector (Invitrogen) according to the manufacturer's protocol and using BglII restriction site-containing primers: nmycF_bgl2 5′-AGATCTATGCCGAGCTGCTCCAC-3′, and nmycR_bgl2 5′-AGATCTAGCAAG-TCCGAGCGTGTT-3′. After 6–8 h the cells were transfected with 40 nM wig1_1 siRNA as above. The cells were cultured for an additional 4 days and were then fixed and mounted for immunofluorescence as described in Vilborg et al.13 and photographed using a bright-field microscope. For the biotin pulldown assay, HCT116 cells were transfected using PEI according to standard protocol with pCMVtag2bhWig-1 or pCMVtag2bhWig-1ZF1pm.10 For ATRA (Sigma Aldrich) treatment, SH-SY5Y cells were incubated with 10 nM ATRA for 5 days. For colony formation assays, SK-N-BE(2) cells were cultured 8 days post transfection, fixed and stained with Giemsa (Sigma Aldrich) according to the manufacturer's protocol. For mouse xenograft experiments, SK-N-BE(2) cells were harvested 24 h post transfection. Inoculations were performed as described below. To ensure transfection efficiency, a few wells of each group were maintained for an additional 24 h, after which the cells were harvested for western blotting as described below. For analysis of p53, N-Myc, and Wig-1 expression after stress, IMR-32 cells were transfected with 40 nM siControl 1 day post plating, treated with 0.4 μM Irn, kept in culture for an additional 2 days, and then harvested for western blotting as described below.
Western blotting
Cells were transfected as indicated, cultured for 48–72 h, harvested, and lysed in NP40 lysis buffer (100 mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 1% Protease Inhibitor Cocktail (Sigma Aldrich)). Tumor samples were washed in PBS and homogenized for 1 min using disposable pellet pestles (Kontes, Vineland, NJ, USA) in 100 to 300 μl of lysis buffer as above. Lysates were cleared by centrifugation, and 30–50 μg of protein were loaded on 10% SDS-PAGE gels (Invitrogen) and run in 1 × MOPS (Invitrogen). Gels were blotted to nitrocellulose membranes using the iBlot (Invitrogen), blocked in 5% milk for 1 h, and incubated with the indicated antibodies. Wig-1 rabbit polyclonal 1 : 1000,10 or 1 : 500 (GeneTex, Irvine, CA, USA), N-Myc: mouse monoclonal (NCM II 100) 1 : 200 Santa Cruz Biotechnology (Santa Cruz, CA, USA), GAPDH: rabbit polyclonal (Fl-335) 1 : 2000 Santa Cruz Biotechnology, p53: mouse monoclonal (DO-1) 1 : 1000 Pharmingen (San Diego, CA, USA), β-actin: mouse monoclonal (Sigma Aldrich) 1 : 5000. Primary antibodies were detected using HRP-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ, USA) and Super Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific, Lafayette, CO, USA) with a CCD camera (Fujifilm, Tokyo, Japan).
Quantigene assays and quantitative real-time PCR
Cells were transfected the day after plating, cultured an additional 48 h, and then lysed and analyzed with the Quantigene 2.0 assay (Panomics, Fremont, CA, USA, QS0008) according to the manufacturer's protocol using probes for N-Myc (SA-10008) and GAPDH (SA-10001) (Panomics). For quantitative real-time PCR, total RNA was extracted using Trizol (Invitrogen). Real-time PCR was carried out in an Applied Biosystems 7500 real-time PCR machine using transcript-specific TaqMan Gene Expression Assays (NPY: Hs00173470_m1, PGK1: Hs99999906_m1) (Applied Biosystems, Foster City, CA, USA).
Luciferase assays and Biotin pulldown
For N-Myc 3′UTR primer sequences see Figure S1. psiCheck2 plasmid constructs were generated by PCR subcloning into the TOPO10 cloning vector (Invitrogen) of the various N-Myc 3′UTR constructs (see Supplementary Figure S1 for details) or, as control, the p53 3′UTR lacking the U-rich region. The latter was cloned using p53 3′UTR_F (5′-CAATGTCTCGAGACATTCTCCACTTCTTGTTCCC-3′) and p53 3′UTR_R (5′TAGTCAGCGGCCGCCACCCCTCAGACACACAGG-3′) using pCS2+ -U13 as template. The 3′UTRs were then excised using Not1 and Xho1, and cloned into the psiCheck2 vector (because of the presence of an Xho1 site in the p53 3′UTR the construct lacks the 346 most 3′ nucleotides of the p53 3′UTR). These plasmids were then transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol in HCT116 cells that had been plated 24 h earlier and transfected with siRNA (siControl or siWig-1_2) 8 hours before DNA transfection. Cells were cultured for an additional 20 h and were then analyzed by Stop and Glo luciferase kit (Promega, Madison, WI, USA) according to the manufacturer's protocol. Each experiment was performed in triplicate.
For biotin pulldown assays, either the psiCheck2 constructs from above were used as templates, or pMirReport vector alone or with the N-Myc 3′UTR downstream of luciferase (kind gift from M. Corcoran) were used. Vectors were linearized with Not1, column purified using columns from gel extraction kit (Qiagen) according to the manufacturer's protocol, and were then used as templates for in vitro transcription using T7 or Sp6 polymerase (Roche, Indianapolis, IN, USA) in the presence of biotin labeling mix containing biotinylated UTPs (Roche) according to the manufacturer's protocol. Approximately 5 μg of probes were then incubated with approximately 500 μg of cell lysate of HCT116 cells transiently transfected with pCMVtag2bhWig-1 or pCMVtag2bhWig-1ZF1pm,10 lysed and used for the pulldown as described in.13
Flow cytometry and propidium iodide staining
SK-N-BE(2) cells were harvested 96 h after transfection, resuspended in 3 ml PBS and fixed in 4 ml 95% ethanol at 4 °C overnight. Cells were pelleted by centrifugation (500 × g for 5 min at 4 °C) resuspended in 100 μl PI staining solution containing 0.05 mg/ml PI and 0.25 mg/ml RNaseA in PBS and stained for 30 min at 37 °C. An additional 300 μl PBS was added to the sample, which was then analyzed by FACS using a calibur flow cytometer (Becton Dickinson, San Diego, CA, USA) using the FCS express v3 software (De Novo Software, Los Angeles, CA, USA).
Animals and in vivo tumorigenicity
Female Sca:NMRI nu/nu mice (5–6 weeks old) (Charles River, Wilmington, MA, USA) were inoculated subcutaneously in the right hind flank with 20 × 106 of SK-N-BE(2) siWig-1 (n=10) or SK-N-BE(2) siControl (n=10) cells under general anesthesia. Tumor growth was assessed with digital calipers daily and up to 80 days after inoculation. Tumor volume was calculated by the formula: (width)2 × length × 0.44. A tumor was considered to be established once it had reached a volume of 0.1 ml (mean 0.13 ml). The animals were killed and the tumors were weighed 7 days after the tumor volume reached 0.2 ml. The animals were housed at a maximum of five per cage in an enriched environment and were provided with sterile food and water ad libitum. The animal experiments were approved by the regional ethics committee for animal research (N304/08) and were in accordance with the Animal Protection Law (SFS 1988 : 534), the Animal Protection Regulation (SFS 1988 : 539), and the Regulation for the Swedish National Board for Laboratory Animals (SFS 1988 : 541).
Microscopy
For immunofluorescence staining, SK-N-BE(2) cells were plated on coverslips, transfected with siRNA as indicated above 24 h past plating, cultured for an additional 4 days, and then washed in PBS, fixed in 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100. Slides were mounted using VECTASHIELD HardSet Mounting Medium with DAPI, H-1500 (Vector Laboratories, Burlingame, CA, USA), and examined under a Axioplan II Imaging (Carl Zeiss, Goettingen, Germany) microscope equipped with a 63 × /1.40 immersion objective (Plan-Apochromat, Carl Zeiss) using an HBO-100 mercury lamp and filter sets for FITC and DAPI or HAL 100 light source for bright-field pictures. Images were captured, processed, and analyzed using an AxioCam MRm camera (Goettingen, Germany) and Axio Vision 4.8 software (Carl Zeiss). For other bright-field imaging, cells were grown in 6-well plates and imaged using an Olympus (Center Valley, PA, USA) CK2 microscope equipped with an LWD CD Arch × 20 objective. Digital images were collected with a DeltaPix (Maalov, Denmark) Invenio camera. All imaging was done at room temperature and images were further processed with Photoshop CS5 (Adobe, San Jose, CA, USA).
Statistical analysis
Time to tumor take was compared using log-rank tests for survival and comparison between animal groups was analyzed by the Mann–Whitney U-test, all tests were two-sided. All other statistical tests were performed using two-sided Student's t-test.
Acknowledgments
This work was supported by grants from the Swedish Cancer Society, Swedish Research Council (VR), the Gustaf V Jubilee Fund, the Robert Lundberg Research Foundation, and Karolinska Institutet.
Glossary
- ARE
AU-rich element
- Irn
irinotecan
- NPY
neuropeptide Y
- Wig-1
wild-type p53-induced gene 1
KGW is cofounder, shareholder and board member of Aprea AB, a company that develops p53-based cancer therapy.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored. Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Varmeh-Ziaie S, Okan I, Wang Y, Magnusson KP, Warthoe P, Strauss M, et al. Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene. 1997;15:2699–2704. doi: 10.1038/sj.onc.1201454. [DOI] [PubMed] [Google Scholar]
- Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, et al. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellborg F, Qian W, Mendez-Vidal C, Asker C, Kost-Alimova M, Wilhelm M, et al. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene. 2001;20:5466–5474. doi: 10.1038/sj.onc.1204722. [DOI] [PubMed] [Google Scholar]
- Varmeh-Ziaie S, Ichimura K, Yang F, Rabbits P, Collins VP. Cloning and chromosomal localization of human WIG-1/PAG608 and demonstration of amplification with increased expression in primary squamous cell carcinoma of the lung. Cancer Lett. 2001;174:179–187. doi: 10.1016/s0304-3835(01)00699-1. [DOI] [PubMed] [Google Scholar]
- Vilborg A, Bersani C, Wilhelm MT, Wiman KG. The p53 target Wig-1: a regulator of mRNA stability and stem cell fate. Cell Death Differ. 2011;18:1434–1440. doi: 10.1038/cdd.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. Stemness': transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Mendez Vidal C, Prahl M, Wiman KG. The p53-induced Wig-1 protein binds double-stranded RNAs with structural characteristics of siRNAs and miRNAs. FEBS Lett. 2006;580:4401–4408. doi: 10.1016/j.febslet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Mendez-Vidal C, Wilhelm MT, Hellborg F, Qian W, Wiman KG. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res. 2002;30:1991–1996. doi: 10.1093/nar/30.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahl M, Vilborg A, Palmberg C, Jornvall H, Asker C, Wiman KG. The p53 target protein Wig-1 binds hnRNP A2/B1 and RNA Helicase A via RNA. FEBS Lett. 2008;582:2173–2177. doi: 10.1016/j.febslet.2008.04.065. [DOI] [PubMed] [Google Scholar]
- Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci USA. 2009;106:15756–15761. doi: 10.1073/pnas.0900862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Wilhelm MT, Wiman KG. Regulation of tumor suppressor p53 at the RNA level. J Mol Med. 2010;88:645–652. doi: 10.1007/s00109-010-0609-2. [DOI] [PubMed] [Google Scholar]
- Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles. Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RA. mRNA stability control: a clandestine force in normal and malignant hematopoiesis. Leukemia. 2007;21:1158–1171. doi: 10.1038/sj.leu.2404656. [DOI] [PubMed] [Google Scholar]
- Manohar CF, Short ML, Nguyen A, Nguyen NN, Chagnovich D, Yang Q, et al. HuD, a neuronal-specific RNA-binding protein, increases the in vivo stability of MYCN RNA. J Biol Chem. 2002;277:1967–1973. doi: 10.1074/jbc.M106966200. [DOI] [PubMed] [Google Scholar]
- Ross RA, Lazarova DL, Manley GT, Smitt PS, Spengler BA, Posner JB, et al. HuD, a neuronal-specific RNA-binding protein, is a potential regulator of MYCN expression in human neuroblastoma cells. Eur J Cancer. 1997;33:2071–2074. doi: 10.1016/s0959-8049(97)00331-6. [DOI] [PubMed] [Google Scholar]
- Johnsen JI, Kogner P, Albihn A, Henriksson MA. Embryonal neural tumours and cell death. Apoptosis. 2009;14:424–438. doi: 10.1007/s10495-009-0325-y. [DOI] [PubMed] [Google Scholar]
- Edsjo A, Holmquist L, Pahlman S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin Cancer Biol. 2007;17:248–256. doi: 10.1016/j.semcancer.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–197. doi: 10.1016/j.bbrc.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart CA, Cheng AJ, Madafiglio J, Kavallaris M, Mili M, Marshall GM, et al. Effects of MYCN antisense oligonucleotide administration on tumorigenesis in a murine model of neuroblastoma. J Natl Cancer Inst. 2003;95:1394–1403. doi: 10.1093/jnci/djg045. [DOI] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH. Outcomes of p53 activation – spoilt for choice. J Cell Sci. 2006;119 (Pt 24:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Keshelava N, Zuo JJ, Chen P, Waidyaratne SN, Luna MC, Gomer CJ, et al. Loss of p53 function confers high-level multidrug resistance in neuroblastoma cell lines. Cancer Res. 2001;61:6185–6193. [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.