Abstract
The effects of co-evolution with lytic phage on bacterial virulence-related traits are largely unknown. In this study we investigate the incidence of the mucoid phenotype of the bacterium Pseudomonas fluorescens SBW25 in response to co-evolution with the lytic phage phi2 (φ2). The mucoid phenotype of Pseudomonas spp. is due to overproduction of alginate and is a considerable virulence factor contributing to the intractability of infections most notably in cystic fibrosis (CF) lung, but also in pathogenic infections of plants. Our data show that this phenotype can evolve as an adaptive response to phage predation and is favoured under specific abiotic conditions, in particular a homogenous spatial structure and a high rate of nutrient replacement. The mucoid phenotype remains partially sensitive to phage infection, which facilitates ‘apparent competition' with phage-sensitive competitors, partially offsetting the costs of alginate production. Although P. fluorescens SBW25 is not a pathogen, several key characteristics typical of Pseudomonas aeruginosa clinical isolates from CF lung were noted, including loss of motility on mucoid conversion and a high rate of spontaneous reversion to the wild-type phenotype. Although the genetic mechanisms of this phenotype remain unknown, they do not include mutations at many of the commonly reported loci implicated in mucoid conversion, including mucA and algU. These data not only further our understanding of the potential role phage have in the ecology and evolution of bacteria virulence in both natural and clinical settings, but also highlight the need to consider both biotic and abiotic variables if bacteriophages are to be used therapeutically.
Keywords: host–parasite antagonistic co-evolution, microbial model system, lytic bacteriophage, mucoid phenotype, alginate, apparent competition
Introduction
Bacteriophages (phages) are known to have important ecological and evolutionary consequences for bacteria, driving, for example, population dynamics and niche specialization (Rodriguez-Valera et al., 2009). Unsurprisingly, phages may also have an important role in the evolution of bacterial pathogenicity, most obviously by transferring virulence genes such as toxins or attachment proteins by lysogenic conversion or genetic transduction (Brussow and Desiere, 2001; Boyd and Brussow, 2002; Canchaya et al., 2003). However, the effects of phage on the ecology and pathogenicity of their bacterial hosts may also be indirect if they select for virulence-related resistance traits in bacteria (Friman et al., 2011). One such virulence trait is mucoid or alginate production in Pseudomonas spp., which may have a protective role against virulent phage infection (Hammad, 1998; Labrie et al., 2010).
The mucoid phenotype of Pseudomonas sp. is considered to be a global adaptive stress response to a range of adverse environmental conditions (Terry et al., 1992; Wood et al., 2006). This phenotype is typically characterized by shiny, raised, opaque, blob-like colonies on agar plates, which is due to overproduction of alginate. This bacterial exopolysaccharide is a considerable virulence factor contributing to the intractability of Pseudomonas aeruginosa infections most notably in the cystic fibrosis (CF) lung (Pedersen, 1992; Ramsey and Wozniak, 2005), but also pathogenic Pseudomonas spp. infections of plants (Fett and Dunn, 1989; Keith et al., 2003). The conversion of P. aeruginosa from a non-mucoid (NM) to a mucoid overproducing phenotype is typically associated with poor patient prognosis and mortality (May et al., 1991), and this phenotypic switch to overproduction of alginate is considered an adaptive response to the lung environment that provides the host cell with a protective capsule that facilitates inter alia salt stress, immune evasion (opsonization and phagocytosis) and diffusion of antibiotics (thus limiting treatment options) (Kharazmi, 1991; Pedersen et al., 1991; Leid et al., 2005). Given the clinical relevance of the mucoid phenotype and the resurgence of interest in phage as a therapeutic option against bacterial infections (Pirnay et al., 2010; Fernebro, 2011), understanding the link between phages and the mucoid phenotype is of interest.
A phenotypic link between capsule formation or exopolysaccharide production and resistance to phage adsorption has been shown for a range of bacteria (Ohshima et al., 1988; Labrie et al., 2010). For example, bacteriophages isolated from mucoid clinical isolates of P. aeruginosa have been shown to convert NM phenotypes to mucoid (Miller and Rubero, 1984). Another study that started as a phage typing experiment reported similar results: mucoid phenotypes were isolated from the edges of lytic phage plaques and this phenotype was only observed in the presence of phage (Martin, 1973). However, the adaptive significance of the mucoid phenotype resulting from phage-imposed selection and whether or not this phenotype is genetically determined remain unclear. Furthermore, the incidence of bacteriophage (Podoviridae)-encoded alginate lyases (Glonti et al., 2010) would suggest that alginate may be a common adaptive response to phage predation, which in turn has selected for this phage counter strategy (Labrie et al., 2010).
To investigate the link between phage-imposed selection and the mucoid phenotype, we used the well-studied Pseudomonas fluorescens SBW25-phi2 system (Buckling and Rainey, 2002a, 2002b), where both species coexist in both nutrient broth and soil (Gómez and Buckling, 2011) environments as a result of extensive co-evolution of resistance and infectivity traits. Crucially, recent in vitro work has shown a link between the presence of mucoid phenotypes and phages (as is the case for P. aeruginosa and other bacteria; Sapelli and Goebel, 1964; Miller and Rubero, 1984; Buckling and Rainey, 2002a; Fischer et al., 2004; Vogwill et al., 2011), although these mucoid phenotypes do not seem to persist for long (Buckling and Rainey, 2002a; Brockhurst et al., 2003). We investigated the evolution and maintenance of the P. fluorescens mucoid phenotype in experimental microcosms resulting from phage-imposed selection under a range of abiotic conditions that can have a major role in the evolution of morphological diversity in this system, that is, spatial heterogeneity (whether tubes are shaken or static (Rainey and Travisano, 1998)) and rate of nutrient replacement (transfer regime (Buckling et al., 2000)).
Materials and methods
Study organisms
Pseudomonas phage phi-2 (Φ2) (GenBank accession number FN594518) is a member of the Podoviridae family of viruses (Paterson et al., 2010). P. fluorescens SBW25 (GenBank accession number AM181176) is a non-pathogenic, Gram-negative bacterium of the Gammaproteobacteria that was originally isolated from the leaf surface of a sugar beet plant grown at the University Farm, Wytham, Oxford, UK (Rainey and Bailey, 1996).
Experimental design
Selection experiments were conducted in liquid microcosms (25-ml glass vials containing 6 ml of King's Medium B (KB) liquid growth media). In addition to manipulating the presence/absence of phages, we manipulated two variables that are known to have a key role in the evolutionary dynamics of this system: spatial structure and rate of nutrient replacement. Eight different treatment regimes were set up, with six replicate populations per treatment (see Table 1). Co-evolving populations were established by inoculating liquid microcosms with 108 clonal ancestral bacteria and 105 clonal ancestral phage obtained from a single plaque of phi2 (φ2) (Buckling and Rainey, 2002b). Control populations of evolving bacteria were established by inoculating microcosms with 108 clonal ancestral bacteria only. Spatially structured populations with a vertical oxygen gradient were established by incubating microcosms with loosely capped lids in a static incubator at 28 °C (Rainey and Travisano, 1998). Populations with no spatial structure and no oxygen gradient were established by incubating microcosms with loosely capped lids at 28 °C, shaking at 170 r.p.m. The time spent in prolonged stationary phase (rate of nutrient replacement) was altered by transferring populations from treatments 1 to 4 every 24 h and from treatments 5 to 8 every 48 h. At the appropriate transfer time (either 24 or 48 h), populations were homogenized by vortexing and 1% of each population was transferred to a fresh microcosm; this procedure was performed for all treatments for a total of 24 days. At every transfer, a sample of each population was stocked at −86 °C in 20% (v/v) glycerol solution and a sample of phage was isolated by adding 10% chloroform to 900 μl of culture, and centrifuging at 13 000 r.p.m. for 3 min, leaving viable phage in the supernatant.
Table 1. Overview of treatments and incidence of mucoid phenotype.
| Treatment | Phage presence (+) or absence (−) | Transfer (h) | Spatial structure | Incidence of mucoid phenotype |
|---|---|---|---|---|
| 1 | + | 24 | Shaking | 5/5a |
| 2 | − | 24 | Shaking | 0/6 |
| 3 | + | 24 | Static | 2/6b |
| 4 | − | 24 | Static | 0/6 |
| 5 | + | 48 | Shaking | 1/6b |
| 6 | − | 48 | Shaking | 0/6 |
| 7 | + | 48 | Static | 1/6b |
| 8 | − | 48 | Static | 0/6 |
By day 10 all populations of Treatment-1 were positive for the mucoid phenotype, which remained a fixed feature of the population for the duration of the experiment.
Mucoid phenotypes were also detected in a small number of populations for these treatments; however, they were not a fixed feature of the populations over time and were only infrequently encountered at a low number relative to total population cells densities.
Measuring densities and colony phenotypes
All replicate populations from each treatment group were serially diluted in 1 × M9 salt solution and plated on KB agar plates in triplicate at regular intervals. Bacterial populations were plated every second day for Treatment-1 (the treatment in which mucoid colonies reached high frequencies) and every 4 to 6 days for all other treatments. The total cell densities (colony-forming units (CFUs ml −1)) and the numbers of each different phenotype were recorded at each time point.
Co-evolution
We wanted to determine whether co-evolution between bacteria and phage (that is, reciprocal evolution of bacterial resistance and phage infectivity) occurred in the treatment where mucoids dominated (Treatment-1). We determined resistance of bacteria to contemporary phage, and phage from two transfers in the past and two transfers in the future; the difference in infectivity over this transfer period provides a measure of phage evolution (Brockhurst et al., 2003). Twenty bacteria were randomly selected from each population from Treatment-1, at days 4, 12 and 20, to test for co-evolution. Bacteria were streaked across a perpendicular line of phage and assayed for resistance or sensitivity after overnight incubation at 28 °C as described elsewhere (Brockhurst et al., 2003; Morgan et al., 2005). The ancestral strain was used as control on all plates.
Bacterial resistance and sensitivity
We used a more sensitive measure than above to assess the resistance of specific bacterial isolates to specific phage populations. In brief, overnight cultures of test bacteria (10% v/v) were added to soft agar (0.3% w/v agar) and this mixture was overlaid on hard agar plates. Ten microlitres of phage (either ancestral or sympatric to the bacteria being assayed) were spotted directly onto the overlay, incubated at 28 °C, and checked after 8 and 24-h incubation.
Growth rate of mucoid phenotypes
We assessed the growth rate of both mucoid and NM colonies (n=15 for each; five from each of three Treatment-1 replicates) according to standard procedure: assays were set up in triplicate and bacterial isolates were reconditioned from stock by growing overnight. Sixty microlitres of overnight culture were used as starting inocula for each assay; microcosms were sampled at regular intervals, serially diluted and plated on KB agar. Total cell densities and any changes in phenotypes were recorded. The growth rate or generation time of each isolate was calculated according to the formula g=t/n, where g is generation time, t=time and n=logNf−logN0/log2, where logNf is final cell density and logN0 is the starting cell density. For assays with phage, 105 particles of sympatric phage were added to the experiment.
Stability of phenotypes
To analyse the reversion rate and phenotypic stability of different phenotypes, single colonies were picked from KB agar plates, serially diluted and re-plated on KB agar. The proportion of each phenotype was recorded for each single colony type.
Competition experiments
Competition experiments were performed according to Lopez-Pascua and Buckling (2008). Microcosms were inoculated with equal densities of a marked strain of ancestral P. fluorescens SBW25-lacZ (Zhang and Rainey, 2007) and the focal competitor phenotype (previously grown overnight to insure all competitors are in the same physiological state), and grown (competed) overnight at 28 °C. Densities of the marked strain and competing phenotype were calculated at the start and end of the assay by plating onto Luria–Bertani medium supplemented with X-gal. Each assay was performed in triplicate and fitness (W) was taken as the ratio of the estimated Malthusian parameters (m) of each competing type, m=ln(Nf/N0), where N0 is the starting density and Nf is the final density (Lenski, 1991).
Alginate assay
Bacteria were assayed for alginate production according to methods outlined by Mathee et al. (1999). In brief, bacterial isolates were grown overnight at 28 °C. Cells were removed and supernatants were collected by centrifugation. Alginate was precipitated using an equal volume of 2% (w/v) cetylpridinium chloride followed by centrifugation at 10 000 r.p.m. for 10 min at room temperature. The purified alginate was mixed with a 1 ml solution of borate/sulphuric acid reagent and 30 μl of carbazole reagent was added. The mixture was heated to 55 °C for 30 min and alginate concentration was determined from absorbance measured at 500 nm using a spectrophotometer.
Motility assays
Swimming (flagella-mediated) motility assays (0.3 w/v agar plates) were performed on a range of phenotypes as outlined by Taylor and Buckling (2010). Plates were assayed after 10-h incubation at 28 °C.
Fluctuation tests
We used modified fluctuation tests to estimate bacterial mutation rates (Pal et al., 2007). Six microcosms per population were inoculated with 100–1000 bacterial cells and were allowed to grow for 24 h shaking in a 28 °C incubator. Final cell density was determined by plating dilutions on non-selective solid medium (KB). Number of mutants was estimated by plating 60 μl of each culture on solid selective medium (KB agar plates supplemented with rifampicin (100 g ml −1) or streptomycin (50 g ml −1)). Jones median estimator was used to calculate mutation rate from the average and median frequency of mutant colonies (Rosche and Foster, 2000).
Genetics of mucoid phenotype
DNA was extracted from 20 bacterial isolates representative of Fixed Mucoid (FM, n=4), Unstable Mucoid (UM, n=6), Wrinkly Spreader (WS, n=5) and SMooth (SM, n=5) phenotypes from Treatment-1 and ancestral SBW25 using the Qiagen DNeasy Blood and Tissue kit (Qiagen Ltd, West Sussex, UK). Based on a comprehensive review of the literature, a candidate gene approach was implemented to identify the most likely sites of mutations involved in conversion to the mucoid phenotype. PCR primers were designed against relevant gene sequences obtained from the database (www.pseudomonas.com) listed in Table 2. PCR products were cleaned using the QIAquick PCR purification kit and sent for commercial sequencing (Geneservice, Oxford, UK). All sequences were analysed using MEGA (Tamura et al., 2007).
Table 2. Overview of genes sequenced and their function.
| Locus tag | Gene | Synonyms | Function |
|---|---|---|---|
| PFLU_0088 | kinB/algB | Regulatory two-component system (2CS) region that is involved in alginate biosynthesis | |
| PFLU_0979 | algA | GMP, PMI, manA | The two enzymatic activities of algA, GMP and PMI, are localized at the N- and C-terminals of the protein, respectively. GMP catalyzes the conversion of mannose-1-phosphage to GDP–mannose and PMI catalyzes the conversion of mannose-1-phosphate to GDP–mannose, which are required for the synthesis of alginate and the A-band of LPS |
| PFLU_0987 | algK | Alginate polymerization | |
| PFLU_0988 | alg44 | Alginate polymerization | |
| PFLU_0990 | algD | GMD GDP–mannose dehydrogenase | The alginate biosynthetic operon is under the transcriptional control of the algD promoter. algD encodes GMD, which is required for A-band LPS synthesis and irreversible oxidation of GDP–D-mannose to GDP–D-mannuronic acid (activated precursor for alginate polymerization) |
| PFLU_1467 | algU | algT or sigma factor-22 (σ22) | Positive regulator of alginate biosynthetic operon |
| PFLU_1468 | mucA | Negative regulator of alginate biosynthetic operon | |
| PFLU_1469 | mucB | Negative regulator of alginate biosynthetic operon | |
| PFLU_1471 | mucD | Negative regulator of alginate biosynthetic operon | |
| PFLU_4443 | FleQ | Regulates flagellum biosynthesis and is negatively regulated by algU | |
| PFLU_4744 | algZ | Alginate polymerization | |
| PFLU_5986 | algC | PMM, PGM, GMP | Encodes the bifunctional enzyme PMM/PGM that is required for both alginate biosynthesis, and addition of glucose and or/rhamnose into the growing core of LPS (O-side chains of LPS) |
Statistical analyses
All statistical analysis was conducted using JMP and Minitab. Where possible, non-normal data were transformed or alternatively a non-parametric test equivalent was implemented. Variables contributing to the incidence of the mucoid phenotype were analysed using a nominal logistic model in JMP fitting incidence of mucoid phenotype (Yes/No) throughout the experiment (the four common time points for all treatments) as the response variable, and spatial structure, presence or absence of phage, and transfer time as nominal factors.
Results
Incidence and emergence of the mucoid phenotype in different treatment groups
The mucoid phenotype of P. fluorescens evolves under specific conditions as shown by the stable emergence of this phenotype for all replicate populations in only one of the eight different treatment groups. Mucoid bacteria were first identified in a single population of Treatment-1 (phage present, daily transfer, shaken) at day 6 and were present at detectable frequencies in all other replicate populations of this treatment group by day 10. Although the mucoid phenotype was observed in other phage treatment groups, this was only infrequently (at single time- points for only 1 or 2 populations within each phage treatment group, that is, 3, 5 and 7) and most importantly no mucoid bacteria were recovered from any of the treatments without phage (Table 1). Although co-evolution with lytic phage is the primary contributory factor in selecting for the mucoid phenotype (χ2=19.9, P<0.00001), mucoids were more common under one-daily than two-daily transfer regimes (χ2=7.67, P<0.01), and under shaken than static conditions (χ2=3.97, P<0.05). The numbers of mucoid phenotypes fluctuated through time for each population of Treatment-1 and were predominantly present at a low frequency relative to total population density (Figure 1). However, once this phenotype emerged, it remained a constant feature of Treatment group-1. Given these results the rest of our study focused on the characterization of mucoid phenotypes isolated from populations of Treatment group-1.
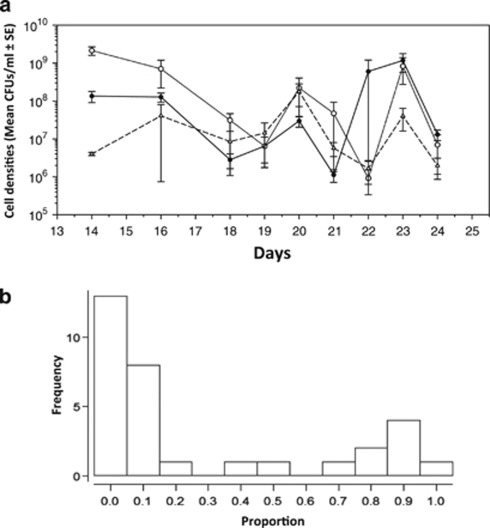
Figure 1.
Dynamics of mucoid evolution through time for Treatment 1 (days 14–24). Averaged bacterial densities of each phenotype for populations from Treatment 1 (a). Closed circles, open circles and open triangles are mucoid, SM, and WS respectively. A frequency histogram showing the proportion of mucoid phenotypes relative to total cell density for all populations of Treatment 1 and all time-points analysed, n=66 (b). The mucoid phenotype was present generally at low frequencies in populations but varied over time from 0.0006 to as high as 0.957, mean 0.3.
Co-evolution
Consistent with previous work on this system, bacteria and phage showed extensive co-evolution in Treatment-1 (shaken and daily transfers), as demonstrated by greater infectivity of phages to past than future bacteria across the three time points where measurements were taken (Supplementary Figure S1; sign test: P<0.0001).
Characteristics of mucoid phenotypes
In vitro instability is a key characteristic of mucoid P. aeruginosa isolates (Pugashetti et al., 1982). In our study this instability was also observed and two different mucoid phenotypes were initially noted when bacterial populations were plated on KB agar. The Unstable Mucoid (UM) phenotype was characterized by a mucoid centre, with the edges of colonies showing an SM phenotype. When single UM colonies were picked and grown overnight in liquid KB, a variable proportion of colonies had an SM phenotype when plated out on KB agar (these SM colonies were called revertants (REVs)). A second mucoid phenotype, Fixed Mucoid (FM), was observed and was characterized by shiny, raised, mucoid, translucent colonies with no SM edges when plated on KB agar (Supplementary Figures S2a–c). This phenotype maintains a stable phenotype when plated after overnight growth in liquid KB (no visible SM edges/or apparent reversion to SM phenotype).
Growth of mucoids in absence of phage
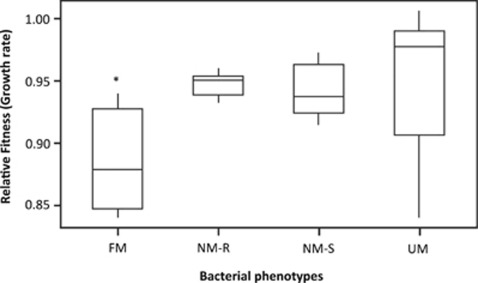
The clear association between the presence of phages and mucoids suggests that the mucoid phenotype confers a cost when phages are absent. Consistent with this view, alginate production is costly as shown by the significant growth rate cost of FM as compared with UM, Non-Mucoid-phage Resistant (NM-R) and Non-Mucoid Sensitive (NM-S) when grown in monoculture in the absence of phage (Kruskal–Wallis; df=3, P=0.0196) (Figure 2).
Figure 2.
Cost of mucoid phenotype. Interquartile range box plots showing the growth rates of FM, UM, non-mucoid resistant (NM-R) and non-mucoid sensitive (NM-S) phenotypes relative to SBW based on monoculture growth rates in the absence of sympatric phage populations (n=30); the asterisk (*) indicates a significant difference compared to control.
Resistance of mucoids to phages
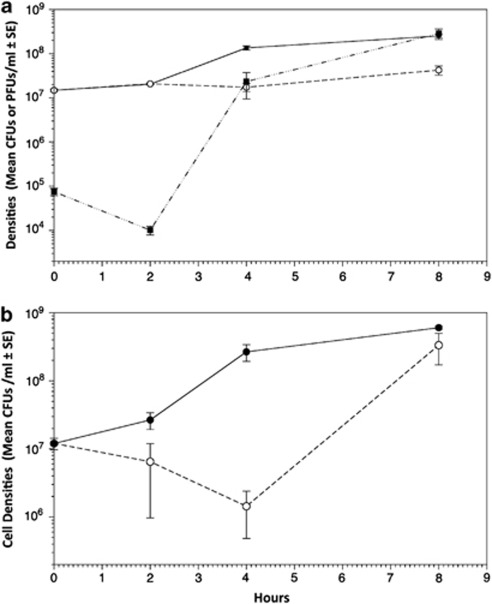
While mucoids showed a fitness cost in the absence of phage, we predicted they would have a fitness advantage relative to other phenotypes in the presence of phage through increased resistance. However, simple binary resistance assays were hard to interpret because FM phenotypes obscured phage plaque formation (turbid plaques). We therefore performed growth rate assays in the presence of their sympatric phages. Both UMs and FMs were sensitive to their sympatric phage populations (100%), as shown by increase in phage density. However, the UM and FM genotypes were more resistant than other genotypes on which phages could grow; cell densities of mucoid phenotypes increased in the presence of phage compared with other NM genotypes, which showed a sharp decrease in cell density in the presence of phages under the same conditions (Figures 3).
Figure 3.
Mean phage production (closed squares) and mean bacterial densities (growth) of UM phenotypes over time with (dashed line, open circles) and without phage (black straight line, closed circles) (a). After two hours only FM phenotypes were detected after co-inoculating initially UM phenotypes with sympatric phage populations (all REVs were readily lysed); however phage production continued, showing that although mucoid phenotypes are sensitive to phage they can continue to grow and increase in cell density in their presence. The effect of phage on non-mucoid bacteria cell densities (growth) through time (b). The ancestral SM phenotype of SBW25 was grown both in the absence (straight black line, closed circles) and in the presence (dashed lines, open circles) of three different populations of phage from Treatment 1. Cell densities are greatly reduced in non-mucoid phenotypes in the presence of phage compared with mucoid phenotypes.
Apparent competition
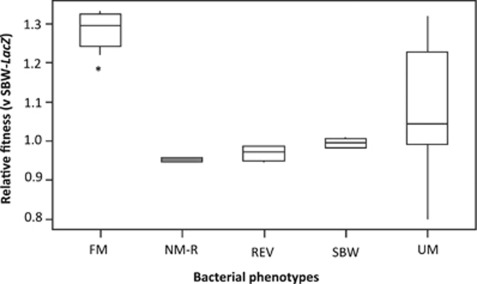
When individual FM and UM colonies were checked for phage by using spot assays, all FM (n=30, 100%) and some UM (n=15, 45%) colonies were found to be phage-positive suggesting that viable phages were physically associated with these bacteria. We therefore speculated that an additional advantage of mucoids might be through ‘apparent competition': competition mediated by a shared enemy (Holt, 1977). When the UM, FM, REV and NM phenotypes were competed with a marked ancestral strain, FM phenotypes have a significant advantage in the presence of this sensitive competitor (Kruskal–Wallis; H=18.03, df=4, P=0.001) (Figure 4). The considerable variability in the fitness of UM is due to whether the UM phenotype is phage-positive or -negative.
Figure 4.
Interquartile range box plots showing the relative fitness (W) of each phenotype when competed against a marked ancestral strain of SBW25 (SBW-LacZ). Mucoid phenotypes engage in apparent competition which confers a signicant fitness advantage in the presence of phage sensitive bacteria; asterisk (*) indicates a significant difference compared to control. The large variability in fitness for UM phenotypes is attributed to the UM phenotype being phage positive or negative.
Revertants
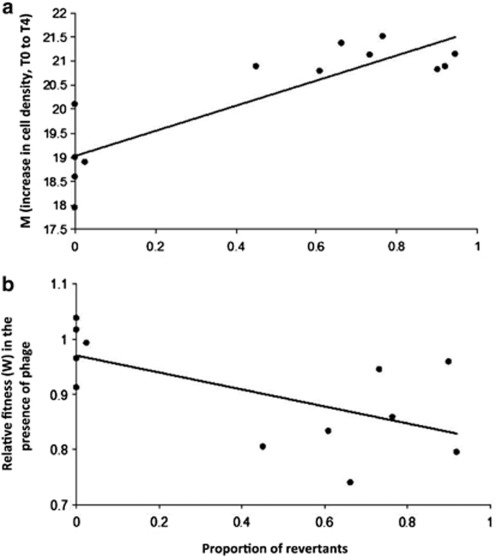
We next attempted to understand the adaptive significance of reversion of mucoids to SM (REV), as apparent in the UM phenotype. Across all mucoids we found a large variation in the rate of reversion when individual colonies were grown in liquid media, with FM showing no REVs. The proportion of REVs for UM grown in liquid monoculture in the absence of phage is also positively correlated with higher growth rate (Pearson's correlation=0.864, P<0.0001); in the absence of phage, bacteria readily revert to an SM phenotype, which has a higher growth rate (Figure 5a). However, the proportion of REVs is negatively correlated with fitness in the presence of phages (Figure 5b, Pearson's correlation=−0.872, P<0.0001); as most REVS are susceptible to phage they are lysed in the presence of sympatric phage populations, thus reducing total population densities. FM phenotypes have an equivalent growth rate in both the presence and absence of sympatric phage populations, and there was no evidence of REVs. Reversion therefore is beneficial in the absence, but costly in the presence, of phages.
Figure 5.
Growth-rate cost of mucoid phenotype in the absence of phage (a). In the absence of virulent phage mucoid colonies readily revert to SM phenotypes and the proportion of REVs for UM grown in liquid monoculture in the absence of phage is positively correlated with higher growth rate (Pearson's correlation=0.864, P<0.0001). The relative fitness (W) of UM and FM phenotypes grown in the presence versus absence of sympatric phage (b). Data was taken from early log growth (T0 to T4 hours) and relative fitness (W) was calculated according to Malthusian parameters using cell densities of each phenotype grown in the presence and absence of phage. The strong negative correlation between the proportion of REVs per phenotype and fitness (W) (Pearsons correlation=−0.872, P<0.0001) explains the high variability in relative fitness in the presence of phage, i.e., all REVs are sensitive to sympatric phage populations and are lysed, which reduces total cell densities.
Variation in frequency of revertants
We next explored the variation in the frequency of revertants. Further analysis showed that UM colonies could be partitioned into those that were phage-positive and phage-negative: 5 out of 11 single UM colony culture broths tested positive for phages. Typically, phage-negative UM showed high rates of reversion, as would be expected if REVs are susceptible but have a higher growth rate than mucoids. Crucially, REVs derived from phage-positive UM parent strains were resistant to phages from the parent culture supernatant, but were sensitive to some members of the co-occurring phage population (that is, phages isolated from the whole community, rather than phages specifically associated with particular mucoids). These data suggest that, both in the absence of phage and where REVs are resistant to phages, UM revert to the SM phenotype owing to growth rate costs associated with mucoid conversion. By contrast, when UM are exposed for 2 h to sympatric phage populations, only FM phenotypes were observed when cultures were plated on solid agar as previously outlined.
Given that phage-imposed selection results in UM producing only FM phenotypes, we speculated that perhaps FM phenotypes could also produce SM revertants, but that these revertants were never observed because they were susceptible to the phages associated with the FM phenotype. We therefore sought to assess reversion rate when phages were diluted out, and hence could not impose any selection. To this end, single UM or FM colonies were picked directly from agar plates, serially diluted and re-plated on agar. Each single colony type is composed of variable proportions of FM, UM and SM phenotypes (Supplementary Figure S3), but there was no significant difference in the number of REV phenotypes per single colonies of UM and FM (Mann–Witney; df=1, P=0.6366). This result highlights that there is no difference in the reversion rate between FM and UM, suggesting that colony heterogeneity of the mucoid phenotype is due to the presence or absence of phage, and whether or not REVs are resistant to the phages associated with mucoid bacteria.
Mechanism of reversion
When REV phenotypes derived from UM and FM parent colonies (which have SM colony morphology) are picked from a plate, serially diluted and plated out, only SM phenotypes are observed. Crucially, these results eliminate the possibility of a bidirectional genetic switch (Beaumont et al., 2009) or contingency loci (Moxon et al., 2006) as a mechanism underlying the phenotypic heterogeneity of UM and FM colonies. Another possibility considered was that the high rate of reversion may have been indicative of the evolution of mutator phenotypes, and indeed previous studies have shown that co-evolution with lytic phage can select for higher mutation rates (Pal et al., 2007). However, no difference in mutation frequencies was evident for the evolved mucoid bacteria and their revertants compared with the ancestral phenotype using fluctuation tests (df=2, F2,13=0.4375, P=0.6564). These data suggest that revertants arise from mucoids at high frequency by simple mutations.
Loss of motility on mucoid conversion
The swimming motility of phenotypes FM, UM NM, REV and wild type (ancestral SBW) differed (H=12.74, df=4, P=0.013), with FM having the lowest motility value. The high variability in UM motility can be explained by the proportion of revertants (Supplementary Figure S4).
Genetic analysis
Conversion to the mucoid phenotype is usually associated with mutations in both the regulatory and biosynthetic operons (Muhammadi and Ahmed, 2007). Despite an extensive sequencing effort, no mutations in any of the candidate genes were detected (see Table 2). As such the genetics underlying these phenotypes is as yet unknown but is independent of mutations that are commonly associated with mucoid conversion, that is, mucA and algU.
Discussion
Effect of environment and spatial structure on the incidence of mucoid phenotype
In this study we have shown that the mucoid phenotype of P. fluorescens SBW25 evolves as an adaptive response during co-evolution with lytic phage. Although mucoids arise under a variety of experimental conditions, they are most favoured under conditions of a homogenous spatial structure and daily transfer. This environment provides a relatively uniform oxygen and nutrient concentration, thus reducing the potential for niche and resource competition that is typically maintained by negative frequency-dependent selection in static microcosms (Rainey and Travisano, 1998; Vogwill et al., 2011). The level of culture aeration is a key factor in the incidence and stability of the mucoid phenotype of P. aeruginosa, and previous experiments have shown that static growth selects for mucoid to NM conversion (Krieg et al., 1986; Hassett, 1996; Wyckoff et al., 2002) as oxygen is required for biosynthesis of alginate (Bayer et al., 1990; Leitao and Sa-Correia, 1997). This, together with the cost associated with the concurrent loss of motility on mucoid conversion observed here and elsewhere (Mahenthiralingam et al., 1994; Tart et al., 2005), will reduce the ability of mucoid phenotypes to compete for oxygen at the air–liquid interface of a microcosm during static incubation (which is typically occupied by the biofilm forming phenotype, WS (Rainey and Travisano, 1998; Koza et al., 2011). Furthermore, the prolonged time spent in stationary phase for the 48-h treatments, where resource competition is much greater, might increase metabolic costs associated with this particular phenotype. As exopolysaccharide and alginate production is metabolically expensive (Linton, 1991; Rehm, 2009), and resources are increasingly depleted through time, the resultant competition between genotypes is greater, resulting in a less favourable environment for the mucoid phenotype. The three other phenotypes that are routinely recovered from this experimental system, namely SM, WS and FS, were also recovered here from treatments with and without phage (Rainey and Travisano, 1998; Buckling and Rainey, 2002a, 2002b), suggesting that the mucoid phenotype of P. fluorescens is the only phenotype exclusively associated with phage predation in this experimental context and is not a general adaption to other experimental variables.
Mucoid conversion and apparent competition
Despite the direct costs associated with the mucoid phenotype, its selective advantage is most likely its ability to reduce the harmful effects of phage predation and to engage in apparent competition with phage-sensitive phenotypes. Apparent competition results from either partial resistance of mucoid phenotypes, which actively produce virulent phage (or produce phage by lysis of revertants), or the presence of phage embedded in the thick exopolysaccharide matrix, which protects the host but serves as a vehicle for contagion dispersal, or both. Although such apparent competition has been reported in the context of lysogenic phages (Brown et al., 2006; Joo et al., 2006), we are not aware of any study reporting use of lytic phages as a potential mechanism to mediate competition. Furthermore, a recent study using this system found that increasing dispersal in the presence of phage favoured the mucoid phenotype (Vogwill et al., 2011), potentially because dispersal allowed the mucoid phenotypes to invade by using their phages on susceptible bacteria in other populations.
Instability of the mucoid phenotype and reversion
It is tempting to speculate that the high rate of reversion to SM is adaptive, that is, a selected trait, as mucoids do well in presence of phage, and revertants do well in the absence of phage. If this behaviour was a genetic switch or a phenotypically plastic trait, this might well be an adaptation, as the same genotype would express both phenotypes. However, the absence of a ‘switch' from REVs to mucoids suggests that REVs are simply mutating from mucoids, and hence selection acting on revertants will not increase the frequency of mucoids. Although the rate of spontaneous reversion to SM is very high, it is clear that mucoids are not mutators, so how then is this rate of reversion feasible? It is probable that the high reversion rate to SM may arise by virtue of the number of genes or sequence space involved in alginate regulation and biosynthesis (Ohman and Chakrabarty, 1981; Muhammadi and Ahmed, 2007), which greatly increases the probability of acquiring reversion or suppressor mutations. As such, it is likely that the mucoid phenotype outlined in this system is an adaptive response to phage predation, but when this selective pressure is removed bacteria readily revert to NM phenotypes that do not incur the costs of alginate production required for protection in the presence of phage.
Clinical relevance of co-evolution with phage and mucoid conversion
Extensive research has detailed the possible causes and consequences of conversion to a mucoid phenotype, including high salt concentrations, free radical damage, antibiotic administration and H2O2. Here we show directly that co-evolution with virulent phage is a key factor in the emergence of this phenotype. Treatment-1 used in this study is analogous to sites in the lung where there is sufficient aeration and high densities of phage, and a review of the literature shows that phages are present in high numbers in sputum (Tejedor et al., 1982; Ojeniyi, 1988; Ojeniyi et al., 1991) and are likely to have a role in the emergence of this phenotype in the CF lung. For example, our results are similar to data from a 1984 study by Miller and Rubero. Clinical isolates of P. aeruginosa were analysed and 21 of the 22 produced virulent phage (between 106 and 109 plaque-forming units per millilitre) and showed a turbid plaque morphology (also observed in this current study) that they understood to be lysogenic conversion. The authors concluded that bacteriophage must provide some selective advantage to the CF mucoid phenotype, and that phages should be considered as a mechanism for mucoid conversion of clinical isolates. Mucoid phenotypes have also been isolated from the edges of zones of lytic phage plaques and these phenotypes maintained sensitivity to the bacteriophage type strain (Martin, 1973).
Genetics of mucoid phenotype
Production of alginate is under complex, hierarchical control and as many as 25 genes have been identified in the process (Muhammadi and Ahmed, 2007). Alginate genes are normally silent but become activated under environmental stress conditions with genotypic switching from NM to mucoid under the transcriptional control of a cluster of genes known as mucA to mucD. mucA acts as a negative regulator of the alternative sigma factor σ22 or algT, which is the key transcription factor required not only for alginate biosynthesis, but also many other important cellular pathways, including flagellum biosynthesis (Tart et al., 2005). Mutations in mucA, and also mucC and mucD, which are negative regulators of alginate production, are concomitant with the constitutive production of alginate with subsequent in vitro conversion to a NM phenotype believed to be due to second site mutations at algU (Ciofu et al., 2008). We investigated these possibilities for our evolved phenotypes, but no mutations relative to the wild type sequence were identified at any of these loci. An intriguing possibility is the redundancy of several enzymes and substrates that function in both the alginate and lipopolysaccharide (LPS) biosynthetic pathways (May and Chakrabarty, 1994; Ye et al., 1994; Olvera et al., 1999; Tavares et al., 1999), which are outlined in Table 2. LPS is the putative receptor-binding site of φ2 phage based on a transposon mutagenesis study of the host bacterium P. fluorescens SBW25 and homologies between T7 phage and phi2. We hypothesized that a switch to alginate production may provide resistance to phage, or delay the adsorption and infection process by two non-mutually exclusive ways: (a) downregulation of LPS production and reduction of the number of available receptor sites for phage to bind to, and/or (b) excess production of alginate may provide a protective coating over LPS receptors, thus impeding or preventing phage attachment to cell-surface receptors. To this end we sequenced three genes that are involved in both pathways in addition to a number of other candidate genes, but no mutations were identified.
Given the role of two-component systems in regulating responses to the environment; the role of regulatory proteins and promoter regions that control phenotypic variation; and the structural and regulatory complexity of LPS and alginate pathways, the genetic basis for the FM/UM phenotype is potentially great (May and Chakrabarty, 1994; Rocchetta et al., 1999; Tavares et al., 1999; Muhammadi and Ahmed, 2007; Oglesby et al., 2008; Damron and Yu, 2011). However, one important result, which the concurrent loss of motility would suggest, is that perhaps some master regulator of these two processes is involved, highlighting the pleiotropic effects of mutations involved in resistance to phage predation.
Conclusions
Up to 80% of all P. aeruginosa isolates from CF patients are mucoid compared with 0.8–2.1% of clinical isolates from the general population (Ohman and Chakrabarty, 1982). This phenotype provides a competitive advantage in the lung environment against a range of stresses, including resistance to host phagocytosis and antibiotic therapy (May et al., 1991; Høiby et al., 2010). Whether co-evolution with virulent phage results in the mucoid phenotype as an adaptive response to phage predation, and in doing so indirectly selects for bacterial virulence in a clinical setting, requires further work, but we have shown here that virulent phages are a key factor in the incidence of the mucoid phenotype in a related Pseudomonas species. We have also demonstrated the impact of phages in selecting for hosts that can engage in apparent competition, which will undoubtedly affect ecological and evolutionary dynamics. In conclusion, the results of this study are not just of interest with respect to the ecology and dynamics of host parasite co-evolution, but may have implications for clinical and applied microbiology. Given the prevalence of bacterial antibiotic resistance in clinical settings, there is a renewed interest in the possible use of phage as a viable therapeutic option. Our data emphasize the importance of considering indirect selection for bacterial virulence caused by lytic phage.
Acknowledgments
We thank the referees and George Kowalchuk for helpful comments. We are grateful to Paul Rainey for SBW25-lacZ. This work was funded by the European Research Council.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bayer AS, Eftekhar F, Tu J, Nast CC, Speert DP. Oxygen-dependent upregulation of mucoid exopolysaccharide (alginate) production in Pseudomonas aeruginosa. Infect Immun. 1990;58:1344–1349. doi: 10.1128/iai.58.5.1344-1349.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- Boyd EF, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–529. doi: 10.1016/s0966-842x(02)02459-9. [DOI] [PubMed] [Google Scholar]
- Brockhurst MA, Morgan AD, Rainey PB, Buckling A. Population mixing accelerates coevolution. Ecol Lett. 2003;6:975–979. [Google Scholar]
- Brown SP, Le Chat L, De Paepe M, Taddei F. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr Biol. 2006;16:2048–2052. doi: 10.1016/j.cub.2006.08.089. [DOI] [PubMed] [Google Scholar]
- Brussow H, Desiere F. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol. 2001;39:213–222. doi: 10.1046/j.1365-2958.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- Buckling A, Kassen R, Bell G, Rainey PB. Disturbance and diversity in experimental microcosms. Nature. 2000;408:961–964. doi: 10.1038/35050080. [DOI] [PubMed] [Google Scholar]
- Buckling A, Rainey PB. The role of parasites in sympatric and allopatric host diversification. Nature. 2002a;420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci. 2002b;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, Hoiby N. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology. 2008;154:103–113. doi: 10.1099/mic.0.2007/010421-0. [DOI] [PubMed] [Google Scholar]
- Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol. 2011;193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernebro J. Fighting bacterial infections—future treatment options. Drug Resist Updat. 2011;14:125–139. doi: 10.1016/j.drup.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Fett WF, Dunn MF. Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol. 1989;89:5–9. doi: 10.1104/pp.89.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CR, Yoichi M, Unno H, Tanji Y. The coexistence of Escherichia coli serotype O157:H7 and its specific bacteriophage in continuous culture. FEMS Microbiol Lett. 2004;241:171–177. doi: 10.1016/j.femsle.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Friman VP, Hiltunen T, Jalasvuori M, Lindstedt C, Laanto E, Örmälä AM, et al. High temperature and bacteriophages can indirectly select for bacterial pathogenicity in environmental reservoirs. PLoS One. 2011;6:e17651. doi: 10.1371/journal.pone.0017651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glonti T, Chanishvili N, Taylor PW. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J Appl Microbiol. 2010;108:695–702. doi: 10.1111/j.1365-2672.2009.04469.x. [DOI] [PubMed] [Google Scholar]
- Gómez P, Buckling A. Bacteria–phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- Hammad AMM. Evaluation of alginate-encapsulated Azotobacter chroococcum as a phage-resistant and an effective inoculum. J Basic Microbiol. 1998;38:9–16. [Google Scholar]
- Hassett DJ. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J Bacteriol. 1996;178:7322–7325. doi: 10.1128/jb.178.24.7322-7325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- Holt RD. Predation, apparent competition, and the structure of prey communities. Theor Pop Biol. 1977;12:197–229. doi: 10.1016/0040-5809(77)90042-9. [DOI] [PubMed] [Google Scholar]
- Joo J, Gunny M, Cases M, Hudson P, Albert R, Harvill E. Bacteriophage-mediated competition in Bordetella bacteria. Proc Biol Sci. 2006;273:1843–1848. doi: 10.1098/rspb.2006.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith RC, Keith LM, Hernandez-Guzman G, Uppalapati SR, Bender CL. Alginate gene expression by Pseudomonas syringae pv. tomato DC3000 in host and non-host plants. Microbiology. 2003;149:1127–1138. doi: 10.1099/mic.0.26109-0. [DOI] [PubMed] [Google Scholar]
- Kharazmi A. Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol Lett. 1991;30:201–205. doi: 10.1016/0165-2478(91)90026-7. [DOI] [PubMed] [Google Scholar]
- Krieg DP, Bass JA, Mattingly SJ. Aeration selects for mucoid phenotype of Pseudomonas aeruginosa. J Clin Microbiol. 1986;24:986–990. doi: 10.1128/jcm.24.6.986-990.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koza A, Moshynets O, Otten W, Spiers AJ. Environmental modification and niche construction: developing O2 gradients drive the evolution of the Wrinkly Spreader. ISME J. 2011;5:665–673. doi: 10.1038/ismej.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Leitao JH, Sa-Correia I. Oxygen-dependent upregulation of transcription of alginate genes algA, algC and algD in Pseudomonas aeruginosa. Res Microbiol. 1997;148:37–43. doi: 10.1016/S0923-2508(97)81898-0. [DOI] [PubMed] [Google Scholar]
- Lenski RE. Quantifying fitness and gene stability in microorganisms. Biotechnology. 1991;15:173–192. doi: 10.1016/b978-0-409-90199-3.50015-2. [DOI] [PubMed] [Google Scholar]
- Linton JD. Metabolite production and growth efficiency. Antonie Van Leeuwenhoek. 1991;60:293–311. doi: 10.1007/BF00430371. [DOI] [PubMed] [Google Scholar]
- Lopez-Pascua LC, Buckling A. Increasing productivity accelerates host–parasite coevolution. J Evol Biol. 2008;21:853–860. doi: 10.1111/j.1420-9101.2008.01501.x. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DR. Mucoid variation in Pseudomonas aeruginosa induced by the action of phage. J Med Microbiol. 1973;6:111–118. doi: 10.1099/00222615-6-1-111. [DOI] [PubMed] [Google Scholar]
- Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145 (Pt 6:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- May TB, Chakrabarty AM. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- May TB, Shinabarger D, Maharaj R, Kato J, Chu L, DeVault JD, et al. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev. 1991;4:191–206. doi: 10.1128/cmr.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RV, Rubero VJ. Mucoid conversion by phages of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J Clin Microbiol. 1984;19:717–719. doi: 10.1128/jcm.19.5.717-719.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AD, Gandon S, Buckling A. The effect of migration on local adaptation in a coevolving host–parasite system. Nature. 2005;437:253–256. doi: 10.1038/nature03913. [DOI] [PubMed] [Google Scholar]
- Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- Muhammadi, Ahmed N. Genetics of bacterial alginate: alginate genes distribution, organization and biosynthesis in bacteria. Curr Genomics. 2007;8:191–202. doi: 10.2174/138920207780833810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby LL, Jain S, Ohman DE. Membrane topology and roles of Pseudomonasaeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonasaeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Chakrabarty AM. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun. 1982;37:662–669. doi: 10.1128/iai.37.2.662-669.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y, Schumacher-Perdreau F, Peters G, Pulverer G. The role of capsule as a barrier to bacteriophage adsorption in an encapsulated Staphylococcus simulans strain. Med Microbiol Immunol. 1988;177:229–233. doi: 10.1007/BF00211222. [DOI] [PubMed] [Google Scholar]
- Ojeniyi B. Bacteriophages in sputum of cystic fibrosis patients as a possible cause of in vivo changes in serotypes of Pseudomonas aeruginosa. APMIS. 1988;96:294–298. doi: 10.1111/j.1699-0463.1988.tb05305.x. [DOI] [PubMed] [Google Scholar]
- Ojeniyi B, Birch-Andersen A, Mansa B, Rosdahl VT, Høiby N. Morphology of Pseudomonas aeruginosa phages from the sputum of cystic fibrosis patients and from the phage typing set. An electron microscopy study. APMIS. 1991;99:925–930. [PubMed] [Google Scholar]
- Olvera C, Goldberg JB, Sanchez R, Soberon-Chavez G. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol Lett. 1999;179:85–90. doi: 10.1111/j.1574-6968.1999.tb08712.x. [DOI] [PubMed] [Google Scholar]
- Pal C, Macia MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen SS. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 1992;28:1–79. [PubMed] [Google Scholar]
- Pedersen SS, Hoiby N, Espersen F, Kharazmi A. Alginate in infection. Antibiot Chemother. 1991;44:68–79. doi: 10.1159/000420300. [DOI] [PubMed] [Google Scholar]
- Pirnay JP, De Vos D, Verbeken G, Merabishvili M, Chanishvili N, Vaneechoutte M, et al. The phage therapy paradigm: pret-a-porter or sur-mesure. Pharm Res. 2010;28:934–937. doi: 10.1007/s11095-010-0313-5. [DOI] [PubMed] [Google Scholar]
- Pugashetti BK, Metzger HM, Jr, Vadas L, Feingold DS. Phenotypic differences among clinically isolated mucoid Pseudomonas aeruginosa strains. J Clin Microbiol. 1982;16:686–691. doi: 10.1128/jcm.16.4.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Bailey MJ. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol Microbiol. 1996;19:521–533. doi: 10.1046/j.1365-2958.1996.391926.x. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Rehm BHA. Alginates: Biology and Applications (Microbiology Monographs) Springer: Germany; 2009. [Google Scholar]
- Rocchetta HL, Burrows LL, Lam JS. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapelli RV, Goebel WF. The capsular polysaccharide of a mucoid variant of E. coli K12. Proc Natl Acad Sci USA. 1964;52:265–271. doi: 10.1073/pnas.52.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol. 2005;187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares IM, Leitao JH, Fialho AM, Sa-Correia I. Pattern of changes in the activity of enzymes of GDP–D-mannuronic acid synthesis and in the level of transcription of algA, algC and algD genes accompanying the loss and emergence of mucoidy in Pseudomonas aeruginosa. Res Microbiol. 1999;150:105–116. doi: 10.1016/s0923-2508(99)80028-x. [DOI] [PubMed] [Google Scholar]
- Taylor TB, Buckling A. Competition and dispersal in Pseudomonas aeruginosa. Am Nat. 2010;176:83–89. doi: 10.1086/652995. [DOI] [PubMed] [Google Scholar]
- Tejedor C, Foulds J, Zasloff M. Bacteriophages in sputum of patients with bronchopulmonary Pseudomonas infections. Infect Immun. 1982;36:440–441. doi: 10.1128/iai.36.1.440-441.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry JM, Pina SE, Mattingly SJ. Role of energy metabolism in conversion of nonmucoid Pseudomonas aeruginosa to the mucoid phenotype. Infect Immun. 1992;60:1329–1335. doi: 10.1128/iai.60.4.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T, Fenton A, Brockhurst MA. Coevolving parasites enhance the diversity-decreasing effect of dispersal. Biol Lett. 2011;7:578–580. doi: 10.1098/rsbl.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- Wyckoff TJ, Thomas B, Hassett DJ, Wozniak DJ. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology. 2002;148:3423–3430. doi: 10.1099/00221287-148-11-3423. [DOI] [PubMed] [Google Scholar]
- Ye RW, Zielinski NA, Chakrabarty AM. Purification and characterization of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa involved in biosynthesis of both alginate and lipopolysaccharide. J Bacteriol. 1994;176:4851–4857. doi: 10.1128/jb.176.16.4851-4857.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XX, Rainey PB. Construction and validation of a neutrally-marked strain of Pseudomonas fluorescens SBW25. J Microbiol Methods. 2007;71:78–81. doi: 10.1016/j.mimet.2007.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.