Abstract
Enhanced biological phosphorus removal (EBPR) is widely used for removal of phosphorus from wastewater. In this study, a metagenome (18.2 Gb) was generated using Illumina sequencing from a full-scale EBPR plant to study the community structure and genetic potential. Quantitative fluorescence in situ hybridization (qFISH) was applied as an independent method to evaluate the community structure. The results were in qualitative agreement, but a DNA extraction bias against gram positive bacteria using standard extraction protocols was identified, which would not have been identified without the use of qFISH. The genetic potential for community function showed enrichment of genes involved in phosphate metabolism and biofilm formation, reflecting the selective pressure of the EBPR process. Most contigs in the assembled metagenome had low similarity to genes from currently sequenced genomes, underlining the need for more reference genomes of key EBPR species. Only the genome of ‘Candidatus Accumulibacter', a genus of phosphorus-removing organisms, was closely enough related to the species present in the metagenome to allow for detailed investigations. Accumulibacter accounted for only 4.8% of all bacteria by qFISH, but the depth of sequencing enabled detailed insight into their microdiversity in the full-scale plant. Only 15% of the reads matching Accumulibacter had a high similarity (>95%) to the sequenced Accumulibacter clade IIA strain UW-1 genome, indicating the presence of some microdiversity. The differences in gene complement between the Accumulibacter clades were limited to genes for extracellular polymeric substances and phage-related genes, suggesting a selective pressure from phages on the Accumulibacter diversity.
Keywords: Accumulibacter, activated sludge, EBPR, metagenomics, microdiversity
Introduction
Enhanced biological phosphorus removal (EBPR) is widely used in wastewater treatment to remove phosphorus from wastewater to protect receiving waters from eutrophication and to improve recovery of the nonrenewable resource phosphorus (Seviour et al., 2003; Oehmen et al., 2007). Even though many EBPR wastewater treatment plants (WWTPs) run well, the phosphorus-removal capacity can at times deteriorate, which is often attributed to competition between beneficial and detrimental bacteria in the plants (Oehmen et al., 2007). Optimization of the EBPR process has so far largely been based on empirical knowledge of plant configuration and process parameters. Hence, further understanding of the microbial community might lead to improved operation of EBPR WWTPs.
The organisms facilitating EBPR are called polyphosphate accumulating organisms (PAOs) and the most important PAOs have been identified using culture independent methods as ‘Candidatus Accumulibacter phosphatis' (hereafter called Accumulibacter) (Hesselmann et al., 1999; Crocetti et al., 2000) and the actinobacterial genus Tetrasphaera (Kong et al., 2005; Nguyen et al., 2011). Little is known about PAO physiology due to the absence of axenic cultures, but a milestone in the understanding of the physiology of Accumulibacter was achieved by obtaining the genome sequence of ‘Candidatus Accumulibacter phosphatis' clade IIA strain UW-1 from an enrichment culture (Garcia Martin et al., 2006). Several studies have subsequently used the genome sequence to understand better the special physiology of Accumulibacter, for example, by applying proteomic (Wexler et al., 2009) and transcriptomic studies on enriched laboratory reactors (He et al., 2010; He and McMahon, 2010).
Information about PAOs in full-scale EBPR plants is, however, more scarce as only few studies have investigated abundance and aspects of the ecophysiology of Accumulibacter and Tetrasphaera (Zilles et al., 2002; Kong et al., 2004, 2005, 2007; Nguyen et al., 2011). Furthermore, hardly anything is known about the flanking community, that is, bacteria providing substrate to the PAOs, such as fermenters, and bacteria involved in hydrolysis of macromolecules; hence there is a great need for more studies of full-scale EBPR plants.
Recently, we performed a survey of 25 Danish EBPR plants using quantitative fluorescence in situ hybridization (qFISH) with an array of oligonucleotide probes and identified a surprisingly stable core community in all EBPR plants consisting of a limited number of probe-defined species (Nielsen et al., 2010). Based on these observations and results from a number of studies of the ecophysiology of some of these core members, we established a conceptual ecosystem model of the EBPR process. The model includes the present knowledge about identity and function of bacteria involved in the different processes in EBPR plants, for example, also nitrification and denitrification. We propose that EBPR is not only industrially relevant but serve as a model system to study microbial ecology because of the relative stable core community and the strict control of input and output to the system.
Studies of microbial ecosystems can be carried out in unprecedented details by metagenomic analysis using next-generation sequencing. Metagenomics now offers the possibility to study the phylogenetic composition and functional potential of a complex community without prior enrichment, exemplified by metagenomics of the human gut microbiome and the cow rumen microbiome (Qin et al., 2010; Hess et al., 2011). Although metagenomics is still not able to provide the same detailed understanding as single genome analysis, it can serve as an entry point into the community-wide profiling of systems by providing a blueprint through which hypotheses can be generated and further studies designed (Kunin et al., 2008a). So far, only a single metagenomic study has been published on a microbial community from a full-scale wastewater treatment system (a conventional activated sludge plant; Sanapareddy et al., 2009), and no information is available from full-scale systems carrying out the EBPR process.
In this study, we generated a metagenome of the microbial community of a full-scale EBPR WWTP and investigated the community structure and genetic potential in detail. The aims were (1) to compare the metagenomic community composition with a DNA-extraction independent method based on qFISH data from the same treatment plant, (2) compare the functional potential with those identified through the conceptual EBPR ecosystem model, (3) compare the functional potential of the metagenome with other metagenomes, (4) investigate whether existing reference genomes are representative for microorganisms present in the community, and finally, (5) if they are, to study the degree of microdiversity of selected species.
Materials and methods
Sampling
Activated sludge was taken from the aeration tank at Aalborg East WWTP, Denmark (57.044565° N, 10.047598° E) on 4 August 2009. The plant has a biodenipho configuration, which includes an anaerobic tank and a tank with alternating denitrifying anoxic and nitrifying oxic conditions. The plant treats mainly domestic wastewater and serves 100 000 person equivalents with an average load of 45 000 person equivalents. The plant had stable EBPR performance for several years prior to the time of sampling removing >95% of the incoming P (10–20 mg P l−1) resulting in effluent concentrations below 0.5 mg P l−1.
Quantitative FISH
qFISH was conducted as described in Nielsen et al. (2010) with an extensive set of 32 oligonucleotide probes that covers most of the diversity usually found in Danish EBPR plants. A list of probes and their target groups can be found in Supplementary Table S1.
Metagenomic DNA extraction and sequencing
Total genomic DNA was extracted from 1.5 ml of activated sludge using the Fast DNA Spin kit for soil (MP Biomedicals, Solon, OH, USA), according to the manufacturer's instructions, except initial cell lysis, which was conducted using a Precellys Homogenizer (Bertin Technologies, Montigny le Bretonneux, France) at 6500 rpm for 3 × 5 s. Following extraction, the integrity of the DNA was verified using gel electrophoresis and concentration measured with a NanoDrop spectrophotometer (Nanodrop Technologies Inc., Wilmington, DE, USA). Following polyacrylamide gel electrophoresis, the 350–400-bp fraction was excised and extracted, and the DNA was afterwards processed according to the genomic DNA sample preparation kit protocol (Illumina Inc., San Diego, CA, USA) using 5 μg of DNA, 8 min of nebulisation and 12 cycles of PCR amplification. Paired End Sequencing (2 × 72 bp) was performed on an Illumina GAII using the Paired End Cluster Generation kit (version 2), and Sequencing kit (version 3), according to the manufacturer's instructions.
Quality filtering and de novo assembly
Base-calling was performed by the Illumina Genome Analyser Pipeline software version 1.5.1 and the resulting files were imported into the CLC Genomics Workbench version 4.5 (CLC Bio, Aarhus, Denmark). The reads were trimmed using a minimum quality score of 20, a minimum read length of 35 bp and allowing no ambiguous nucleotides. The reads were assembled using CLC's de novo assembly algorithm and contigs ⩾300 bp were retained for further analysis. The remaining contigs were clustered using cd-hit-est v.4.2.1 (Li and Godzik, 2006), with the following parameters: −c 0.95 and –r 1. The read coverage of the individual contigs was calculated by aligning the reads to the contigs using CLC's reference mapping algorithm, requiring 95% identity over 90% of the read length. The set of nonredundant contigs was uploaded to the MG-RAST v2 server (Meyer et al., 2008) (accession number: 4463936.3) and used in local database searches.
Gene prediction
Open reading frames (ORFs) were predicted using MetaGeneAnnotator (Noguchi et al., 2008) with the –m parameter for multiple species. The predicted ORFs were translated to proteins using the NCBI translation table 11. BLASTP v. 2.2.24+ (Altschul et al., 1997) was used to search the predicted proteins against a subset of the NCBI nr database containing all bacterial, viral and archaeal proteins (downloaded 13 October 2010). The following parameters were used in all BLASTP searches: −outfmt 5, e-value 1 × e−5, −num_descriptions 10, −num_alignments 10.
Taxonomic binning and functional assignment
Taxonomic binning of the predicted proteins was made using MEGAN v. 4.30 (Huson et al., 2007) based on the BLASTP searches. MEGAN uses a lowest common ancestor approach that assigns a sequence to the lowest common ancestor if it cannot be assigned uniquely to a given species. To be uniquely assigned to a species a 10% bit score difference to the closest relative was required. A custom made program was used to calculate the percentage genome coverage, individual gene coverage, percentage protein identity and construct recruitment plots on the basis of the BLASTP searches. Functional assignments were conducted through the SEED subsystems of the MG-RAST v2 server, using an e-value of minimum 1 e−5. The results were compared with 26 metagenomes from six different biomes selected from Dinsdale et al. (2008) using the MG-RAST v2 server and the metagenome from a non-EBPR WWTP (Sanapareddy et al., 2009) (see Supplementary Table S2 for details).
Polyphosphate kinase 1 (ppk1) analysis
Total genomic DNA was extracted from the same sludge sample by the use of the Fast DNA Spin kit for soil (MP Biomedicals). PCR amplification of ppk1 fragments was done using the primers ACCppk1-254F and ACCppk1-1376R (McMahon et al., 2007) and HotStar Taq polymerase (Qiagen AB, Sollentuna, Sweden) with 25 cycles of: 95 °C for 30 s, 66 °C for 1 min and 72 °C for 2 min. To obtain a product, the annealing temperature was 2 °C lower than published. The product was cloned (TOPO-XL; Invitrogen, Helleup, Denmark) and sequenced (Macrogen, Seoul, Korea). Publicly available full-length ppk1 sequences representing the diversity of the Betaproteobacteria were globally aligned using t-coffee (Notredame et al., 2000) and imported into ARB v. 5.2 (Ludwig et al., 2004). Publically available environmental sequences and the sequences obtained in the current study were aligned locally against the global alignment. A phylogenetic tree was constructed using RAxML, v. 7.0.3 (Stamatakis, 2006) implemented in ARB, comparing the 362 amino acid positions encoded by the Accumulibacter ppk1 PCR product. The calculation used Dayhoff substitution matrix, rapid hill climbing and PROTMIX rate distribution model. In silico ppk1 probes were designed to discriminate clade II (IIA: 5′-ACCAGAGCTTCAATCCGG-3′ IIB: 5′-ATCAGAGCTTCAACCCGG-3′ IIC+D: 5′-ACCAGAGCTTCAATTCGG-3′ and IIx: 5′-ACCAGAGTTTCAATCCGG-3′) from clade I (5′-ACCAGAGCTTCAATCCGA-3′) in the same region of the ppk1 gene.
Accumulibacter microdiversity
To investigate the microdiversity of Accumulibacter in the metagenome, a reference mapping was conducted against the Accumulibacter clade IIA strain UW-1 genome (NC_013194) using CLC's reference mapping algorithm (minimum 85% similarity over 70% of the read length). A custom-made program was used to visualize the mapping. Circos v. 0.54 (Krzywinski et al., 2009) was used to construct circular genome visualizations. The specificity and sensitivity of the reference mapping was investigated by reference mapping to 87 ppk1 genes from Accumulibacter and closely related species (see Supplementary Text and Supplementary Figure S7).
Results and discussion
The metagenome sequencing of the activated sludge sample from Aalborg East WWTP resulted in 205 million reads after quality filtering which were assembled using CLC's de novo assembly algorithm, resulting in 269 385 contigs with a minimum length of 300 bp, an average length of 540 bp and a maximum contig length of 32 884 bp (Table 1, Supplementary Figures S1 and S2). The minimum contig length of 300 bp was chosen as it ensured a reasonable length for ORF prediction and resulted in a dataset of a manageable size. The percentage of reads assembled into contigs over 300 bp (16%) was lower than reported for the human gut metagenome by Qin et al. (2010), where they were able to assemble 20–55% of reads (of similar length) into contigs over 500 bp. This difference might be a combination of (i) a greater diversity in the EBPR metagenome compared with the human gut microbiome, (ii) limitations of the DNA assembly algorithms to effectively deal with sequence microdiversity and (iii) a large difference in species abundance that results in an uneven representation of genome sequences from different species (Pop, 2009). In addition, the CLC de novo assembly algorithm does not scaffold, which might contribute to some of the difference compared with other algorithms. Nonetheless, the overall assembly encompassed 145 Mb of nonredundant sequence equivalent to approximately 30–40 full bacterial genomes. In total, 338 863 ORFs were predicted in the assembled contigs and 61% could be assigned a putative function and species (Table 1).
Table 1. Assembly and sequencing statistics.
| Sequenced reads (72 bp) | 255 511 260 (18.2 Gb) |
| Reads after quality filtering | 205 144 342 (12.5 Gb) |
| Contigs ⩾300 bp | 269.385 |
| Total assembly length | 145 294 146 |
| Reads useda | 32 025 362 |
| Average contig coveragea | 14 |
| N50b | 522 |
| Average contig lengthb | 540 |
| Maximum contig length | 32 884 |
| Predicted ORFs | 338 863 |
| Average ORF length (s.d.; bp) | 363±262 |
| Assigned putative function | 209 413 |
Abbreviation: ORF, open reading frame.
N50 is the length of the smallest contig in the set that contains the fewest (largest) contigs whose combined length represents at least 50% of the assembly (Miller et al., 2010). The number of ORF-assigned putative functions was calculated on the basis of a BLASTP search against a subset of the NCBI nr database containing all bacterial, archaeal and viral proteins using an expect value (e-value) cutoff of 1e−5.
Based on a reference mapping of reads to contigs with the criteria of 95% identity over 90% of the read length.
Calculated on the basis of contigs ⩾300 bp.
Community structure—comparison of qFISH and metagenome results
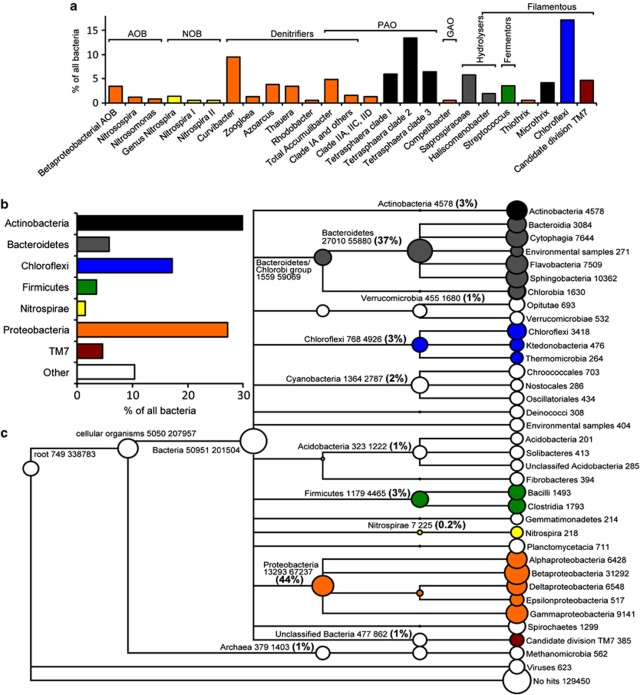
qFISH using many specific and some broader oligonucleotide probes (Supplementary Table S1) allowed us to identify and quantify approximately 90% of all bacteria targeted by the EUBmix probe. The primary function in the treatment process is known for most of these bacteria (that is, nitrifiers, denitrifiers, PAOs and so on; Nielsen et al., 2010). The abundances of probe-defined populations in these functional groups are shown in Figure 1a. As the number of available reference genomes is still low and does not represent the phylogenetic diversity of EBPR (see below), it was rarely possible to identify the same genera in the metagenome as with qFISH. Therefore, a comparison was made at the phylum level (Figures 1b and c). As the specific FISH probes used cover most of the diversity within their respective phyla, the error of summing to the phylum level is relatively small. As an example, the specific probes used for Tetrasphaera and Microthrix covers most of the diversity within actinobacteria found in Danish EBPR treatment plants (Nielsen et al., 2010; Nguyen et al., 2011). The most abundant phyla identified using qFISH also dominated the metagenome with many proteobacteria and bacteroidetes, and a notable number of ORFs assigned to firmicutes, chloroflexi and actinobacteria (Figure 1b). However, significant quantitative differences were observed. It is evident that actinobacteria (3% vs 30%), chloroflexi (3% vs 17%) and TM7 (0.3% vs 5%) were underrepresented in the metagenome compared with the qFISH data, whereas bacteroidetes (6% vs 37%) was overrepresented. There could be a number of explanations for this: specificity of the FISH probes or incomplete probe coverage, biovolume measurement versus DNA abundance, intact cells (FISH) versus all DNA (including extracellular DNA, Dominiak et al., 2011), lack of suitable reference genomes in the database to facilitate taxonomic assignment of metagenomic contigs, relative difference in genome sizes, and in the case of short read metagenomics, relative difference in the sequence assembly itself, and potentially a DNA extraction bias.
Figure 1.
Comparison of the metagenome and qFISH results for Aalborg East EBPR plant. (a) Detailed qFISH results. ‘% of all bacteria' is the specific probe compared with the EUBmix probes. The probe-defined groups in the top left panel have been grouped according to functional class and colored according to their phylogenetic classification. (b) qFISH results summed to phylum level to facilitate comparison with the metagenome data. (c) Overview of the metagenome binning based on MEGANs lowest common ancestor approach. ORFs were assigned based on a 10% bitscore difference to other BLAST hits; only nodes with over 200 ORFs assigned are shown. The numbers after descriptions denotes, respectively, the number of ORFs assigned to the particular node and the sum of ORFs assigned below the particular node. AOB, ammonia-oxidizing bacteria; GAO, glycogen-accumulating organism; NOB, nitrite-oxidizing bacteria; PAO, polyphosphate-accumulating organism.
We expected to identify a high number of reads assigned to actinobacteria in the metagenome as the actinobacterial genera Tetrasphaera were present in high numbers according to the qFISH data. To investigate if the low number of ORFs annotated to actinobacteria was due to lack of closely related Tetrasphaera genomes, we added four draft genomes of pure cultures from this genus to the database (unpublished data). However, this resulted in only 400 (0.2%) additional ORFs assigned to the actinobacteria. As an unknown microdiversity might prevent de novo assembly of Tetrasphaera metagenome reads, we also conducted recruitment of the raw metagenome reads directly to the four Tetrasphaera genomes. In total, 0.4% of all reads recruited to the four genomes confirmed the low abundance of Tetrasphaera in the metagenome. Therefore, the underrepresentation of actinobacteria (Tetrasphaera) in the metagenome is not due to shortcomings of the databases available. Instead, as Tetrasphaera are gram positive bacteria, a DNA extraction bias against gram positive bacteria would explain at least some of the underrepresentation of actinobacteria in the metagenome. A DNA extraction bias might also explain some of the underrepresentation of chloroflexi, where several genomes are available in the database, as chloroflexi can have complex layered cell envelopes (Sutcliffe, 2010). The low abundance of TM7 in the metagenome is also likely to be influenced by an extraction DNA bias as TM7 is gram positive (Hugenholtz et al., 2001) or because of lack of representative genomes as only four partial genomes were available for the TM7 phylum at the time of analysis. Overrepresentation of bacteroidetes has also been observed in other studies based on DNA extraction, which might be attributed to the ease of lysing some gram negative bacteria compared with gram positive bacteria (Salonen et al., 2010; Dethlefsen and Relman, 2011).
In this study, we used the FastDNA kit for DNA extraction, which was previously assessed as the best available for activated sludge communities (Vanysacker et al., 2010). In our experience, it provides the highest and most consistent DNA yield compared with other DNA extraction procedures we have tested (data not shown). However, the method may be supplemented with enzymatic lysis to increase the amount of gram positive bacteria. Other studies have also seen quantitative differences between FISH data and 16S rRNA gene clone libraries that are believed to be primarily due to extraction bias (Juretschko et al., 2002; Kong et al., 2007). In addition, a recent metagenome study of an in vitro-simulated microbial community compared different extraction protocols and sequencing techniques, and found that the extraction protocols had a profound effect on the metagenomic microbial community structure (Morgan et al., 2010). Therefore, quantitative metagenome results must be interpreted with caution and they cannot readily be used to prove absence of species or functions, and if possible, they have to be complemented with methods independent of DNA extraction such as FISH.
Similarity of metagenome species to known genomes
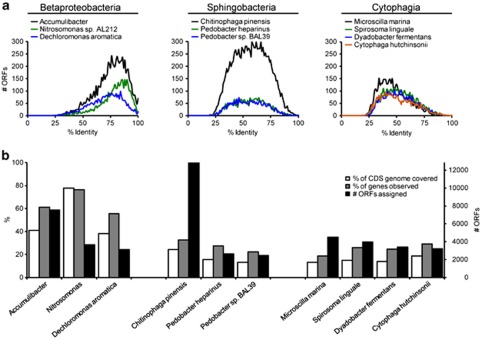
Only few publicly available reference genome sequences had a high number of ORFs assigned (Supplementary Figure S3). Figure 2 provides an overview of the ten genomes with the highest number of ORFs assigned. Of these, only Accumulibacter clade IIA, Dechloromonas aromatica RCB and Nitrosomonas sp. AL212 had a high number of ORFs assigned with high identity (>75%) and a relatively high coverage of the protein coding regions (40–80%), indicating that these species have closely related strains present in this activated sludge. These three genera are also part of the core microbiome in EBPR plants (for example, Kong et al., 2007), and the Accumulibacter and Nitrosomonas sp. AL212 genomes were retrieved from wastewater treatment systems. However, a closer look at the genomes and their ability to recruit reads from the metagenome revealed that only Accumulibacter could be used for further studies of the microdiversity, see below.
Figure 2.
Analysis of the 10 species with the highest number of ORFs assigned. ORFs were assigned based on the best BLASTP hit. (a) The number of ORFs assigned to a particular species as a function of percent identity of the OFRs compared with the species. (b) The number of ORFs assigned, the percent of the coding genome observed and percent of genes observed.
The high number of ORFs assigned to the phylum bacteroidetes (Figure 1) is likely to be related to bacteria identified by the qFISH-probes targeting the family of saprospiraceae (class sphingobacteria, ≈5%) including the genus Haliscomenobacter (2%) and the genus ‘Candidatus Epiflobacter' (Xia et al., 2008). However, at the time of analysis, no reference genomes within the family saprospiraceae were available; the closest related full genomes were the more distantly related Chitinophaga and Pedobacter within the class of sphingobacteria. In agreement with this hypothesis, the ORFs assigned to the three genome sequences from sphingobacteria had a relatively low percent identity compared with the reference genomes and a relatively low genome coverage (13–24%), indicating that these genomes recruited ORFs from more distantly related species (Figure 2).
Interestingly, many of the bacteria targeted by rather specific gene probes in the EBPR community had low similarity and low abundance compared with genomes of species from same genus in the reference databases. Examples are Thauera (1099 ORFs assigned, 69% average ORF similarity and 1 sequenced genome in the database), Azoarcus (1201 ORFs, 68% similarity, 1 genome) and Streptococcus (171 ORFs, 51% similarity, 54 genomes). Thus, the isolated sequenced species in the database are still not sufficiently closely related to the strains in the EBPR community for reliable taxonomic assignment. This genetic difference likely reflects considerable physiological differences between the sequenced isolates and the strains present in the EBPR plant.
The present metagenome also has the potential to direct further development of FISH probes to target a larger fraction of the community. Based on high abundance in the metagenome, additional FISH-probes targeting the orders of burkholderiales (betaproteobacteria), cytophagacea (bacteroidetes) and sphingomonadales (alphaproteobacteria) could complement the existing FISH probes.
The functional potential of the metagenome reflected EBPR selection pressure
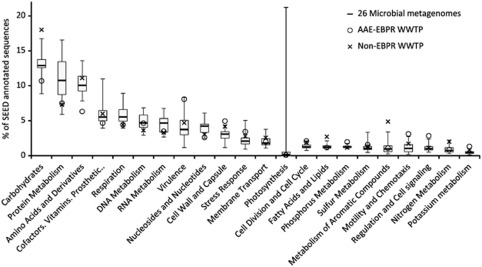
The overall functional potential of the metagenome was compared with an average of 26 microbial metagenomes from six distinct biomes (selected from Dinsdale et al., 2008) and with the metagenome of a non-EBPR WWTP (Sanapareddy et al., 2009) through the use of the SEED subsystems (Figure 3). No proteins were assigned to photosynthesis in the EBPR community, as was expected for a WWTP. A high abundance of phosphorus metabolism was detected, which can be attributed to the selection in EBPR treatment plants for microorganisms with the ability to store and use polyphosphate. A relatively high number of proteins was assigned to cell wall and capsule formation. This might be attributed to the high abundance of bacteria in treatment plants involved in the production and degradation of extracellular macromolecules as they grow in flocs or biofilms. As an example, 167 proteins in the metagenome were assigned to alginate metabolism, a polysaccharide that has been hypothesized to play an important role in biofilm formation in activated sludge systems (Lin et al., 2010). The high number of proteins attributed to the production of extracellular macromolecules may also be attributed to the strong selective pressure on activated sludge bacteria for effective floc/biofilm formation that leads to the good settling properties that are required to retain the biomass in the plant. In comparison with our community, the non-EBPR treatment plant had a higher number of proteins assigned to the metabolism of aromatic compounds. This may be attributed to the high industrial and medical wastewater load (Sanapareddy et al., 2009), in comparison with Aalborg East WWTP, which mainly treats municipal wastewater. Future investigations of different EBPR metagenomes may reveal other functions specific for the EBPR process.
Figure 3.
SEED subsystem comparison of the functional potential of the Aalborg East EBPR metagenome with a non-EBPR WWTP metagenome (Sanapareddy et al., 2009) and an average of 26 microbial metagenomes from six distinct biomes (Dinsdale et al., 2008). The data for the 26 microbial metagenomes is shown in a box plot with median, 25th percentile, 75th percentile, minimum and maximum values depicted.
Phages and transposases were abundant in the metagenome
In line with previous metagenomic studies, we found that transposases were prevalent in the metagenome, accounting for 1.6%. of all annotated sequences (3478 identified) (Brazelton and Baross, 2009; Konstantinidis et al., 2009; Sanapareddy et al., 2009; Aziz et al., 2010). In addition, a high number of phage proteins were identified. As an example, all ORFs predicted in the largest contig (32 884 bp) were annotated as either hypothetical or phage-related proteins, indicating that we have assembled at least part of a phage (Supplementary Figure S4). However, the host-specificity of the phage could not be suggested based on sequence similarity to prophages in sequenced genomes. Phages have recently been found to affect population dynamics of Accumulibacter in laboratory-scale enrichments (Kunin et al., 2008b; Barr et al., 2010), and the presence of phages in the metagenome of the full-scale plant suggests that deeply-sequenced metagenomic studies show promise for identifying the active phages in full-scale WWTPs.
Microdiversity and core genes of Accumulibacter
The Accumulibacter ppk1 sequences have been used as a phylogenetic marker for the genus and the known sequences appear to cover much of the in situ diversity (He et al., 2007). We amplified and sequenced 25 partial ppk1 sequences from Aalborg East WWTP and the sequences fell largely within the known diversity (Supplementary Figure S5). However, it is interesting that all ppk1 sequences were assigned to Accumulibacter clade II (IIA, IIB and IIx) and none to Accumulibacter clade I, even though the qFISH results indicated that clade I was the dominant clade. An extraction bias against clade I seems unlikely as this would infer that the clades have markedly different cell wall composition. Therefore, to further investigate the clade discrepancy, putative clade I and II 16S rRNA reads were identified in the metagenome by searching the clade I and II FISH probes against the 205 million raw metagenome reads, which resulted in 38 clade I and 23 clade II putative 16S rRNA reads. Applying the same principle using a single region discriminating clade I and II ppk1 sequences, we identified 25 clade II (9 × IIA, 4 × IIB, 6 × IIC+IID and 6 × IIE) and 0 clade I putative ppk1 reads. Hence, the observed discrepancy might be a factor of incomparable tree topology between ppk1 and 16S rRNA gene sequences or the available ppk1 sequences (and primers) do not represent the clade I diversity found in the treatment plant investigated in this study. This is contrary to Kim et al. (2010), who concluded that ppk1 and 16S rRNA tree topology seemed congruent. However, more full genomes (or isolates) are needed to compare 16S rRNA and ppk1 gene topologies definitively.
After assembly of the short metagenome reads into longer contigs, we were able to bin 7313 ORFs to Accumulibacter with a relatively high similarity (average 76% amino-acid similarity). However, no clear observations could be made about the microdiversity. The high similarity made it possible to utilize the full metagenome dataset instead of the subset of reads that could be assembled into contigs. This effectively increased the entire dataset, a factor ∼100 from 145 to 12 500 Mb of sequence, thereby enabling a more comprehensive investigation of the genome-wide microdiversity within the Accumulibacter species present in the metagenome.
In short, a reference assembly was made against the genome of Accumulibacter clade IIA strain UW-1 with all 205 million metagenome reads. The short read length required rather strict mapping criteria (85% identity over 70% of the read) to include Accumulibacter sequences without including reads from other bacteria, compared with other studies using longer reads (Rusch et al., 2007). In total, 1 428 946 reads (0.71% of all) were recruited to the Accumulibacter genome with the rRNA and tRNA genes masked.
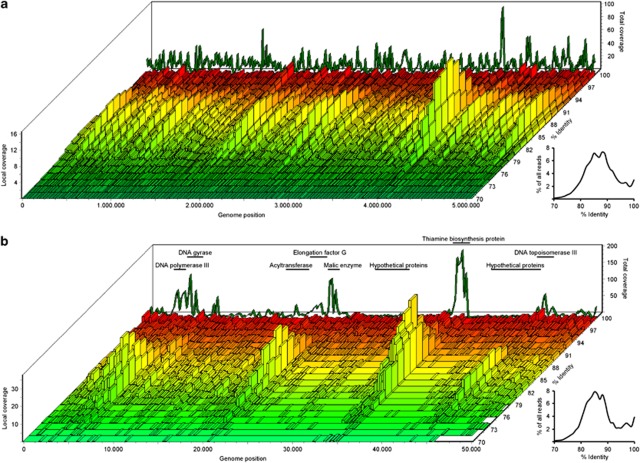
A graphical overview of the reference mapping is shown in Figure 4a. It illustrates that the reads had a distribution that facilitated grouping into matches at a high identity (179 741 reads ≈95–100% nucleotide identity) and a 7-time more abundant pool of reads with a lower identity (1 249 205 reads ≈85% nucleotide identity). The high-identity reads were evenly distributed along the entire genome, including hypothetical proteins (Figure 4b). In contrast, reads with lower identity had a more discrete distribution with markedly higher coverage of functional annotated genes compared with hypothetical proteins. The two, somewhat separated, read pools coincide with the sequence-discrete populations observed in other studies (Rusch et al., 2007; Konstantinidis and Delong, 2008). The relative low percentage of reads mapped to Accumulibacter (0.71%) compared with the biomass fraction of 4.8% determined by qFISH might be a consequence of disproportionally recruiting reads to the core genes of Accumulibacter and missing a considerable number of reads from genes in the pan-genome of Accumulibacter.
Figure 4.
Graphical overview of the mapping of the short metagenome reads to the genome of Accumulibacter clade IIA strain UW-1 as a function of percent identity of each read. rRNA and tRNA genes have been masked. In the back of the figure, the summed coverage for a given genome position is shown. To the right of each figure, an insert presents the overall read identity compared with the genome. (a) Overview of the full genome. Each bar represents an average read coverage of 10 000 bases. (b) Zoom on the first 50 000 bases. Each bar represents an average read coverage of 200 bases. Selected core and hypothetical genes have been marked on the figure to highlight the difference in coverage of functional annotated genes versus hypothetical proteins.
If we assume that the two read pools represent different Accumulibacter clades, then the smaller pool with >95% identity is putatively closely related to clade IIA (15% of all Accumulibacter reads), and the <95% identity pool of reads putatively represent the other Accumulibacter clades that are present. The high number of reads enables a comparison of the relative abundance in the two read pools of the genes they share with the clade IIA strain. We chose to use the fraction of the gene covered by metagenome reads (percent covered gene length) as a measure of the prevalence of a specific gene instead of the average number of metagenome reads recruiting to the specific gene (average gene coverage). The percent covered gene length mitigates the effect of highly conserved areas in genes, which would otherwise have a significant effect on the average gene coverage. In order to compare which genes that differed between the high (>95%) and low (⩽95%) identity read pools, the read-pool size of the low-identity group was normalized to the same size as the high-identity read pool (by subsampling), thereby effectively comparing the prevalent genes in both read pools (Supplementary Figure S6). Genes were classified as prevalent in one read pool compared with the other by having more than 50% more sequence covered in one of the read pools. In total, 353 genes were classified as exclusively to the high-identity read pool and 22 genes were classified as exclusive to the low-identity read pool. However, the main differences besides hypothetical proteins (51% of all) seemed to be phage-related and no major differences in essential gene content was found between the putative different groups of Accumulibacter.
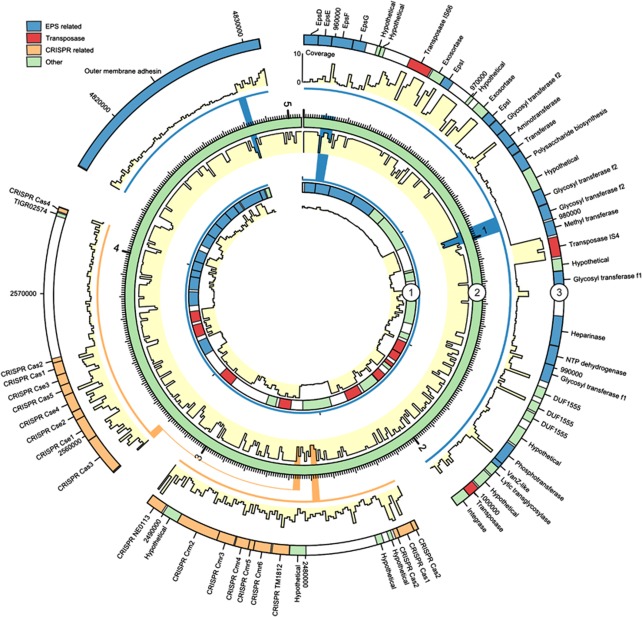
By classifying genes that were <10% covered by metagenome reads as unique to Accumulibacter clade IIA strain UW-1, 203 genes could be identified. To investigate these genes further, a circular genome plot was constructed to visualize genes potentially missing from the Accumulibacter strains present in the metagenome compared with the reference genome (Figure 5). A general trend was that the missing genes were localized in clusters and the main differences were related to phage predation and defense. The two EPS (extracellular polymeric substances) cassettes identified in the Accumulibacter genome (Garcia Martin et al., 2006) were not found within the genomes of the strains present in this metagenome, although some coverage was seen in very confined places owing to local sequence conservation. The mobility of EPS cassettes has been proposed to be involved in defense against phage predation (Kunin et al., 2008b). It is interesting that a high number of transposable elements are located within both of these clusters, which might enable the observed hyper variability compared with the rest of the genome. In addition, the observed lack of coverage within the two clustered regularly interspaced short palindromic repeats loci suggests that phage defense is highly active within the Accumulibacter species in the WWTP.
Figure 5.
Circular visualization of gene clusters unique to Accumulibacter clade IIA strain UW-1. Ring two represents the full Accumulibacter genome and in ring one and three selected areas of the genome have been expanded. Below each ring, the coverage profile is shown (yellow fill). The coverage profiles were calculated on the basis of the reference mapping and binned in either 10-kb (ring two) or 200-bp fragments (ring one and three). The coverage profile maximum was set at 10 to facilitate visualization (average coverage). Small marks represent 10 kb of sequence in all rings.
In conclusion, the diversity of Accumulibacter strains in the full-scale plants had considerable sequence divergence from the sequenced clade IIA strain, but shared majority of the genes (metabolic potential), exceptions being only typically variable elements such as surface proteins and phage-related genes. This suggests a considerable functional redundancy within the diversity of these process critical organisms. However, we are only able to investigate genes that are present in the sequenced Accumulibacter clade IIA strain, which was assembled from a highly enriched lab-scale reactor. Other Accumulibacter strains may possess genes that enable ecological differentiation, which has been shown to exist between different clades in lab-scale reactors (Flowers et al., 2009) and hypothesized to be present in nature (Peterson et al., 2008).
The EBPR ecosystem—a future model system in microbial ecology
The full-scale EBPR system investigated in this study is one of the best studied complex microbial ecosystems described to date (Nielsen et al., 2010) in addition to, for example, the acid mine drainage (Denef et al., 2010). So far, more traditional approaches have been applied to study the full-scale EBPR system (for example, the full cycle 16S rRNA approach and single cell microbiology), and this study clearly underline that metagenome-based work is needed for a deeper understanding of these engineered and highly complex communities. Clear differences to biomes from other ecosystems, also the only one existing from a WWTP, were demonstrated on the overall phylogenetic and functional level. However, the importance of not relying on a single approach in microbial ecology was demonstrated with the discrepancies between qFISH counts and metagenomic read numbers, although both are considered to be among the most reliable approaches currently available for quantitative microbial community analyses. We stress that these methods should be used in parallel whenever possible. The metagenome showed its great potential for mining functions of interest, such as the potential for alginate biosynthesis and possible phage-controlled microdiversity in Accumulibacter. However, the study also demonstrated that the lack of proper reference genomes is a real limitation of our approach to study these plants in detail in accordance with findings for other complex ecosystems (Nelson et al., 2010), and single cell genomics or deep sequencing is necessary to reveal the true diversity and at the same time increase the suitability for further transcriptomic or proteomic analyses. Although only one proper reference genome was available, we could demonstrate the great possibilities in the use of deep sequencing for investigations of genomic plasticity even in highly complex environments as exemplified through the Accumulibacter genome-wide gene-complement analysis. As key organisms in EBPR plants, Accumulibacter is a well-suited model organism for PAO metabolism, and further insights into their microdiversity, shared core genes and defense mechanisms (for example, to phages) are needed for unraveling in depth the structure–function relationships in EBPR systems with a high resolution (even to strain or ecotype). This is of interest beyond wastewater research as EBPR plants are more accessible and much easier to manipulate than most natural microbial ecosystems, and are therefore perfect as model systems in microbial ecology.
Acknowledgments
The study was funded by the Danish Strategic Research Council (EcoDesign-MBR). We thank Phil Hugenholtz and anonymous reviewers for helpful comments.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JJ, Slater FR, Fukushima T, Bond PL. Evidence for bacteriophage activity causing community and performance changes in a phosphorus-removal activated sludge. FEMS Microbiol Ecol. 2010;74:631–642. doi: 10.1111/j.1574-6941.2010.00967.x. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Baross JA. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 2009;3:1420–1424. doi: 10.1038/ismej.2009.79. [DOI] [PubMed] [Google Scholar]
- Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D, et al. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantification. Appl Environ Microbiol. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Relman D. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Dominiak DM, Nielsen JL, Nielsen PH. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol. 2011;13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD. Denitrification capabilities of two biological phosphorus removal sludges dominated by different ‘CandidatusAccumulibacter'clades. Environ Microbiol Rep. 2009;1:583–588. doi: 10.1111/j.1758-2229.2009.00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- He S, Gall DL, McMahon KD. ‘CandidatusAccumulibacter' population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl Environ Microbiol. 2007;73:5865–5874. doi: 10.1128/AEM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Kunin V, Haynes M, Garcia Martin H, Ivanova N, Rohwer F, et al. Metatranscriptomic array analysis of ‘Candidatus Accumulibacter phosphatis'-enriched enhanced biological phosphorus removal sludge. Environ Microbiol. 2010;12:1205–1217. doi: 10.1111/j.1462-2920.2010.02163.x. [DOI] [PubMed] [Google Scholar]
- He S, McMahon KD. ‘CandidatusAccumulibacter' gene expression in response to dynamic EBPR conditions. ISME J. 2010;5:329–340. doi: 10.1038/ismej.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- Hesselmann RPX, Werlen C, Hahn D, van der Meer JR, Zehnder AJB. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst Appl Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretschko S, Loy A, Lehner A, Wagner M. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. System Appl Microbiol. 2002;25:84–99. doi: 10.1078/0723-2020-00093. [DOI] [PubMed] [Google Scholar]
- Kim JM, Lee HJ, Kim SY, Song JJ, Park W, Jeon CO. Analysis of the fine-scale population structure of ‘Candidatus Accumulibacter phosphatis' in enhanced biological phosphorus removal sludge, using fluorescence in situ hybridization and flow cytometric sorting. Appl Environ Microbiol. 2010;76:3825–3835. doi: 10.1128/AEM.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2004;70:5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2005;71:4076–4085. doi: 10.1128/AEM.71.7.4076-4085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Xia Y, Nielsen JL, Nielsen PH. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology. 2007;153:4061–4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician's guide to metagenomics. Microbiol Mol Biol Rev. 2008a;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, et al. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008b;18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lin Y, de Kreuk M, van Loosdrecht MCM, Adin A. Characterization of alginate-like exopolysaccharides isolated from aerobic granular sludge in pilot-plant. Water Res. 2010;44:3355–3364. doi: 10.1016/j.watres.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KD, Yilmaz S, He S, Gall DL, Jenkins D, Keasling JD. Polyphosphate kinase genes from full-scale activated sludge plants. Appl Microbiol Biotechnol. 2007;77:167–173. doi: 10.1007/s00253-007-1122-6. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:e386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Darling AE, Eisen JA. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One. 2010;5:e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PH, Mielczarek AT, Kragelund C, Nielsen JL, Saunders AM, Kong Y, et al. A conceptual ecosystem model of microbial communities in enhanced biological phosphorus removal plants. Water Res. 2010;44:5070–5088. doi: 10.1016/j.watres.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Nguyen HTT, Le VQ, Hansen AA, Nielsen JL, Nielsen PH. High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol Ecol. 2011;76:256–267. doi: 10.1111/j.1574-6941.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Oehmen A, Lemos PC, Carvalho G, Yuan Z, Keller J, Blackall LL, et al. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 2007;41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Peterson SB, Warnecke F, Madejska J, McMahon KD, Hugenholtz P. Environmental distribution and population biology of CandidatusAccumulibacter, a primary agent of biological phosphorus removal. Environ Microbiol. 2008;10:2692–2703. doi: 10.1111/j.1462-2920.2008.01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinform. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajiliæ-Stojanoviæ M, Kekkonen RA, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Meth. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Sanapareddy N, Hamp TJ, Gonzalez LC, Hilger HA, Fodor AA, Clinton SM. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl Environ Microbiol. 2009;75:1688–1696. doi: 10.1128/AEM.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seviour RJ, Mino T, Onuki M. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol Rev. 2003;27:99–127. doi: 10.1016/S0168-6445(03)00021-4. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2290. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010;18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Vanysacker L, Declerck SAJ, Hellemans B, De Meester L, Vankelecom I, Declerck P. Bacterial community analysis of activated sludge: an evaluation of four commonly used DNA extraction methods. Appl Microbiol Biotechnol. 2010;88:299–307. doi: 10.1007/s00253-010-2770-5. [DOI] [PubMed] [Google Scholar]
- Wexler M, Richardson DJ, Bond PL. Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ Microbiol. 2009;11:3029–3044. doi: 10.1111/j.1462-2920.2009.02007.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Kong Y, Thomsen TR, Nielsen PH. Identification and characterization of epiphytic protein-hydrolyzing Saprospiraceae (Candidatus Epiflobacter spp.) in activated sludge. Appl Environ Microbiol. 2008;74:2229–2238. doi: 10.1128/AEM.02502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles JL, Peccia J, Kim M-W, Hung C-H, Noguera DR. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl Environ Microbiol. 2002;68:2763–2769. doi: 10.1128/AEM.68.6.2763-2769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.