The world is home to more than 1 billion pigs, which produce large quantities of feces. We know that some organisms in pig feces can cause human disease, and now we might have another to add to the list. Little is known about where the common intestinal parasite Dientamoeba fragilis comes from and how it is spread. However, recent molecular analysis confirmed that the organism found in pigs is indeed the same as the one found in humans. Therefore, pigs (or their feces) might be a source of this parasitic infection in humans.
Keywords: Parasites, Dientamoeba fragilis, pigs, zoonoses, protozoa, humans, genotypes
Abstract
Dientamoeba fragilis is a common intestinal parasite in humans. Transmission routes and natural host range are unknown. To determine whether pigs are hosts, we analyzed 152 fecal samples by microscopy and molecular methods. We confirmed that pigs are a natural host and harbor genotypes found in humans, suggesting zoonotic potential.
The flagellated protozoan Dientamoeba fragilis is one of the most common parasites in the intestinal tract of humans (1). Infection is highly prevalent in economically developing regions and in industrialized countries (1,2). Infected persons often show no symptoms, but a pathogenic role for this parasite has been reported recently in humans and gorillas (2–4). Little is known about transmission routes of this parasite, and a transmissible stage (e.g., a cyst) has not been described (1,5). Molecular characterization of human isolates based on sequence analysis of ribosomal genes revealed 2 genotypes (1 and 2), with genotype 1 predominating worldwide (6,7).
Other than humans, few animal hosts of D. fragilis have been reported. Surveys of mammals and birds have identified only nonhuman primates (gorillas, macaques, and baboons) as natural hosts (8,9). Recently, however, a high prevalence of infection (43.8%) has been reported in pigs in Italy (10). To determine whether pigs are a host of D. fragilis, we analyzed fecal samples from 152 pigs in Italy by microscopy and molecular methods.
The Study
During June–August 2010, a total of 152 fecal samples were collected from the rectums of piglets (age 1–3 months; weight 6–24 kg), fattening pigs (age 3–4 months; weight 25–50 kg), and sows (age 1–2 years; weight 180–250 kg). The pigs were raised in 6 farrow-to-finish farms, 2 fattening farms, and 1 weaner indoor farm of central Italy (7 farms in the Umbria region and 2 farms in the Marche region). Pig fecal samples from 7 of the 9 farms were available for molecular analysis. Fecal samples from 21 pig farmers were collected from 5 of the 9 farms, 17 of which were available for molecular analysis.
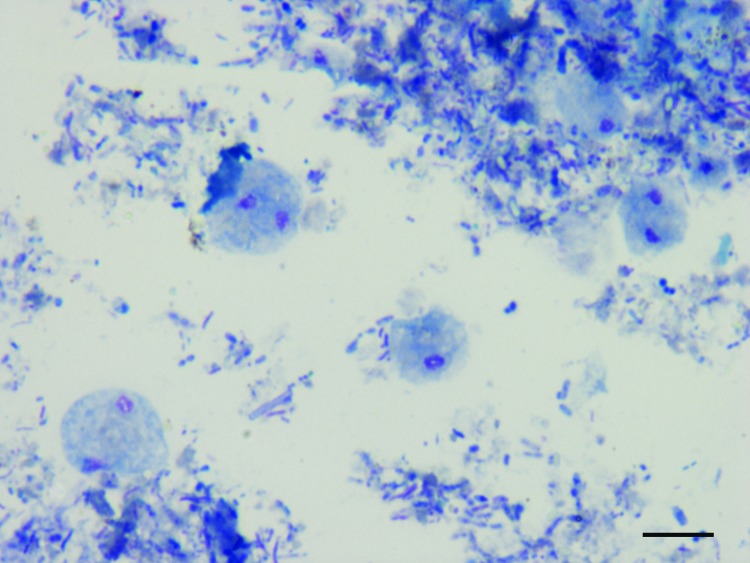
Microscopic diagnosis of D. fragilis was based on visualization of pleomorphic trophozoites, ranging in size from 4 µm to 20 µm, with fragmented chromatin and pale gray-blue finely vacuolated cytoplasm after Giemsa staining (Figure 1). DNA was extracted directly from 200 mg of feces by using the QIAamp DNA stool minikit (QIAGEN, Hilden, Germany). Reference D. fragilis DNA of genotype 1 (strains 379 and 1085) was used as a positive control.
Figure 1.
Dientamoeba fragilis trophozoites in a smear of pig feces after Giemsa staining, Italy, 2010–2011. Scale bar = 10 μm.
A TaqMan real-time PCR that targets the 5.8 S ribosomal locus was performed in a LightCycler 480 apparatus (Roche Diagnostics GmbH, Mannheim, Germany) as described (11). For the 18S rRNA gene, a published assay (12) was used to amplify a 662-bp fragment, followed by amplification of a 366-bp fragment with newly designed primers DF322For (5′-GAGAAGGCGCCTGAGAGATA-3′) and DF687Rev (5′-TTCATACTGCGCTAAATCATT-3′). For the internal transcribed spacer 1 (ITS1) region, a nested PCR protocol was developed. In the primary reaction, the forward primer ssu2 (13) and the reverse primer Df-ITSRev (5′-GCGGGTCTTCCTATATAAACAAGAACC-3′) were used, whereas the forward primer Df-ITSnesFor (5′-ATACGTCCCTGCCCTTTGTA-3′) and the reverse primer Df-ITSnesRev (5′-GCAATGTGCATTCAAAGATCGAAC-3′) were used in the nested PCR. PCR products were purified and sequenced on both strands. The sequences were assembled by using SeqMan II (DNASTAR, Madison, WI, USA) and compared with those available in public databases by using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences from this study were submitted to GenBank under accession nos. JQ677147–JQ677168.
The microscopic examination showed that 52 of the 74 piglets, 11 of the 14 fattening pigs, and 8 of the 64 sows were positive for D. fragilis (Table 1). More trophozoites were observed in fecal samples from piglets, suggesting a higher susceptibility of young animals to infection (data not shown). The microscopic analysis also showed Blastocystis spp. (in 42% of pigs), Endolimax nana protozoa (32%), Iodoamoeba buetschli protozoa (25%), and other flagellates (4.5%). Of the 21 samples from pig farmers, 4 from farmers working on 2 farms were positive for D. fragilis by microscopy (Table 1).
Table 1. Prevalence of Dientamoeba fragilis protozoa in pig and human fecal samples after microscopy and Giemsa staining, Italy, 2010–2011.
| Farm | Herd type | No. samples positive/no. tested* |
|||
|---|---|---|---|---|---|
| Piglet | Fattening pig | Sow | Human | ||
| 1 | Weaner production | 10/10 | NA | 1/10 | NA |
| 2 | Farrow-to-finish | 9/10 | NA | 3/10 | 0/4 |
| 3 | Farrow-to-finish | 10/10 | 7/10 | 0/10 | 2/8 |
| 4 | Farrow-to-finish | 1/10 | NA | 0/10 | NA |
| 5 | Farrow-to-finish | 4/10 | NA | 0/10 | 0/2 |
| 6 | Farrow-to-finish | 4/10 | NA | 1/10 | NA |
| 7 | Fattening | NA | NA | 3/4 | NA |
| 8 | Fattening | 10/10 | NA | NA | 2/3 |
| 9 | Farrow-to-finish | 4/4 | 4/4 | NA | 0/4 |
| Total | 52/74 | 11/14 | 8/64 | 4/21 | |
*NA, sample not available.
Molecular techniques were applied to 38 pig fecal samples, specifically 24 samples positive by microscopy from 6 farms and 14 samples negative by microscopy from 2 farms, and to all 17 human fecal samples. A comparison of human and pig samples collected from the same farm was possible for farms 2, 3, and 5 (Table 2). Using real-time PCR, all 24 positive pig samples were amplified, with cycle threshold values ranging from 30 to 34, whereas none of the 14 negative samples were positive to this assay (Table 2). However, because no inhibition controls were run, false-negative results cannot be ruled out. Of the 17 human fecal samples, 13 were positive with cycle threshold values of 29–40. The sequence analysis of 15 amplified products (11 from pigs and 4 from humans) showed 100% homology with D. fragilis genotype 1 (Table 2). Genotype 2 was not found in any of the samples from pigs or humans.
Table 2. Results of the molecular tests for Dientamoeba fragilis applied to pig and human fecal samples, Italy, 2010–2011*.
| Sample | Farm | Microscopy | 5.8S |
18S |
ITS1 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Real-time PCR | Sequence | PCR | Sequence | PCR | Sequence | |||||
| P21 | 2 | + | + | ND | – | ND | – | ND | ||
| P26 | 2 | + | + | D. fragilis | + | D. fragilis† | + | D. fragilis‡ | ||
| P27 | 2 | + | + | D. fragilis | – | ND | + | unclassified | ||
| P37 | 2 | + | + | ND | – | ND | – | ND | ||
| P42 | 3 | + | + | D. fragilis | + | D. fragilis† | + | D. fragilis§ | ||
| P44 | 3 | + | + | D. fragilis | – | ND | + | flagellate | ||
| P45 | 3 | + | + | ND | – | ND | – | ND | ||
| P50 | 3 | + | + | ND | – | ND | – | ND | ||
| P52 | 3 | + | + | D. fragilis | – | ND | + | unclassified | ||
| P54 | 3 | + | + | D. fragilis | – | ND | + | flagellate | ||
| P56 | 3 | + | + | D. fragilis | + | D. fragilis† | + | unclassified | ||
| P59 | 3 | + | + | D. fragilis | + | D. fragilis† | + | unclassified | ||
| P60 | 3 | + | + | ND | – | ND | – | ND | ||
| P71 | 4 | + | + | ND | – | ND | – | ND | ||
| P75 | 4 | + | + | ND | – | ND | – | ND | ||
| P91 | 5 | + | + | ND | – | ND | – | ND | ||
| P93 | 5 | + | + | D. fragilis | + | D. fragilis† | + | unclassified | ||
| P 97 | 5 | + | + | D. fragilis | + | D. fragilis† | + | flagellate | ||
| P111 | 6 | + | + | ND | – | ND | – | ND | ||
| P113 | 6 | + | + | ND | – | ND | – | ND | ||
| P116 | 6 | + | + | ND | – | ND | – | ND | ||
| P122 | 6 | + | + | D. fragilis | – | ND | + | flagellate | ||
| P131 | 6 | + | + | ND | – | ND | – | ND | ||
| P133 | 7 | + | + | ND | – | ND | – | ND | ||
| Pig 1 | 1 | – | – | ND | + | Trichomitus¶ | – | ND | ||
| Pig 2 | 1 | – | – | ND | – | ND | – | ND | ||
| Pig 3 | 1 | – | – | ND | + | Trichomitus¶ | – | ND | ||
| Pig 4 | 1 | – | – | ND | – | ND | – | ND | ||
| Pig 5 | 1 | – | – | ND | + | Trichomitus¶ | – | ND | ||
| Pig 6 | 1 | – | – | ND | – | ND | – | ND | ||
| Pig 7 | 1 | – | – | ND | – | ND | – | ND | ||
| Pig 8 | 1 | – | – | ND | – | ND | – | ND | ||
| Pig 9 | 1 | – | – | ND | – | ND | – | ND | ||
| Pig 10 | 1 | – | – | ND | – | ND | – | ND | ||
| DF-P1 | 6 | – | – | ND | – | ND | – | ND | ||
| DF-P2 | 6 | – | – | ND | – | ND | – | ND | ||
| DF-P3 | 6 | – | – | ND | – | ND | – | ND | ||
| DF-P4 | 6 | – | – | ND | – | ND | – | ND | ||
| H1 | 2 | – | + | D. fragilis | + | D. fragilis† | – | ND | ||
| H2 | 2 | – | – | ND | – | ND | – | ND | ||
| H3 | 2 | – | + | ND | – | ND | – | ND | ||
| H4 | 2 | – | + | ND | + | D. fragilis† | – | ND | ||
| H5 | 5 | – | + | ND | – | ND | – | ND | ||
| H6 | 5 | – | + | ND | + | D. fragilis† | – | ND | ||
| H7 | 3 | – | + | ND | + | D. fragilis† | – | ND | ||
| H8 | 3 | – | + | ND | – | ND | – | ND | ||
| H9 | 3 | – | – | ND | – | ND | – | ND | ||
| H10 | 3 | + | + | ND | + | D. fragilis† | – | ND | ||
| H11 | 3 | + | + | D. fragilis | + | D. fragilis† | – | ND | ||
| H12 | 3 | – | + | ND | – | ND | – | ND | ||
| H13 | 3 | – | + | ND | – | ND | – | ND | ||
| H14 | 3 | – | – | ND | – | ND | – | ND | ||
| H15 | 8 | + | + | D. fragilis | + | D. fragilis† | + | D. fragilis# | ||
| H16 | 8 | + | – | ND | – | ND | – | ND | ||
| H17 | 9 | – | + | D. fragilis | + | D. fragilis† | + | D. fragilis# | ||
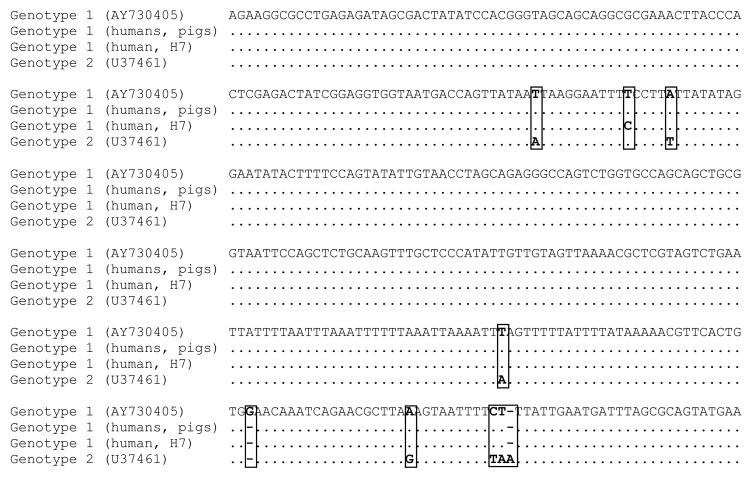
Next, a 366-bp fragment of the 18S rRNA gene was analyzed. In this fragment, genotypes 1 and 2 can be distinguished by 8 substitutions or insertions or deletions (Figure 2), which were further confirmed by sequencing the entire 18S rRNA gene from 2 reference isolates and 2 human isolates from this study. Amplification was obtained from 6 of the 24 positive pig samples and from 8 of the 17 human samples. Genotype 1 was identified in all samples (Figure 2). One human isolate (H7) showed a single nucleotide substitution in the fragment sequenced (Figure 2). Sequences from 3 microscopically negative pig samples (all from farm 1) had a high homology (96%) with Trichomitus batrachorum, a flagellate of reptiles, although the sequence could originate from T. rotunda, a flagellate of pigs that has not been described at the molecular level.
Figure 2.
Multiple alignment of the 366-bp fragment of the 18S rRNA gene from Dientamoeba fragilis genotypes 1 and 2. Dot indicates identical nucleotides. Dashes indicate insertion or deletion. Nucleotide differences are presented in boxes.
Last, we studied the more variable ITS1 locus. Amplification was obtained from 11 of the 24 pig samples (Table 2), but only 2 sequences could be clearly identified as D. fragilis. Four sequences showed homology (80%) with flagellates from different vertebrate classes whereas the remaining 5 sequences were excluded because of insufficient quality. The 2 D. fragilis sequences from pigs showed 100% homology with sequences from human isolates from the United Kingdom (Table 2), further supporting the presence of genotype 1 in these 2 hosts. A direct comparison of ITS1 sequences from humans and pigs from a single farm in Italy was not possible because D. fragilis was amplified from only 2 human samples from 2 farms from which no pig samples were available. The analysis of ITS1 from the 2 human isolates showed full identity to human isolates from the Netherlands and the United Kingdom (Table 2).
Conclusions
Considering the size of the world’s pig population (>1 billion), the close contact between pigs and humans in many parts of the world, and the difficulties in the proper management of pig fecal waste, the role of these animals as reservoirs of zoonotic pathogens must be carefully evaluated. We demonstrated that pigs are hosts of D. fragilis, on the basis of molecular analysis of 3 fragments in the ribosomal cluster. Sequence analyses of fragments of the 18S and 5.8S rRNA genes showed genotype 1 in isolates collected in the same farm from humans and pigs, suggesting the potential for zoonotic transmission of this parasite. If a transmissible cyst stage exists, then environmental contamination with pig feces should be considered a key factor in the transmission of this parasite. Pigs also are a fascinating animal model to elucidate the life cycle of this elusive parasite.
Acknowledgments
We thank Daniele Tonanzi, Sonia Salamida, and Silvia Crotti for their excellent technical contribution. We also thank J. Windsor for supplying the reference DNA used in this study.
This study was supported by a research grant from the Italian Ministry of Health (IZSUM 16/09 RC) and by the European Commission (contract SANCO/2006/FOODSAFETY/032).
Biography
Dr. Simone Cacciò is a senior researcher at the Department of Infectious, Parasitic, and Immunomedioated Diseases of the Istituto Superiore di Sanità in Rome, Italy. His main research interest is the molecular epidemiology of intestinal parasites, particularly Cryptosporidium and Giardia.
Footnotes
Suggested citation for this article: Cacciò SM, Sannella AR, Manuali E, Tosini F, Sensi M, Crotti C, et al. Pigs as natural hosts of Dientamoeba fragilis genotypes found in humans. Emerg Infect Dis [serial on the Internet]. 2012 May [date cited]. http://dx.doi.org/10.3201/eid1805.111093
References
- 1.Johnson EH, Windsor JJ, Clark CG. Emerging from obscurity: biological, clinical, and diagnostic aspects of Dientamoeba fragilis. Clin Microbiol Rev. 2004;17:553–70. 10.1128/CMR.17.3.553-570.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barratt JL, Harkness J, Marriott D, Ellis JT, Stark D. A review of Dientamoeba fragilis carriage in humans: several reasons why this organism should be considered in the diagnosis of gastrointestinal illness. Gut Microbes. 2011;2:3–12. 10.4161/gmic.2.1.14755 [DOI] [PubMed] [Google Scholar]
- 3.Stark D, Barratt J, Roberts T, Marriott D, Harkness J, Ellis J. A review of the clinical presentation of dientamoebiasis. Am J Trop Med Hyg. 2010;82:614–9. 10.4269/ajtmh.2010.09-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lankester F, Kiyang JA, Bailey W, Unwin S. Dientamoeba fragilis: initial evidence of pathogenicity in the western lowland gorilla (Gorilla gorilla gorilla). J Zoo Wildl Med. 2010;41:350–2. 10.1638/2009-0190.1 [DOI] [PubMed] [Google Scholar]
- 5.Barratt JL, Harkness J, Marriott D, Ellis JT, Stark D. The ambiguous life of Dientamoeba fragilis: the need to investigate current hypotheses on transmission. Parasitology. 2011;138:557–72. 10.1017/S0031182010001733 [DOI] [PubMed] [Google Scholar]
- 6.Johnson JA, Clark CG. Cryptic genetic diversity in Dientamoeba fragilis. J Clin Microbiol. 2000;38:4653–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Windsor JJ, Macfarlane L, Clark CG. Internal transcribed spacer dimorphism and diversity in Dientamoeba fragilis. J Eukaryot Microbiol. 2006;53:188–92. 10.1111/j.1550-7408.2006.00092.x [DOI] [PubMed] [Google Scholar]
- 8.Stark D, Phillips O, Peckett D, Munro U, Marriott D, Harkness J, et al. Gorillas are a host for Dientamoeba fragilis: an update on the life cycle and host distribution. Vet Parasitol. 2008;151:21–6. 10.1016/j.vetpar.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 9.Windsor JJ, Johnson EH. Dientamoeba fragilis: the unflagellate human flagellate. Br J Biomed Sci. 1999;56:293–306. [PubMed] [Google Scholar]
- 10.Crotti D, Sensi M, Crotti S, Grelloni V, Manuali E. Dientamoeba fragilis in swine population: a preliminary investigation. Vet Parasitol. 2007;145:349–51. 10.1016/j.vetpar.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 11.Verweij JJ, Mulder B, Poell B, van Middelkoop D, Brienen EA, van Lieshout L. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol Cell Probes. 2007;21:400–4. 10.1016/j.mcp.2007.05.006 [DOI] [PubMed] [Google Scholar]
- 12.Vandenberg O, Peek R, Souayah H, Dediste A, Buset M, Scheen R, et al. Clinical and microbiological features of dientamoebiasis in patients suspected of suffering from a parasitic gastrointestinal illness: a comparison of Dientamoeba fragilis and Giardia lamblia infections. Int J Infect Dis. 2006;10:255–61. 10.1016/j.ijid.2005.05.011 [DOI] [PubMed] [Google Scholar]
- 13.Bart A, van der Heijden HM, Greve S, Speijer D, Landman WJ, van Gool T. Intragenomic variation in the internal transcribed spacer 1 region of Dientamoeba fragilis as a molecular epidemiological marker. J Clin Microbiol. 2008;46:3270–5. 10.1128/JCM.00680-08 [DOI] [PMC free article] [PubMed] [Google Scholar]