Abstract
Myostatin (MSTN) is a negative regulator of skeletal muscle growth and development. There are two types of MSTNs in fish, but little is known about their gene regulation. Here, the 5′ flanking fragments of 1029 bp from MSTN-1 and 643 bp from MSTN-2 were cloned, sequenced, and analyzed in Larimichthys crocea. Both fragments contained CAAT box and several putative cis-regulatory elements. However, putative TATA box, MyoD, MEF3, SP1, USF, and GH-CSE sites were identified only in the L. crocea MSTN-1 (lcMSTN-1) promoter. Transcriptional activities of four fragments (1013, 841, 514, and 261 bp) truncated from lcMSTN-1 upstream region and two fragments (643 and 296 bp) from lcMSTN-2 upstream region were examined in vitro, using transient transfection in CIK and L6 cells. In CIK cells, the promoter activity correlated positively with the length of truncated fragments in both MSTN-1 and 2. The lcMSTN-2 promoter showed a higher activity than lcMSTN-1 in the corresponding region, which was consistent with MSTN gene expression in vivo. In L6 cells, lcMSTN-2 upstream showed an extremely high luciferase activity. These data indicated that both cloned 5′ flanking sequences contained functional promoters, and that transcription regulation of lcMSTN-1 and 2 promoters was significantly different between mammalian and fish cells.
Introduction
Myostatin (MSTN), also known as growth and differentiation factor-8, was first identified in mice (McPherron et al., 1997). It is a member of the transforming growth factor-beta superfamily, and a potent negative regulator of skeletal muscle growth and development in mammals (Lee, 2004). Both naturally occurring and experimentally induced mutations that result in lower MNST activity induce remarkable muscle mass increase in a number of vertebrates, including cattle (Grobet et al., 1997; Kambadur et al., 1997; McPherron and Lee, 1997; Smith et al., 2000; Marchitelli et al., 2003), sheep (Clop et al., 2006), dogs (Mosher et al., 2007), mice (McPherron et al., 1997; Szabo et al., 1998; Zhu et al., 2000), humans (Schuelke et al., 2004), medaka (Sawatari et al., 2010), and zebrafish (Xu et al., 2003; Acosta et al., 2005; Lee et al., 2009). In cattle, because of its beneficial effects on meat production, several double-muscling breeds have been selectively bred for this trait (Rodgers and Garikipati, 2008). Conversely, MSTN overexpression in the skeletal muscle of the transgenic mice is associated with decreased body weight and skeletal muscle mass (Reisz-Porszasz et al., 2003). Overexpression of MSTN-2 in zebrafish reduces the expression of dystrophin associated protein complex, which leads to muscle dystrophy (Amali et al., 2008). These growth-inhibitory actions have also been demonstrated in vitro using different mammalian cell lines (Thomas et al., 2000; Rios et al., 2001, 2002; Taylor et al., 2001; Langley et al., 2002; Joulia-Ekaza et al., 2003; McCroskery et al., 2003). In fish, soluble recombinant MSTN-1 prodomain can improve body growth (Lee et al., 2010).
The key role of MSTN in skeletal muscle growth and its potential application prompted the study of MSTN from fishes with important commercial value. These studies have shown that fish MSTN is different in comparison with mammals. First, two or more types of MSTNs have been recently found in some teleosts (Garikipati et al., 2006; Helterline et al., 2007; Ostbye et al., 2007). However, only one MSTN has been found so far in mammals. Second, fish MSTN has a wider expression range than mammals. In mammals, MSTN expression was observed in the skeletal muscle (McPherron et al., 1997; Gonzalez-Cadavid et al., 1998), and at a lower level, in the adipose tissue (McPherron et al., 1997), cardiac muscle (Sharma et al., 1999), and mammary gland (Ji et al., 1998). In contrast to mammals, fish MSTN is differentially expressed in many tissues. In seabream, MSTN-1 is expressed in the muscle, brain, eye, intestine, heat, and kidney, while MSTN-2 is expressed mainly in the brain (Maccatrozzo et al., 2001a, 2001b). In rainbow trout, MSTN-1a mRNA was present ubiquitously in trout tissues, while MSTN-1b mRNA expression was restricted to the muscle and brain (Rescan et al., 2001). The expression of rainbow trout MSTN-2a was detected in all sampled tissues, including the brain, skin, gill, heart, kidney, spleen, intestine, stomach, liver, eye, and muscle (Garikipati et al., 2007). Diverse expression patterns in fish suggest that the biological action of MSTN may not be restricted to the skeletal muscle, but may additionally influence other fish tissues as well (Kerr et al., 2005).
In mammals, sequence alignment of the human, goat, sheep, pig, and cow MSTN promoters revealed five conserved sequences (Allen and Du, 2008). The regulation of MSTN expression under various physiological conditions has been extensively studied (Joulia-Ekaza and Cabello, 2006). Compared with higher vertebrates, the transcription regulation of MSTN promoters in teleosts is poorly understood. The MSTN promoters have been sequenced in some teleost species such as zebrafish (Danio rerio) (AY323521, DQ451548), largemouth bass (Micropterus salmoides) (EF071854), Atlantic salmon (Salmo salar) (EF392862), barramundi (Lates calcarifer) (EF672685), gilthead sea bream (Sparus aurate) (EU881511), olive flounder (Paralichthys olivaceus) (DQ997779), rainbow trout (Oncorhynchus mykiss) (DQ136028, DQ138301), brook trout (Salvelinus fontinalis) (AY227656), sea perch (Lateolabrax japonicas) (AY965685), and channel catfish (Lctalurus punctatus) (AF396747), but only gilthead sea bream MSTN-1 promoter activity has been functionally analyzed (Funkenstein et al., 2009). Functional analysis of the MSTN-2 promoter has not been reported so far. Large yellow croaker (Larimichthys crocea) is a major marine-cultured species in China. We have cloned MSTN-1 (Xue et al., 2006) and 2 (EU571244) genes from L. crocea. To better understand their promoter structure and regulatory mechanisms, we cloned and functionally analyzed the promoters of MSTN-1 and 2 from L. crocea.
Materials and Methods
Sample
L. crocea was collected at the Ningbo Bay Breeding Center of Aquatic Animals. Skeletal muscle tissue was dissected from living large yellow croakers, rapidly frozen in liquid nitrogen, and stored at −70°C.
Genomic DNA extraction
Genomic DNA was extracted from skeletal muscle tissue. Five hundred microliters lysis buffer (1% sodium dodecyl sulfate, 100 mM/L ethylenediaminetetraacetic acid, and 10 nM/L Tris, pH 8.0) was added to 0.15 g muscle tissue and incubated for 2 h with proteinase K (20 mg/mL) at 65°C. Three consecutive extractions, phenol, phenol: chloroform: isoamyl alcohol (25:24:1), and chloroform: isoamyl alcohol (24:1), were then performed. The precipitated DNA was washed with 70% ethanol, dried, resuspended in 100 μL ddH2O, and stored at −20°C.
Cloning of L. crocea MSTN-1 and 2 promoters
Based on the cloned MSTN genes of L. crocea (AY842933, EU571244), six specific forward primers were designed to amplify the 5′ flanking regions of MSTN-1 and 2 genes. Three forward primers, 1SP1, 1SP2, and 1SP3, were used in three round polymerase chain reaction (PCR) for L. crocea MSTN-1 (lcMSTN-1) promoter cloning, respectively. The other three primers, 2SP1, 2SP2, and 2SP3, were used in three-round PCR for lcMSTN-2 promoter cloning. The primer sequences were listed in Table 1. The nested PCR of three rounds was performed to isolate lcMSTN-1 and -2 promoters from genomic DNA using the Genome Walking Kit (TaKaRa Biotechnology) according to the manufacturer's instructions. The reverse primer was AP4 in all PCR reactions, which was supplied in the kit. For lcMSTN-1 promoter cloning, the first round PCR conditions were 1 cycle (94°C 1 min, 98°C 1 min), 5 cycles (94°C 30 s, 65°C 1 min, and 72°C 2 min), 1 cycle (94°C 30 s, 25°C 3 min, and 72°C 2 min), and 15 cycles (94°C 30 s, 65°C 1 min, and 72°C 2 min; 94°C 30 s, 65°C 1 min, and 72°C 2 min; 94°C 30 s, 44°C1 min, and 72°C 2 min), and 1 cycle (72°C 10 min). The second-round PCR conditions were 15 cycles (94°C 30 s, 60°C 1 min, and 72 2 min; 94°C 30 s, 60°C 1 min, and 72°C 2 min; 94°C 30 s, 44°C 1 min, and 72°C 2 min), and 1 cycle (72°C 10 min). The third-round PCR conditions were 15 cycles (94°C 30 s, 60°C 1 min, and 72°C 2 min; 94°C 30 s, 60°C 1 min, and 72°C 2 min; 94°C 30 s, 44°C 1 min, and 72°C 2 min), and 1 cycle (72°C 10 min). The cloning protocol for the lcMSTN-2 promoter was the same as the lcMSTN-1 promoter. The detailed PCR conditions are listed in Supplementary Table S1. The PCR products were run on a 1% agarose gel, and then purified by EZ-10 Spin Column DNA Gel Extraction Kit (Shenggong).The purified PCR products were cloned into pMD18-T vector (TaKaRa Biotechnology), and the recombinant vector was transformed into competent DH5α cells. The recombinant plasmids were isolated from the transformed DH5α cells with EZ-10 Spin Column Plasmid Mini-Preps Kit (Shenggong). The inserted PCR fragments were sequenced on an ABIPRISM 3700 DNA sequencer.
Table 1.
Nucleotide Sequences of Primers used in Polymerase Chain Reaction for lcMSTN Promoter Cloning and Characterization
| Assigned name | Sequence | Purpose |
|---|---|---|
| 1SP1 | 5′ CTGGTGCGTCTCTTGGTCAC 3′ | Cloning of lcMSTN-1 promoter |
| 1SP2 | 5′ GAGACAGATGCATTGTCTCTC 3′ | Cloning of lcMSTN-1 promoter |
| 1SP3 | 5′ GGACTGGGTTTGGATTAATGTC 3′ | Cloning of lcMSTN-1 promoter |
| 2SP1 | 5′ AATGGTCTCTGTCGTAGCGTGGT 3′ | Cloning of lcMSTN-2 promoter |
| 2SP2 | 5′ TTGATACTATGGAGCCGCATCTGTTT 3′ | Cloning of lcMSTN-2 promoter |
| 2SP3 | 5′ CCCATTGAAAAGCCCGCAGAGAA 3′ | Cloning of lcMSTN-2 promoter |
| 1F1 | 5′ GGGGTACCTGGTGTTCACACTTTAGAGT 3′ | Construction of lcMSTN-1 promoter reporter plasmids |
| 1F2 | 5′ GGGGTACCCTAATGTGAATGACGTGAAC 3′ | Construction of lcMSTN-1 promoter reporter plasmids |
| 1F3 | 5′ GGGGTACCGTCTGATGGATTTATTGTG 3′ | Construction of lcMSTN-1 promoter reporter plasmids |
| 1F4 | 5′ GGGGTACCTCACAGTCTCCATCCCTTTAT 3′ | Construction of lcMSTN-1 promoter reporter plasmids |
| 1R | 5′ CCCAAGCTTTGTCTCTCAGGTGTGAAG 3′ | Construction of lcMSTN-1 promoter reporter plasmids |
| 2F1 | 5′ GGGGTACCGCAGTGCAGCACATCCA 3′ | Construction of lcMSTN-2 promoter reporter plasmids |
| 2F2 | 5′ GGGGTACCGAAGTAAAATGCGAAC 3′ | Construction of lcMSTN-2 promoter reporter plasmids |
| 2R | 5′ CCCAAGCTTCCTGGAGCGATGAGGA 3′ | Construction of lcMSTN-2 promoter reporter plasmids |
Restriction enzyme recognition sites introduced in some oligonucleotides are underlined.
Sequence analysis
Similarity searches of the sequenced lcMSTN promoters were done by Blastn (www.ncbi.nlm.nih.gov/blast/). The transcription factor binding sites were analyzed with TFSEARCH with a threshold score of 90 (Heinemeyer et al., 1998), TESS (www.cbil.upenn.edu/cgi-bin/tess/tess), and MatInspector (Cartharius et al., 2005). A multiple-sequence alignment was performed using ClustalW (Thompson et al., 1994). Phylogenetic trees were constructed using the neighbor-joining tree of MEGA version 4.0 (Tamura et al., 2007). The species in the sequence alignment include D. rerio (AY323521, DQ451548), M. salmoides (EF071854), S. salar (EF392862,), L. calcarifer (EF672685), S. aurate (EU881511, GQ379809), P. olivaceus (DQ997779), O. mykiss (DQ136028, DQ138301), S. fontinalis (AY227656), L. japonicas (AY965685), and L. punctatus (AF396747).
Construction of lcMSTN-1-Luc and lcMSTN-2-Luc transgenes
Four fragments, 1013, 841, 514, and 261 bp, of lcMSTN-1 5′ flanking sequence were amplified with primers 1F1, 1F2, 1F3, 1F4 (forward), and 1R (reverse), and two fragments, 643 and 296 bp, of lcMSTN-2 5′ flanking sequence with primers 2F1, 2F2, and 2R, respectively. The forward primers introduced a 5′ KpnI restriction site, and the reverse primers, a 5′ HindIII restriction site (Table 1). The PCR condition was as follows: 94°C for 3 min followed by 35 cycles of 40 s at 94°C, 40 s at 60°C, 2 min at 72°C, and 72°C extended for 8 min. All PCR products were run on a 1% agarose gel and then gel purified by EZ-10 Spin Column DNA Gel Extraction Kit. Both the purified fragments and the promoterless reporter plasmid pGL3-Basic which contains a luciferase gene (Promega), were double digested with KpnI and HindIII (TaKaRa Biotechnology). The resultant fragments were directly inserted into the pGL3-Basic, and the constructed plasmids were named pGL3-1-1013, pGL3-1-841, pGL3-1-514, pGL3-1-261, pGL3-2-643, and pGL3-2-296, respectively. All recombinant constructs were sequenced to verify the orientation of the promoter relative to the luciferase reporter gene.
Cell culture, transfection, and dual-luciferase reporter assay
The CIK cell line, derived from Ctenopharyngodon idellus kidney tissue, was provided by Zhejiang Institute of Freshwater Fisheries, and maintained in RPMI 1640 containing 10% fetal bovine serum at 26°C. CIK cells are heterogeneous cell culture, including fiber cell (98.3%), polygon cell (1.4%), and giant cell (0.3%). L6, a rat skeletal muscle cell line purchased from BOSTER, was maintained in DMEM with 10% fetal bovine serum in a 5% CO2 incubator at 37°C.
For the transfection, 1.0×104 cells/well were plated in 96-well plates and grown to ∼90% confluence. The cells were co-transfected at 26°C with 0.2 μg of the constructed reporter plasmid vector containing the MSTN promoter fragment and 1 ng of the Renilla luciferase reporter plasmid pRL-CMV (Promega) using the Lipofectamine 2000 system (Invitrogen) for each well according to the manufacturer's protocol. The cells were cultured for 6 h, and then, the transfection medium was replaced with fresh medium and incubation was continued. Cells were collected 48 h after transfection. Firefly and Renilla luciferase activities were measured in cell lysates using the Dual-Luciferase Assay System (Promega) in an SHG-D Luminometer (Shanghai Shangli Detecting Instrument Factory). The relative luciferase activities of promoters were normalized to the control reporter. The final data were from three independent experiments, and each experiment was performed in triplicate.
Mutation of MyoD binding site in the lcMSTN-1 promoter
The E-box6 (MyoD binding site) in the lcMSTN-1 promoter was mutated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the instruction manual. Two complementary primers containing the desired mutation to the MyoD binding site were used as primers for PCR to introduce mutations in pGL3-1-1013 using Pfu DNA polymerase. The two primers have the following sequences: P1 5′ CACTCACTCAGGTTGCCGCTTTGACGTCTCTAATG 3′, and P2 5′ CATTAGAGACGTCAAAGCGGCAACCTGAGTGAGTG 3′ (the mutated positions are underlined). The PCR product was then digested with the Dpn I restriction enzyme that specifically cuts the original methylated template plasmid but was unable to cut the newly synthesized DNA. The digested DNA was then transformed into XL-1 blue supercompetent cells. The white colonies were selected for sequencing analysis to identify the correct clone with desired mutations. The pGL3-1-1013 with MyoD binding site mutations were used in dual-luciferase reporter assay.
Expression analysis of lcMSTN-2
Total RNA was extracted using Trizol Reagent (Sangon) from ten different tissues and organs, spleen, heart, brain, adipose, kidney, gill, eye, intestine, liver, and skeletal muscle of large yellow adult croakers. Reverse transcription (RT)-PCR was used to analyze the tissue-specific expression of lcMSTN-2. Primers for amplifying MSTN-2 are Myo2F 5′ CCATAGTATCAAGTCCCAGATCC 3′, and Myo2R 5′ TGAACAGGCAGCAGGAGGACAAC 3′. β-actin gene served as a positive control. The primers of β-actin were 5′ CCAGATCATGTTCGAGACCTTC 3′, and 5′ GAACCTCTCATTGCCAATGGTG 3′. The RT-PCR products from each tissue were electrophoresed on a 1% agarose gel.
Results
Cloning of lcMSTN-1 and 2 5′ flanking regions
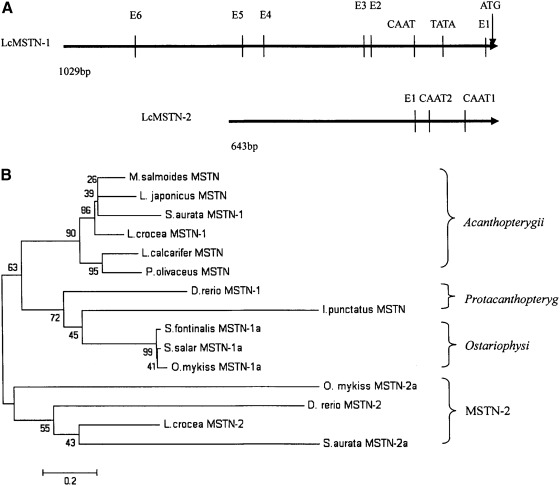
One thousand twenty-nine and 643 bp upstream sequences from the translation start codon of lcMSTN-1 and 2 were cloned by the genome walking method. Their nucleotide sequences are shown in Figure 1A and B. The upstream sequence of lcMSTN-1 had a homology of more than 90% with the reported sequences of Sparus aurata and L. japonicas in the same region. A multiple-sequence alignment showed that the CAAT box and TATA box were conserved among teleost MSTN-1 upstream sequences (Fig. 2). However, the upstream sequence of lcMSTN-2 showed a low conservation compared with that of other fish MSTN-2. The conserved sequence could not be identified among MSTN-2, and between MSTN-1 and 2 promoter sequences.
FIG. 1.
Promoter sequences of Larimichthys crocea MSTN-1 (A) and 2 (B). The first nucleotide in the translation start codon ATG is designated +1. The TATA box and CAAT box are in boldface. E-box (CANNTG) is underlined. MSTN, myostatin.
FIG. 2.
The alignment of 11 teleost MSTN-1 upstream sequences (partial). The conserved regions are CAAT box and TATA box. Consenus nucleotides in all aligned sequences are indicated with asterisks.
Sequence alignment revealed a big difference between 5′ flanking sequences of lcMSTN-1 and 2. More transcription factor recognition sites existed in lcMSTN-1 5′ flanking region than that of lcMSTN-2 (Fig. 3A). A phylogenetic tree showed that 11 fish MSTN-1 upstream sequences formed a subgroup, and three fish MSTN-2 upstream sequences formed the other group (Fig. 3B).
FIG. 3.
Comparative analysis of major elements in 5′ flanking regions of L. crocea MSTN-1 (lcMSTN-1) and 2 (A) and neighbor-joining tree based on 5′ flanking regions of MSTN genes (B). Numbers at tree nodes refer to the percentage bootstrap values after 1000 replicates.
Transcription factor binding sites in the 5′ flanking regions of lcMSTN-1 and 2 genes
One TATA box, one CAAT box, and six putative E-boxes were found in lcMSTN-1. The TATA box and CAAT box were located at -131 and -175 upstream from the ATG initiation codon, respectively. In the lcMSTN-2, two CAAT boxes and only one putative E box were found. The E box was located at -155, and two CAAT boxes at -76 and-146 bp upstream from the ATG initiation codon, respectively (Fig. 1A, B). However, no TATA box was found in the lcMSTN-2 promoter.
Sequence analysis identified several putative muscle-specific transcriptional factor binding sites and cis-regulatory elements in both sequences. These regulatory elements include myocyte enhancer factor 2 (MEF2), activator protein 1 (AP1), cAMP response element (CRE), POU3F2 (N-Oct-3), POU1F1a (Pit1a), CCAAT/enhancer-binding protein, and nuclear transcription factor Y. However, the myoblast determining factor (MyoD), MEF3, SP1, USF, and growth hormone cell specific element (GH-CSE) sites were found only in the lcMSTN-1 and SRF only in the lcMSTN-2 promoter. The detailed distribution of transcriptional response elements on lcMSTN-1 and 2 promoters is listed in Supplementary Table S2.
lcMSTN-1 and 2 promoter activities in CIK and L6 cells
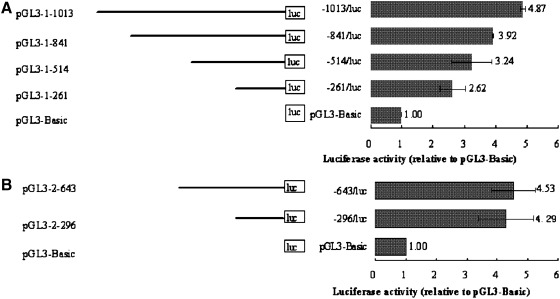
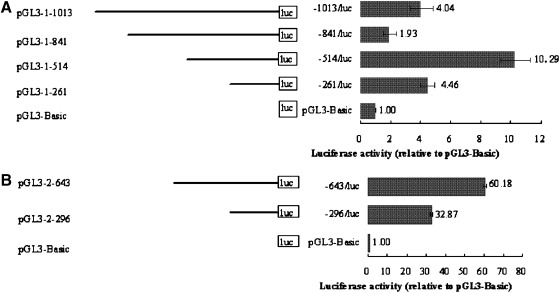
To determine whether the genomic 5′ flanking regions of lcMSTN-1 and 2 contain a functional promoter, six genomic fragments were truncated from the 5′ ends of lcMSTN-1 and 2, 1013, 841, 514, and 261 bp for lcMSTN-1 and 643, and 296 bp for lcMSTN-2. The fragments were then subcloned into the promoterless plasmid pGL3-Basic, respectively. The different fragments had a diverse effect on the luciferase activities in CIK cells (Fig. 4) and L6 cells (Fig. 5) in vitro. Both results from CIK and L6 cells indicate that the cloned sequences contain functional promoters.
FIG. 4.
Promoter activity analysis of the 5′ flanking region of lcMSTN-1 (A) and 2 (B) in CIK cells. The promoterless plasmid, pGL3-Basic, and plasmids containing 5′ flanking fragments were shown on the left. The relative luciferase activities (fold-increase) expressed as mean±SE of three experiments were shown in the right, which was normalized against the activity of the pGL3-Basic plasmid. SE, standard error.
FIG. 5.
Promoter activity analysis of the 5′ flanking region of lcMSTN-1 (A) and 2 (B) in L6 cells. The promoterless plasmid, pGL3-Basic, and plasmids containing 5′ fragments were shown on the left. The relative luciferase activities (fold-increase) expressed as mean±SE of three experiments were shown in the right, which was normalized against the activity of the pGL3-Basic plasmid.
In CIK cells, the 1013 bp fragment of the lcMSTN-1 upstream sequence showed the strongest activity, 4.87-fold relative to pGL3-Basic. The lowest increase in luciferase activity was with the 261 bp fragment, 2.62-fold relative to pGL3-Basic. Luciferase activity rose as the fragment length and the number of E-boxes increased. The construct pGL3-1-261, which includes one CAAT box and one TATA box, exhibited about 54% of the promoter activity of pGL3-1-1013, indicating that these motifs alone are not sufficient to drive a significant amount of promoter activity. In the lcMSTN-2 upstream sequence, the 296 bp fragment exhibited 4.29-fold, and 643 bp 4.53-fold relative to pGL3-Basic. There is no significant difference in promoter activity between the 296 and 643 bp of the MSTN-2 5′ flanking region. Notably, the lcMSTN-2 promoter showed a higher activity than the lcMSTN-1 promoter in CIK cells in the corresponding 5′ flanking region, which reflected the expression of lcMSTN-1 and 2 in the kidney tissue of L. crocea.
Promoter activity was significantly different between MSTN-1 and 2 in L6 cells and between CIK and L6 cells. In L6 cells, the 514 bp fragment of the lcMSTN-1 upstream sequence showed the strongest activity, 10.29-fold relative to pGL3-Basic. The lowest increase in luciferase activity was with the 841 bp fragment, 1.93-fold relative to pGL3-Basic. The activities of the shortest construction pGL3-1-261 and the longest construction pGL3-1-1013 were in the middle, exhibiting 4.46- and 4.04-fold relative to pGL3-Basic, respectively. In the lcMSTN-2 upstream sequence, the 643 bp fragment showed extremely strong activity, reaching 60.18-fold relative to pGL3-Basic, which is 5.85-fold of the activity of the lcMSTN-1 514 bp fragment. The short fragment construction pGL3-2-296 also exhibited a relatively high activity, 32.87-fold relative to pGL3-Basic.
In order to determine the effect of the MyoD binding site on promoter activity, the E-box6 (MyoD binding site) of the lcMSTN-1 promoter was mutated. The results showed that the activity of the construction pGL3-1-1013 with the MyoD binding site mutation was 0.65-fold in L6 cells, and 0.93-fold in CIK cells, respectively, relative to the construction pGL3-1-1013. Mutation of E-box6 had an obvious effect on the expression of the GFP reporter gene in L6 cells, and had little or no effect on the expression of GFP in CIK cells.
Tissue-specific expression of lcMSTN-2
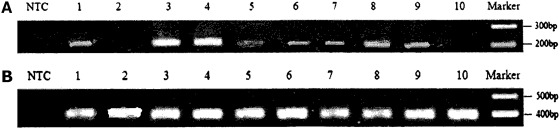
Previous studies identified MSTN-1 expression in the liver, kidney, brain, intestine, and skeletal muscle of large yellow croakers. MSTN-2 also had a wide distribution of expression, and was expressed in the spleen, brain, adipose, kidney, gill, eye, intestine, and liver. The strongest expression was detected in the brain and adipose, and weak in the kidney (Fig. 6). However, MSTN-2 mRNA transcript was not detected in the heart and skeletal muscle. No alternative splicing variant of MSTN-2 was found in all tested tissues.
FIG. 6.
The expression pattern of MSTN-2 in the tissues of L. crocea. (A) MSTN-2; (B) β-actin. Lines from 1 to 10 represent spleen, heart, brain, adipose, kidney, gill, eye, intestine, liver, and skeletal muscle, respectively, and NTC represents negative control.
Discussion
Blast searches indicated that lcMSTN-1 5′ flanking sequence had a high homology with the reported sequences in Perciformes. In this region, putative TATA box, CAAT box, and several putative transcriptional factor binding sites such as CRE, MEF2, MEF3, POU3F2, GH-CSE, and E-box were identified. These putative cis-regulatory elements also existed in the MSTN-1 promoter regions of other fishes, including brook trout (Roberts and Goetz, 2003), zebrafish (Xu et al., 2003; Kerr et al., 2005), rainbow trout (Garikipati et al., 2006), Atlantic salmon (Ostbye et al., 2007), sea perch (Ye et al., 2007), and gilthead sea bream (Funkenstein et al., 2009), of which some elements, such as MEF2 and MyoD, are muscle-specific transcription factor binding sites. CAAT box and TATA box were highly conserved among teleost MSTN-1 upstream sequences. MyoD and MEF2 are critical to the differentiation of skeletal muscle and the regulation of MSTN gene expression in mammals (Cornelison et al., 2000; Ma et al., 2001; Spiller et al., 2002; Salerno et al., 2004; Du et al., 2005).
The alignment of MSTN-2 promoter sequences among large yellow croakers, zebrafish (DQ451548), and rainbow trout (DQ138301) revealed a very low conservation. The cloned fragment of the lcMSTN-2 promoter contains only one E-box, two CAAT boxes, and several putative transcriptional factor binding sites such as CRE, MEF2, POU3F2, and SRF. No putative TATA box was found when 90 was set as a threshold score of TFSEARCH. However, a putative TATA box can be found when a threshold score is 85. Two TATA boxes were found in rainbow trout MSTN-2a (Garikipati et al., 2007) and one TATA box in zebrafish MSTN-2 (Kerr et al., 2005). These results may mean that the MSTN-2 promoter region is more variable than that of MSTN-1.
Previous studies demonstrated that differences between two types of MSTNs existed in both coding sequence and gene expression patterns (Rodgers et al., 2007). In addition, their 5′ flanking regions are different. Three MSTN-2 promoter sequences formed a subgroup, and all MSTN-1 promoter sequences formed the other subgroup in the phylogenetic tree (Fig. 2B). A sequence analysis of their respective promoter regions revealed significant differences in the quantity and functional nature of the putative cis regulatory elements. The MSTN-1 promoter had five E-boxes, and the MSTN-2 promoter had only one in the corresponding region. Some important elements, such as MyoD, MEF3, SP1, USF, and GH-CSE sites, were found only in the lcMSTN-1 promoter. These results suggest that lcMSTN-1 might play a more important role in the skeletal muscle development of L. crocea than lcMSTN-2. Some putative cis regulatory elements, such as MEF2, POU1F1a, POU3F2, CRE, and AP1, were indentified in both promoters of lcMSTN-1 and 2, but their locations were different. The divergence of two MSTN promoters supports the idea that there was early genome duplication during the fish lineage evolution.
Recently, several fish MSTN promoters were reported (Roberts and Goetz, 2003; Xu et al., 2003; Garikipati et al., 2006; Ostbye et al., 2007; Ye et al., 2007; Funkenstein et al., 2009). The transient expression of GFP in zebrafish muscle fibers could be directed by zebrafish and sea perch MSTN promoters (Xu et al., 2003; Ye et al., 2007). However, a quantitative assay for functional analysis of fish MSTN-1 promoter activity has been done only in gilthead sea bream, using the reporter gene luciferase and A204 cell line derived from a human rhabdomyosarcoma (Funkenstein et al., 2009). Our previous works showed that the lcMSTN-1 gene expressed in both the skeletal muscle and kidney (Xue et al., 2006), and lcMSTN-2 expressed in the kidney, but not in the skeletal muscle of L. crocea. In this study, CIK, a grass carp kidney cell line, and L6, a rat skeletal muscle cell line, were used to analyze the promoter activity of lcMSTN-1 and 2. In CIK cells, the shortest 261 bp fragment of the lcMSTN-1 5′ flanking region exhibited the lowest luciferase activity, and the longest 1013 bp fragment exhibited the highest luciferase activity. Luciferase activity correlated positively with the length of lcMSTN-1 5′ flanking fragment. These results confirm that the lcMSTN-1 5′ flanking region within the 1013 bp upstream from the translation initiation codon ATG contains a functional promoter. It also suggests that negative regulatory elements might not exist in the 1013 bp upstream. Similar results were obtained in the gilthead sea bream MSTN-1 promoter, in which negative regulatory elements were found between 1127 and 1369 bp upstream (Funkenstein et al., 2009). In 5′ flanking region of lcMSTN-2, the 643 bp fragment had a little higher luciferase activity than 296 bp, and both were over fourfold relative to pGL3-Basic, which indicates that a functional promoter exists in this region. In the corresponding 5′ flanking region, the lcMSTN-2 promoter showed a higher activity than the lcMSTN-1 promoter in the CIK cells, which was consistent with the MSTN gene expression in vivo. In the kidney tissue of L. crocea, the quantity of lcMSTN-2 mRNA was larger than the lcMSTN-1.
E-boxes are involved in regulating the muscle-specific expression of genes. In our study, mutation of E-box6 (MyoD binding site) had a greater effect on the expression of the GFP reporter gene in L6 cells than in CIK cells. In zebrafish embryos, mutation of a single E-box (E1), MEF2, or MEF3 site alone had little or no effect on the muscle-specific expression of GFP reporter genes. However, when both E-boxes in the myogenin promoter were mutated, there was a significant reduction in the muscle-specific expression of GFP reporter genes. Mutation of the MEF2 binding site together with MEF3 binding sites or with E-boxes could reduce the activity of the promoter (Du et al., 2003). In the lcMSTN-1 upstream sequence, there are five E-boxes, one EMF2 and one EMF3 sites. These sites may play an important role together in regulating MSTN-1 expression in the skeletal muscle of large yellow croakers.
Surprisingly, there is a significant difference between CIK and L6 cells. In L6 cells, the fragments from the lcMSTN-2 5′ flanking region showed an extremely high luciferase activity, of which the 643 bp fragment reached 60.18-fold relative to pGL3-Basic, and was 13.28-fold in CIK cells. In vivo, the expression of MSTN-2 was significantly lower than that of MSTN-1, and hardly detectable in the skeletal muscle of L. crocea. The MSTN-2 gene has not been found in mammals, and functional analysis of other fish MSTN-2 promoters has not been reported so far. The reason that the MSTN-2 promoter has such a high activity in L6 cells is unknown. For the lcMSTN-1 5′ flanking region, the highest activity was found with the fragment of 514 bp, while the lowest activity was with the fragment of 841 bp. It suggests that the upstream sequence of lcMSTN-1 might have a negative regulatory element between −514 and −841 in L6 cell experiments. These data indicated that transcription regulation of lcMSTN-1 and 2 promoters was significantly different between mammalian and fish cells or in vivo and in vitro. In gilthead sea bream, no reporter gene activity was found on transient transfection of the recombinant plasmids into the murine myoblast C2 cell line and a mammalian nonmuscle cell line CHO-K1 (Funkenstein et al., 2009). These results showed that it was important to choose an appropriate cell line for a quantitative assay of the fish MSTN promoter function.
Our work revealed that both lcMSTN-1 and 2 promoters contained numerous potential transcription factor binding sites, and had the function of transcription regulation. It lays a foundation for the functional analysis of fish MSTN-1 and 2 promoters. Further studies are needed to understand the regulation of fish MSTN-1 and 2 gene expression under various physiological conditions as well as the effect of transcription factors and hormones.
Conclusion
The 5′ flanking fragments of 1029 bp from lcMSTN-1 and 643 bp from lcMSTN-2 were cloned. The upstream sequence of lcMSTN-1 had a homology of more than 90% with the reported sequences of S. aurata and L. japonicas, while the upstream sequence of lcMSTN-2 showed low conservation. Both fragments contained a CAAT box and several putative cis-regulatory elements. However, some elements such as a putative TATA box, MyoD, MEF3, USF, and GH-CSE sites were identified only in the lcMSTN-1 upstream region. Functional analysis of promoters revealed that the different fragments had diverse effects on luciferase activities in CIK and L6 cells. In CIK cells, luciferase activity rose as the length of truncated fragments and the number of E-boxes increased. The lcMSTN-2 promoter showed a higher activity than lcMSTN-1 in the corresponding region, which was consistent with the expression of lcMSTN-1 and 2 genes in the kidney tissue of L. crocea. In L6 cells, functional analysis of promoters suggested that the upstream sequence of lcMSTN-1 might have a negative regulatory element between -514 and -841. The lcMSTN-2 upstream showed an extremely high luciferase activity. Mutation of E-box6 (MyoD binding site) had a greater effect on GFP expression in L6 cells than in CIK cells. These results indicated that both cloned fragments contained functional promoters, and that the transcription regulation mechanism of two promoters differed between mammalian and fish cells. It suggests that a cell line, derived from the tissue in which both MSTN-1 and 2 genes are expressed, may be appropriate for a quantitative assay for fish MSTN promoter activity.
Supplementary Material
Acknowledgments
The project was supported by National Natural Science Foundation of China (Grant No. 30871916) and the Scientific Research Foundation of Graduate School of Ningbo University (Grant No. G10JA028). This article is sponsored by K. C. Wong Magna Fund in Ningbo University. Xue thanks Pao Yu-Kong and Pao Zhao-Long Scholarship for Chinese Students Studying Abroad for providing financial support.
Disclosure Statement
No competing financial interests exist.
References
- Acosta J. Carpio Y. Borroto I. Gonzalez O. Estrada M.P. Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J Biotechnol. 2005;119:324–331. doi: 10.1016/j.jbiotec.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Allen D.L. Du M. Comparative functional analysis of the cow and mouse myostatin genes reveals novel regulatory elements in their upstream promoter regions. Comp Biochem Phys B. 2008;150:432–439. doi: 10.1016/j.cbpb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Amali A.A. Lin C.J. Chen Y.H. Wang W.L. Gong H.Y. Rekha R.D. Wu J.L. Overexpression of Myostatin2 in zebrafish reduces the expression of dystrophin associated protein complex (DAPC) which leads to muscle dystrophy. J Biomed Sci. 2008;15:595–604. doi: 10.1007/s11373-008-9250-2. [DOI] [PubMed] [Google Scholar]
- Cartharius K. Frech K. Grote K. Klocke B. Haltmeier M. Klingenhoff A. Frisch M. Bayerlein M. Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Clop A. Marcq F. Takeda H. Pirottin D. Tordoir X. Bibe B. Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Cornelison D.D.W. Olwin B.B. Rudnicki M.A. Wol B.J. MyoD +/− satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol. 2000;224:122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- Du R. Chen Y.F. An X.R. Yang X.Y. Ma Y. Zhang L. Qin J. Cloning and sequence analysis of myostatin promoter in sheep. DNA Seq. 2005;16:412–417. doi: 10.1080/10425170500226474. [DOI] [PubMed] [Google Scholar]
- Du S.J. Gao J. Anyangwe V. Muscle-specific expression of myogenin in zebrafish embryos is controlled by multiple regulatory elements in the promoter. Comp Biochem Phys B. 2003;134:123–134. doi: 10.1016/s1096-4959(02)00194-x. [DOI] [PubMed] [Google Scholar]
- Funkenstein B. Balas V. Rebhan Y. Pliatner A. Characterization and functional analysis of the 5′ flanking region of Sparus aurata myostatin-1 gene. Comp Biochem Phys A. 2009;153:55–62. doi: 10.1016/j.cbpa.2008.09.031. [DOI] [PubMed] [Google Scholar]
- Garikipati D.K. Gahr S.A. Roalson E.H. Rodgers B.D. Characterization of Rainbow Trout Myostatin-2 genes (rtMSTN-2a and -2b): genomic organization, differential expression, and pseudogenization. Endocrinology. 2007;148:2106–2115. doi: 10.1210/en.2006-1299. [DOI] [PubMed] [Google Scholar]
- Garikipati D.K. Gahr S.A. Rodgers B.D. Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J Endocrinol. 2006;190:879–888. doi: 10.1677/joe.1.06866. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid N.F. Taylor W.E. Yarasheski K. Sinha-Hikim I. Ma K. Ezzat S. Bhasin S. Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proc Natl Acad Sci U S A. 1998;95:14938–14943. doi: 10.1073/pnas.95.25.14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobet L. Martin L.J. Poncelet D. Pirottin D. Brouwers B. Riquet J. Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T. Wingender E. Reuter I. Hermjakob H. Kel A.E. Kel O.V. Ignatieva E.V. Ananko E.A. Podkolodnaya O.A. Kolpakov F.A. Podkolodny N.L. Kolchanov N.A. Databases on Transcriptional Regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:364–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helterline D.L. Garikipati D. Stenkamp D.L. Rodgers B.D. Embryonic and tissue-specific regulation of myostatin-1 and -2 gene expression in zebrafish. Gen Comp Endocrinol. 2007;151:90–97. doi: 10.1016/j.ygcen.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S. Losinski R.L. Cornelius S.G. Frank G.R. Willis G.M. Gerrard D.E. Spurlock M.E. Myostatin expression in porcine tissues: tissue specificity and developmental and postnatal regulation. Am J Physiol. 1998;275:1265–1273. doi: 10.1152/ajpregu.1998.275.4.R1265. [DOI] [PubMed] [Google Scholar]
- Joulia-Ekaza D. Bernardi H. Garandel V. Rabenoelina F. Vernus B. Cabello G. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp Cell Res. 2003;286:263–275. doi: 10.1016/s0014-4827(03)00074-0. [DOI] [PubMed] [Google Scholar]
- Joulia-Ekaza D. Cabello G. Myostatin regulation of muscle development: molecular basis, natural mutations, physiopathological aspects. Exp Cell Res. 2006;312:2401–2414. doi: 10.1016/j.yexcr.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Kambadur R. Sharma M. Smith T.P. Bass J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Kerr T. Roalson E.H. Rodgers B.D. Phylogenetic analysis of the myostatin gene sub-family and the differential expression of a novel member in zebrafish. Evol Dev. 2005;7:390–400. doi: 10.1111/j.1525-142X.2005.05044.x. [DOI] [PubMed] [Google Scholar]
- Langley B. Thomas M. Bishop A. Sharma M. Gilmour S. Kambadur R. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J Biol Chem. 2002;277:49831–49840. doi: 10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]
- Lee C.Y. Hu S.Y. Gong H.Y. Chen M.H. Lu J.K. Wu J.L. Suppression of myostatin with vector-based RNA interference causes a double-muscle effect in transgenic zebrafish. Biochem Biophys Res Commun. 2009;387:766–771. doi: 10.1016/j.bbrc.2009.07.110. [DOI] [PubMed] [Google Scholar]
- Lee S.B. Kim Y.S. Oh M.Y. Jeong I.H. Seong K.B. Jin H.J. Improving rainbow trout (Oncorhynchus mykiss) growth by treatment with a fish (Paralichthys olivaceus) myostatin prodomain expressed in soluble forms in E. coli. Aquaculture. 2010;302:270–278. [Google Scholar]
- Lee S.J. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- Ma K. Mallidis C. Artaza J. Taylor W. Gonzalez-Cadavid N. Bhasin S. Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol Endocrinol Metab. 2001;281:1128–1136. doi: 10.1152/ajpendo.2001.281.6.E1128. [DOI] [PubMed] [Google Scholar]
- Maccatrozzo L. Bargelloni L. Cardazzo B. Rizzo G. Patarnello T. A novel second myostatin gene is present in teleost fish. FEBS Lett. 2001a;509:36–40. doi: 10.1016/s0014-5793(01)03124-6. [DOI] [PubMed] [Google Scholar]
- Maccatrozzo L. Bargelloni L. Radaelli G. Mascarello F. Patarnello T. Characterization of the myostatin gene in the gilthead seabream (Sparus aurata): sequence, genomic structure, and expression pattern. Mar Biotechnol. 2001b;3:224–230. doi: 10.1007/s101260000064. [DOI] [PubMed] [Google Scholar]
- Marchitelli C. Savarese M.C. Crisa A. Nardone A. Marsan P.A. Valentini A. Double muscling in Marchigiana beef breed is caused by a stop codon in the third exon of myostatin gene. Mamm Genome. 2003;14:392–395. doi: 10.1007/s00335-002-2176-5. [DOI] [PubMed] [Google Scholar]
- McCroskery S. Thomas M. Maxwell L. Sharma M. Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron A.C. Lawler A.M. Lee S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron A.C. Lee S.J. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher D.S. Quignon P. Bustamante C.D. Sutter N.B. Mellersh C.S. Parker H.G. Ostrander E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:779–786. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostbye T.K. Wetten O.F. Tooming-Klunderud A. Jakobsen K.S. Yafe A. Etzioni S. Andersen O. Myostatin (MSTN) gene duplications in Atlantic salmon (Salmo salar): evidence for different selective pressure on teleost MSTN-1 and -2. Gene. 2007;403:159–169. doi: 10.1016/j.gene.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Reisz-Porszasz S. Bhasin S. Artaza J.N. Shen R. Sinha-Hikim I. Hogue A. Gonzalez-Cadavid N.F. Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am J Physiol Endocrinol Metab. 2003;285:876–888. doi: 10.1152/ajpendo.00107.2003. [DOI] [PubMed] [Google Scholar]
- Rescan P.Y. Jutel I. Ralliere C. Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss) J Exp Biol. 2001;204:3523–3529. doi: 10.1242/jeb.204.20.3523. [DOI] [PubMed] [Google Scholar]
- Rios R. Carneiro I. Arce V.M. Devesa J. Myostatin regulates cell survival during C2C12 myogenesis. Biochem Biophys Res Commun. 2001;280:561–566. doi: 10.1006/bbrc.2000.4159. [DOI] [PubMed] [Google Scholar]
- Rios R. Carneiro I. Arce V.M. Devesa J. Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol. 2002;282:993–999. doi: 10.1152/ajpcell.00372.2001. [DOI] [PubMed] [Google Scholar]
- Roberts S.B. Goetz F.W. Myostatin protein and RNA transcript levels in adult and developing brook trout. Mol Cell Endocrinol. 2003;210:9–20. doi: 10.1016/j.mce.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Rodgers B.D. Garikipati D.K. Clinical, agricultural, and evolutionary biology of myostatin: a comparative review. Endocr Rev. 2008;29:513–534. doi: 10.1210/er.2008-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers B.D. Roalson E.H. Weber G.M. Roberts S.B. Goetz F.W. A proposed nomenclature consensus for the myostatin gene family. Am J Physiol Endocrinol Metab. 2007;292:371–372. doi: 10.1152/ajpendo.00395.2006. [DOI] [PubMed] [Google Scholar]
- Salerno M.S. Thomas M. Forbes D. Watson T. Kambadur R. Sharma M. Molecular analysis of fiber type-specific expression of murine myostatin promoter. Am J Physiol Cell Physiol. 2004;287:1031–1040. doi: 10.1152/ajpcell.00492.2003. [DOI] [PubMed] [Google Scholar]
- Sawatari E. Seki R. Adachi T. Hashimoto H. Uji S. Wakamatsu Y. Kinoshita M. Overexpression of the dominant-negative form of myostatin results in doubling of muscle-fiber number in transgenic medaka (Oryzias latipes) Comp Biochem Physiol A. 2010;155:183–189. doi: 10.1016/j.cbpa.2009.10.030. [DOI] [PubMed] [Google Scholar]
- Schuelke M. Wagner K.R. Stolz L.E. Hubner C. Riebel T. Komen W. Lee S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. New Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- Sharma M. Kambadur R. Matthews K.G. Somers W.G. Devlin G.P. Conaglen J.V. Bass J.J. Myostatin, a transforming growth factor-beta superfamily member, is expressed in heart muscle and is upregulated in cardiomyocytes after infarct. J Cell Physiol. 1999;180:1–9. doi: 10.1002/(SICI)1097-4652(199907)180:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Smith J.A. Lewis A.M. Wiener P. Williams J.L. Genetic variation in the bovine myostatin gene in UK beef cattle: allele frequencies and haplotype analysis in the South Devon. Anim Genet. 2000;31:306–309. doi: 10.1046/j.1365-2052.2000.00521.x. [DOI] [PubMed] [Google Scholar]
- Spiller M.P. Kambadur R. Jeanplong F. Thomas M. Martyn J.K. Bass J.J. Sharma M. The myostatin gene is a downstream target gene of basic helix-loop-helix transcription factor MyoD. Mol Cell Biol. 2002;22:7066–7082. doi: 10.1128/MCB.22.20.7066-7082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G. Dallmann G. Muller G. Patthy L. Soller M. Varga L. A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome. 1998;9:671–672. doi: 10.1007/s003359900843. [DOI] [PubMed] [Google Scholar]
- Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taylor W.E. Bhasin S. Artaza J. Byhower F. Azam M. Willard D.H. Gonzalez-Cadavid N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab. 2001;280:221–228. doi: 10.1152/ajpendo.2001.280.2.E221. [DOI] [PubMed] [Google Scholar]
- Thomas M. Langley B. Berry C. Sharma M. Kirk S. Bass J. Kambadur R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J Biol Chem. 2000;275:40235–40243. doi: 10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- Thompson J.D. Higgins D.G. Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. Wu G. Zohar Y. Du S.J. Analysis of myostatin gene structure, expression and function in zebrafish. J Exp Biol. 2003;206:4067–4079. doi: 10.1242/jeb.00635. [DOI] [PubMed] [Google Scholar]
- Xue L. Qian K. Qian H. LI L. Yang Q. Li M. Molecular cloning and characterization of the myostatin gene in croceine croaker, Pseudosciaena crocea. Mol Biol Rep. 2006;33:129–136. doi: 10.1007/s11033-006-0015-6. [DOI] [PubMed] [Google Scholar]
- Ye H.Q. Chen S.L. Sha Z.X. Liu Y. Molecular cloning and expression analysis of the myostatin gene in sea perch (Lateolabrax japonicus) Mar Biotechnol. 2007;9:262–272. doi: 10.1007/s10126-006-6093-6. [DOI] [PubMed] [Google Scholar]
- Zhu X. Hadhazy M. Wehling M. Tidball J.G. McNally E.M. Dominant negative myostatin produces hypertrophy without hyperplasia in muscle. FEBS Lett. 2000;474:71–75. doi: 10.1016/s0014-5793(00)01570-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.