Abstract
Human umbilical cord Wharton's jelly stem cells (HWJSCs) are gaining attention as a possible clinical source of mesenchymal stem cells for cell therapy and tissue engineering due to their high accessibility, expansion potential, and plasticity. We employed a combination of highly sensitive techniques to determine the average cell viability levels and proliferation capabilities of 10 consecutive cell passages of cultured HWJSCs and then used RNA microarrays to identify genes associated with changes in cell viability levels. We found an initial decrease in cell viability from the first to the third cell passage followed by an increase until the sixth passage and a final decrease from the sixth to tenth cell passages. The highest cell viability levels corresponded to the fifth and sixth passages. The intracellular ionic contents of potassium, sodium, and chlorine suggest that the lower cell viability levels at passages 2, 3, and 8–10 may be associated with apoptotic cell death. In fact, gene expression analysis revealed that the average cell viability was significantly associated with genes with a function in apoptotic cell death, especially pro-apoptotic FASTKD2, BNIP3L genes and anti-apoptotic TNFAIP8 and BCL2L2 genes. This correlation with both pro-apoptotic and anti-apoptotic genes suggests that there may be a complex live-death equilibrium in cultured HWJSCs kept in culture for multiple cell passages. In this study, the highest cell viability levels corresponded to the fifth and sixth HWJSC passages, suggesting that these passages should be preferentially employed in cell therapy or tissue engineering protocols using this cell type.
Introduction
The umbilical cord is a major source of nonembryonic stem cells with high proliferation and differentiation capabilities.1–3 Among these, mesenchymal stem cells (MSCs) can be isolated for culture from umbilical cord blood4 or from Wharton's jelly. Human Wharton's jelly stem cells (HWJSCs) are considered to have an elevated differentiation potential5 and telomerase activity6 and a low expression of class I and II major histocompatibility complex antigens,7,3 making them excellent candidate cells for the generation of artificial tissues by tissue engineering and for use in cell therapy protocols.
Various in vivo studies have examined the usefulness of HWJSC to treat different diseases,5,8 but the results have been controversial. One possibility is that the stem cells used in some of these studies did not have adequate cell viability levels, which are not usually measured by authors using highly sensitive methods. Only viable cells are suitable for clinical or research utilization, and the effectiveness of most cell therapy and tissue engineering protocols is strongly dependent on the availability of an adequate source of viable and functional cells.9,10 One of the key factors in the viability of cultured cells is the cell passage at which they are clinically utilized; thus, some cell cultures established from newborn tissues tend to show a slow proliferation rate beyond the fifth passage.11 However, the cell viability of HWJSCs kept in culture has not been determined to date, and sequential changes that may take place in consecutive cell passages are poorly understood.
Various methods have been used to determine the viability of different cultured cell types. They include techniques that determine permeability alterations in the cell membrane by using trypan blue or other vital dyes,12 but these are not sufficiently accurate to detect and predict early cell damage, only identifying cell alterations once they have become irreversible. One of the most sensitive and accurate viability assays uses electron-probe X-ray microanalysis to quantify the intracellular contents of the major cell elements, especially potassium (K), sodium (Na), and chlorine (Cl).13–16 This highly sensitive technique also allows the simultaneous analysis of the intracellular ionic concentrations and ultrastructure of the cells.15,17,18 The use of this method has permitted accurate measurement of the intracellular ionic composition of several cell types, including U937,19 MCF720 and K56218 cell lines, and primary cell cultures of human oral mucosa keratinocytes,21 umbilical cord endothelial cells,10 and rabbit cornea endothelial cells.9
The recent development of microarray techniques that quantify the expression levels of thousands of genes in a single experiment allows cell viability to be evaluated from a genetic standpoint. Specific cell properties, including toxic stress response and cell viability, can be quantified by using microarrays to measure gene expression levels.22–26
With this background, the aim of this study was to identify the most viable subcultures of HWJSC for use in cell therapy and tissue engineering by means of classical and highly sensitive methods.
Materials and Methods
Isolation and culture of HWJSC
Human umbilical cords were obtained from full-term newborns delivered by cesarean section (n=10). After delivery, the umbilical cords were handled under sterile conditions in a tissue bank. Arteries and veins were surgically removed to prevent red blood cell contamination, and small explants of Wharton's jelly were obtained. These explants were then digested in type I collagenase (Gibco BRL Life Technologies) and trypsin 0.5 g/L-ethylenediaminetetraacetic acid (EDTA) 0.2 g/L solution (Gibco BRL) for isolation of the stromal cells. Once isolated, HWJSCs were cultured in 75 cm2 culture flasks in Amniomax™ culture medium (Gibco BRL) as previously described.1 Cell cultures were maintained at 37°C in a humidified incubator with a 5% CO2 atmosphere. The culture medium was changed once every 3 days, and cell growth was monitored every 12 h by ocular inspection using a phase-contrast light microscope. Once the cells reached subconfluence (42,000±13,500 cells per cm2), cell cultures were trypsinized using trypsin 0.5 g/L-EDTA 0.2 g/L solution at 37°C for 8 min. The number of days between consecutive passages depended on the time taken by the cells to reach subconfluence (mean 5.8±2.6 days between passages), plating subcultured cells in new flasks at a density of 13,333 cells per cm2 (1,000,000 cells per flask of 75 cm2 of surface). We used identical conditions (culture plates, culture medium, and cell incubator) for all subcultures. The cells were kept in culture for ten consecutive passages (P1 to P10).
Ethical approval for this experimental protocol was granted by the Institutional Ethical and Research Review Board. Parents or guardians of all newborns provided written consent for their inclusion in this study.
Cell proliferation analysis
First, cell proliferation was assessed by means of a highly sensitive Real-Time Cell Analyzer (RTCA; XCELLigence, Roche Applied Science). This analyzer uses sensor electrodes that measure the electronic impedance of cultured cells, yielding the Cell Index (unit-less parameter) to measure the relative change in electrical impedance and represent the cell status. The Cell Index is calculated by dividing the impedance change by a background value and is dependent on the number of cultured cells, because the impedance increases with more cells on the electrodes. In our assays, 30,000 HWJSCs per cm2 were cultured on 96-well plates with attached microelectrodes and analyzed for 100 h. All determinations were carried out using duplicates. Data were analyzed using XCELLigence software.
Cell proliferation and viability were then measured by using the water-soluble tetrazolium salt-1 (WST-1) colorimetric assay on 10 consecutive cell passages of HWJSC (Cell Proliferation Reagent WST-1, Roche Diagnostics). WST-1 is a tetrazolium dye containing an electron coupling reagent that is cleaved by the mitochondrial dehydrogenase enzyme to a formazan dye. The reaction directly correlates with the number of metabolically active proliferating cells and can serve as a marker of cell viability and cell proliferation. For this analysis, 20,000 cells were seeded per well in a 96-well plate with 100 μL of culture medium. After 24 h, the medium was replaced with fresh medium containing 1/10 volume of cell proliferation reagent WST-1 and incubated for 4 h at 37°C before the absorbance at 450 nm was quantified in a spectrophotometer. Cells incubated for 15 min in 1% triton-X were used as controls. The data presented are from five separate experiments.
Cell viability analysis
The percentage of live and dead cells in each cell passage was determined by means of the trypan blue dye exclusion test. Briefly, each subconfluent cell culture was trypsinized with trypsin 0.5 g/L-EDTA 0.2 g/L solution for 8 min at 37°C, and the detached cells were harvested by centrifugation and resuspended in 1 mL of culture medium. Then, 100 μL of the cell suspension were mixed with 0.4% trypan blue solution (Sigma-Aldrich ref. T8154) and incubated for 5 min. at room temperature. Finally, we quantified the number of white and blue cells by using a Neubauer chamber in a Nikon Eclipse 90i light microscope to count a minimum of 200 cells per cell passage, enabling calculation of the percentage of live and dead cells. All cell counts were done in quadruplicate, and median values and means (with standard deviations) were calculated for each cell passage.
For the LIVE/DEAD® Cell Viability Assay in each cell passage, 10,000 HWJSCs were cultured on chamber slides (Lab-Tek Chamber Slides, Nunc) and allowed to attach to the bottom for 24 h. Then, cells were washed twice with PBS and stained with the LIVE/DEAD® assay reagents (calcein/AM and ethidium homodimer-1; Invitrogen) following the protocols supplied by Invitrogen. Finally, the number of live (green) and dead (red) cells was determined by using a fluorescence microscope. These experiments were carried out in quadruplicate, and the mean percentage of live cells was calculated for each cell passage.
For electron-probe X-ray microanalysis, subconfluent HWJSCs were subcultured using trypsin-EDTA on plated gold grids coated with a thin layer of Pioloform (polyvinyl butyral) (Ted Pella, Inc.) following previously published methods.9,21,27 Cells were seeded at a density of 5000 cells per grid and cultured in Amniomax medium. After 24 h of culture on the Pioloform-coated gold grids, support grids containing the HWJSCs were washed in ice-cold distilled water for 5 s to remove the extracellular medium. After washing, excess water was drained from the surface, and the grids were immediately plunge-frozen in liquid nitrogen.18 After cryofixation, the grids were placed in a precooled aluminum specimen holder at liquid nitrogen temperature and freeze dried at increasing temperatures for 24 h in an E5300 Polaron freezer-drier apparatus equipped with a vacuum rotatory pump system. Freeze-dried gold grids were carbon coated in a high-vacuum coating system and microanalyzed within 6 h. Electron probe X-ray microanalysis of the specimens was performed with a Philips XL30 scanning electron microscope (SEM) equipped with an EDAX DX-4 microanalytical system and a solid-state backscattered electron (BSE) detector. The samples were examined by SEM with a combination of secondary electron and BSE imaging modes. For X-ray microanalysis, the analytical conditions were tilt angle 08, take-off angle 61.348, and working distance 10 mm. The acceleration voltage was 10 kV. All spectra were collected in the spot mode at 10,000×(equivalent to 50 nm spot diameter) for 200 s live time, and the number of counts per second recorded by the detector was around 500. All determinations were performed on the central area of the cell nucleus. To determine total ion content, we used the peak-to-local-background ratio method28 with reference to standards composed of 20% dextran containing known amounts of inorganic salts.29 In this study, we analyzed the ionic content of 25 HWJSCs corresponding to each cell passage (P1 to P10) using cells from four different umbilical cords. Controls were human fibroblasts isolated from oral mucosa biopsies (highly viable cells) and the same cells treated with 10 mM HEMA, a known inductor of cell apoptosis30 (cells with low viability levels).
Flow cytometry
To evaluate the expression of key MSC markers on the different HWJSC cultures used in this study, cells corresponding to passages P1, P3, P6, and P9 were analyzed using anti-CD90-Allophycocyanin conjugated (CD90-APC) and anti-CD45-Phycoerythrin conjugated (CD45-PE) antibodies (R & D Systems, Inc.). Samples containing 0.5×106 cells were placed in flow cytometry tubes, washed with 2 mL of staining buffer (R & D Systems, Inc.), and Fc receptors were then blocked by incubating the cells for 5 min with 2 mL of PBS 1× containing 0.1% BSA and 0.1% FBS. Next, cells were stained with the antibodies and isotype controls in different tubes and incubated for 45 min at 4°C in darkness. Cells were washed with 2 mL of staining buffer, and samples were immediately acquired on an FACS CALIBUR Flow Cytometer.
Multilineage cell differentiation analysis
The multilineage differentiation potential of isolated HWJSCs was confirmed by subculturing primary HWJSC cultures on chamber slides and inducing differentiation to osteogenic, chondrogenic, and adipogenic cell lineages by using specific conditioning media as previously described.31 Then, cell differentiation was assessed by cytochemical histological staining, staining samples with 2% Alizarin Red S for 5 min at room temperature to detect the osteoblastic phenotype and identify any signs of mineralization. Chondrogenesis was confirmed by mucopolysaccharide staining using Alcian Blue solution (1% alcian blue 8GX and 3% glacial acetic acid, pH adjusted to 2.5). Adipogenic differentiation was identified by Oil Red O staining (0.7 mg in 100 mL of propylene glycol) for 5 min at 60°C. All these cell differentiation assays were carried out at selected passages (P1, P3, P6 and P9) to ascertain that HWJSCs retain their stem cell nature throughout the ten cell passages.
Microarray analysis
Total RNA was isolated from three different samples corresponding to three different individuals at each HWJSC passage (P1 to P10) by using the Qiagen RNeasy System™. Total RNA was converted into cDNA using a reverse-transcriptase (Superscript II, Life Technologies, Inc.) and T7-oligo(dT) primer. Subsequently, biotinilated cRNA was generated by using a T7 RNA polymerase and biotin-11-uridine-5′-triphosphate (Enzo Diagnostics). Labeled cRNA were chemically fragmented to facilitate the hybridization process and hybridized to Affymetrix Human Genome U133 plus 2.0 oligonucleotide arrays for 6 h at 45°C. Finally, all expression values were normalized using the software provided by Affymetrix to obtain comparable data and to identify present (expressed) and absent (unexpressed) genes at each cell passage. All expression data are publically available at the public functional genomics data repository supporting minimum information about a microarray experiment (MIAME)-compliant data submissions GeneExpressionOmnibus (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30391).
Once expression levels were obtained for all probe sets, all genes with apoptosis-related function were selected by using the information supplied by Affymetrix. In addition, all MSC markers proposed by the Mesenchymal and Tissue Stem Cell Committee of the International Society for Cellular Therapy32 were selected and analyzed at each cell passage (CD105, CD73, CD90, CD11B, CD14, CD19, CD34, CD45, CD79A, and HLADR) along with the cell proliferation markers, proliferating cell nuclear antigen (PCNA) and MKI67.
Statistical analysis
The nonparametric Friedman test was used to evaluate the statistical significance of differences among the 10 cell passages analyzed. Pair-wise comparisons between two successive cell subcultures were carried out using the nonparametric Wilcoxon's test. A Bonferroni-adjusted significance level of ≤0.001 was considered for multiple comparisons, because up to 48 statistical comparisons were performed simultaneously using the Wilcoxon's test. The Spearman correlation test was used to establish correlations between distributions corresponding to cell viability and cell proliferation. The Statistical Package for the Social Sciences (SPSS) 16.00 software package was used for the statistical analysis.
In order to calculate average cell viability levels (ACVLs) for each cell passage, the raw cell viability values determined by trypan blue, WST-1, LIVE/DEAD,® and K/Na index were first normalized to z-scores (mean=0 and standard deviation=1) using the formula: Z=(X−μ)/σ, where μ is the average cell viability obtained for each method, X is the specific cell viability for a particular cell passage, and σ is the standard deviation for each method. Then, mean z-scores values were obtained for each of the 10 cell passages analyzed.
For the microarray analysis, we used the Pearson (r) correlation test to determine the correlation between each probe set and the ACVL for each cell passage. All genes with an absolute r correlation coefficient >0.700 were selected as being statistically significant. Gene Ontology (GO) analysis of the selected genes was performed using BiNGO (www.psb.ugent.be/cbd/papers/BiNGO/), a plug-in for the Cytoscape program.33 The set of selected genes was tested for enrichment of any GO category in comparison to all annotated genes represented in the array. Scores were evaluated based on the hypergeometric distribution and using Bonferroni correction for multiple testing.34 To identify apoptosis-related genes whose expression was significantly associated with the ACVL at the different cell passages, we first selected all probe sets with a role in apoptosis or anti-apoptosis by using the information provided by Affymetrix. These apoptosis-related genes were then analyzed with version 3.09b of the significance analysis of microarrays (SAM) software of Stanford University, using a δ value that permitted a false discovery rate of 0 (i.e., no genes are falsely named). A multiclass analysis of genes associated with the ACVLs was performed. The program is available at www-stat.stanford.edu/∼tibs/SAM/.
Results
Culture and characterization of HWJSCs isolated from human umbilical cord
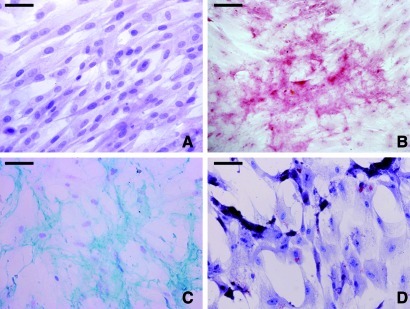
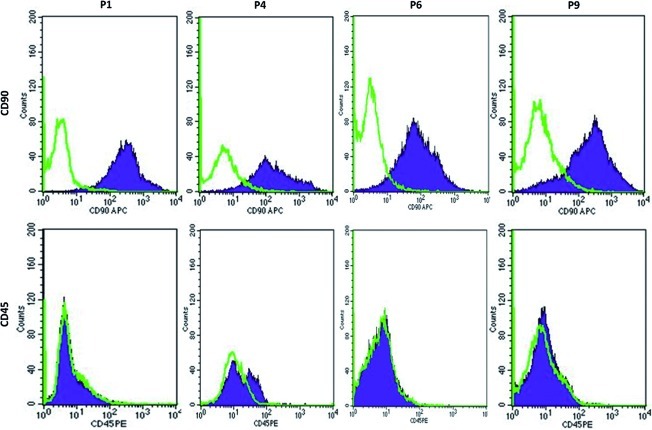
Histological evaluation of HWJSC cultures from human umbilical cord revealed that the cells were adherent and displayed a typical mesenchymal morphology, with spindle and star-shaped cells. Specific conditioning media were used to efficaciously differentiate the isolated HWJSCs in this study to osteogenic, chondrogenic, and adipogenic cell lineages, as determined by alizarin red S, alcian blue, and oil red O staining, respectively, at all cell passages analyzed (P1, P3, P6, and P9). Illustrative examples of the multi-lineage differentiation capability of HWJSCs at the first passage are shown in Figure 1. Microarray analysis of specific MSC markers showed high expression of CD105, CD73, and CD90 and no expression of CD11B, CD14, CD19, CD34, CD45, CD79A, and HLADR at all 10 cell passages (P1 to P10; Table 1). Flow cytometry study confirmed the high expression of CD90 and the absence of CD45 expression in HWJSCs corresponding to the first, third, sixth, and ninth cell passages (Fig. 2).
FIG. 1.
Multi-lineage differentiation capabilities of the cultured HWJSCs at cell passage P1. (A) Undifferentiated control HWJSCs (H&E staining). (B) HWJSCs differentiated to the osteogenic cell type showing positive calcium deposits by alizarin red staining. (C) Differentiation to the chondrogenic phenotype (alcian blue staining). (D) Adipogenic differentiation (oil red O). Scale bars: 50 μm. HWJSCs, Wharton's jelly stem cells. Color images available online at www.liebertonline.com/tec
Table 1.
mRNA Expression of Relevant MSC Markers as Determined by Microarray in Wharton's Jelly Stem Cells at Passages 1 to 10 (P1 to P10)
| GENE | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CD105 | 143.0 (P) | 138.2 (P) | 155.2 (P) | 176.0 (P) | 177.3 (P) | 132.1 (P) | 150.2 (P) | 245.4 (P) | 198.5 (P) | 243.7 (P) |
| CD73 | 598.1 (P) | 847.5 (P) | 1052.5 (P) | 1167.4 (P) | 1205.9 (P) | 892.3 (P) | 1184.5 (P) | 1519.1 (P) | 1553.1 (P) | 1785.2 (P) |
| CD90 | 2126.9 (P) | 1438.8 (P) | 1572.0 (P) | 1783.5 (P) | 1564.7 (P) | 1416.3 (P) | 2237.6 (P) | 1874.2 (P) | 1735.8 (P) | 1732.5 (P) |
| CD11B | 8.5 (A) | 10.9 (A) | 7.0 (A) | 9.5 (A) | 10.9 (A) | 9.9 (A) | 12.6 (A) | 4.5 (A) | 11.6 (A) | 16.9 (A) |
| CD14 | 23.6 (A) | 10.8 (A) | 5.3 (A) | 7.7 (A) | 3.9 (A) | 3.6 (A) | 1.5 (A) | 1.6 (A) | 4.7 (A) | 5.8 (A) |
| CD19 | 5.1 (A) | 6.9 (A) | 6.9 (A) | 6.4 (A) | 4.9 (A) | 4.7 (A) | 7.3 (A) | 7.3 (A) | 7.0 (A) | 3.1 (A) |
| CD34 | 12.0 (A) | 9.3 (A) | 11.0 (A) | 9.2 (A) | 14.1 (A) | 8.6 (A) | 9.7 (A) | 8.1 (A) | 14.6 (A) | 24.3 (A) |
| CD45 | 3.4 (A) | 3.4 (A) | 2.5 (A) | 2.8 (A) | 3.0 (A) | 4.4 (A) | 2.9 (A) | 5.0 (A) | 5.3 (A) | 3.4 (A) |
| CD79A | 28.5 (A) | 33.7 (A) | 22.7 (A) | 38.2 (A) | 29.0 (A) | 31.5 (A) | 38.2 (A) | 23.5 (A) | 25.7 (A) | 24.2 (A) |
| HLADR | 7.9 (A) | 11.2 (A) | 8.6 (A) | 8.0 (A) | 7.5 (A) | 8.9 (A) | 7.1 (A) | 9.2 (A) | 7.8 (A) | 8.3 (A) |
It shows the mean expression of each marker at each cell passage. A, absent gene expression; P, present gene expression (by Affymetrix array).
FIG. 2.
Flow cytometry analysis of CD90 (positive MSC marker) and CD45 (negative MSC marker) in HWJSCs at passages P1, P4, P6, and P9. Cells showing positive expression of each marker are shown in blue; green lines show the expression of cells labeled with the control isotypes. Color images available online at www.liebertonline.com/tec
Cell proliferation analysis of ten consecutive cell passages of HWJSC
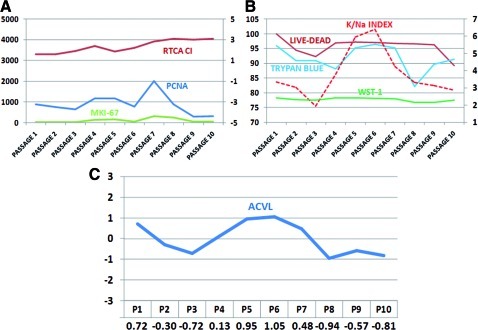
Microarray expression analysis of PCNA and MKI67 genes showed no significant differences from the first to the tenth cell passage (Fig. 3).
FIG. 3.
Analysis of cell proliferation and cell viability of 10 successive passages of HWJSC. (A) Cell proliferation as determined by RTCA Real-Time Cell Analysis Cell Index (RTCA CI) and microarray analysis of PCNA and MKI-67. (B) Cell proliferation as determined by electron-probe X-ray microanalysis (potassium/sodium index), trypan blue exclusion analysis, LIVE/DEAD® Cell Viability Assay, and WST-1. (C) Average cell viability level (ACVL). WST-1, water-soluble tetrazolium salt-1. Color images available online at www.liebertonline.com/tec
Application of highly sensitive RTCA revealed a highly similar Cell Index in all 10 subcultures analyzed (Table 2 and Fig. 3). According to the WST-1 assay results, the HWJSC proliferation index tended to remain stable over the 10 cell passages, with no statistically significant differences among the passages (Table 2 and Fig. 3).
Table 2.
Analysis of Cell Proliferation and Cell Viability at Successive Human Umbilical Cord Wharton's Jelly Stem Cell Passages (P1 to P10) Water-Soluble Tetrazolium Salt-1: Mitochondrial Function Determined by Water-Soluble Tetrazolium Salt-1
| Cell passage | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WST-1 | Mean | 2.41 | 2.31 | 2.29 | 2.42 | 2.42 | 2.41 | 2.37 | 2.15 | 2.15 | 2.28 |
| Median | 2.43 | 2.37 | 2.30 | 2.43 | 2.42 | 2.41 | 2.39 | 2.15 | 2.23 | 2.27 | |
| Q1-Q3 | 2.39 2.44 | 2.37 2.38 | 2.26 2.34 | 2.41 2.43 | 2.41 2.43 | 2.40 2.42 | 2.31 2.46 | 2.10 2.21 | 2.19 2.23 | 2.25 2.31 | |
| RTCA CI | Mean | 1.60 | 1.62 | 1.94 | 2.42 | 1.89 | 2.22 | 2.86 | 3.11 | 3.03 | 3.11 |
| Median | 1.61 | 1.56 | 1.94 | 2.79 | 1.94 | 2.31 | 2.90 | 3.21 | 3.16 | 2.97 | |
| Q1-Q3 | 1.01 2.21 | 1.05 2.18 | 1.30 2.50 | 1.43 3.22 | 1.59 2.39 | 1.61 2.92 | 2.50 3.26 | 2.98 3.30 | 2.80 3.25 | 2.71 3.60 | |
| Trypan blue | Mean | 95.91 | 90.98 | 90.91 | 88.15 | 95.19 | 96.40 | 95.14 | 82.13 | 89.71 | 91.30 |
| Median | 96.69 | 92.64 | 91.15 | 90.05 | 95.14 | 96.16 | 94.98 | 82.09 | 89.78 | 91.23 | |
| Q1-Q3 | 94.67 97.57 | 89.00 94.67 | 89.87 93.64 | 82.23 94.28 | 93.43 96.78 | 95.24 97.25 | 93.26 98.04 | 79.81 84.42 | 89.04 90.45 | 88.18 94.36 | |
| LIVE/DEAD® | Mean | 100 | 94.43 | 92.35 | 96.95 | 97.17 | 97.04 | 96.73 | 96.66 | 96.22 | 89.35 |
| Median | 100 | 96.49 | 92.31 | 97.52 | 96.34 | 97.20 | 97.39 | 97.16 | 96.32 | 89.25 | |
| Q1-Q3 | 100 100 | 93.15 96.88 | 90.23 95.73 | 95.10 98.87 | 95.42 98.99 | 94.24 100 | 95.78 99.17 | 95.41 97.59 | 96.16 96.48 | 89.13 89.36 |
WST-1, water-soluble tetrazolium salt-1; RTCA CI, real-time cell analyzer cell index. Trypan blue: percentage of live cells in each cell passage by trypan blue exclusion test. LIVE/DEAD®: percentage of live cells in each cell passage by calcein/AM and ethidium homodimer-1. Mean and median values and first and third quartiles (Q1–Q3) are shown for each passage and each technique.
Sequential cell viability analysis of HWJSCs
Trypan blue staining showed that the cell viability of the successive HWJSC subcultures was in general very high, with >82% viable cells at each of the 10 cell passages analyzed (Fig. 3 and Table 2). No significant differences were observed among the different cell passages (p>0.001, Wilcoxon test).
LIVE/DEAD® Cell Viability Assay results showed a nonsignificant decrease in the percentage of live cells from the first to the third passage and a nonsignificant increase at the fourth, with the percentage then remaining stable until the ninth passage followed by a decrease to the tenth passage, when the lowest percentage of live cells was found (Fig. 3 and Table 2). Although statistical significance was not reached, there was a tendency for these percentages to correlate with the K/Na index of the cells as determined by electron-probe X-ray microanalysis (p=0.0022 and r=0.8428, Spearman correlation test, for mean K/Na index values vs. mean LIVE/DEAD® Cell Viability) and with cell viability and cell proliferation levels as determined by WST-1 assay (p=0.0117 and r=0.7538, Spearman correlation test).
Microanalytical analysis of 10 consecutive HWJSC subcultures showed generally high K/Na ratios for these stem cells, ranging from 1.94 for the third cell passage to 6.40 for the sixth passage (Table 3 and Fig. 4). The Friedman test for multiple samples revealed significant differences (p<0.001) among the 10 cell passages in the intracellular ionic concentration of all elements analyzed (Ca, Na, K, Mg, P, Cl, and S; Table 4). Pairwise comparisons between consecutive cell subcultures (Wilcoxon test) demonstrated a tendency for several elements to increase or decrease their intracellular concentrations with a higher number of passages. Intracellular K concentrations tended to decrease from the first to the third passage, including a statistically significant difference between the second and the third passages, followed by an increase until the seventh passage and then a statistically significant decrease to the eighth passage and a continuing decline to the tenth passage (Tables 3 and 4 and Fig. 4). In contrast, intracellular Na concentrations showed a nonsignificant increase from the first to the second and third passages, a decrease from the third to the fourth passage, and an increase from the fifth to the seventh passage, decreasing thereafter (Tables 3 and 4 and Fig. 4). Intracellular Cl concentrations significantly decreased from the first to the second cell passage and from the sixth to the seventh passage and then significantly increased from the seventh to the eighth passage. Intracellular P concentrations significantly decreased from the first to the second cell passages and from the seventh to the eighth passages. Intracellular Ca significantly decreased from the third to the fourth passage and significantly increased from the seventh to the eighth passage. Intracellular S significantly increased from the fifth to the sixth passage and then significantly decreased from the sixth to the seventh passages, and intracellular Mg significantly increased from the seventh to the eighth passages.
Table 3.
Intracellular Ionic Concentrations of Calcium, Chlorine, Potassium, Magnesium, Sodium, Phosphorous, and Sulfur, and Potassium/Sodium Ratio at HWJSC Passages (P1 to P10)
| Cell passage | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca | Mean | 11.85 | 17.00 | 14.21 | 4.74 | 3.72 | 6.48 | 5.26 | 20.69 | 12.33 | 14.13 |
| Median | 11.70 | 16.57 | 12.18 | 3.90 | 3.70 | 6.00 | 5.20 | 17.30 | 12.67 | 15.35 | |
| Q1-Q3 | 4.87 14.62 | 10.72 23.39 | 6.82 19.49 | 0.00 8.28 | 0.01 6.00 | 0.03 11.00 | 0.00 9.26 | 14.13 23.76 | 6.70 18.76 | 8.04 18.76 | |
| Cl | Mean | 202.55 | 141.46 | 137.56 | 176.37 | 172.19 | 195.33 | 74.75 | 203.15 | 171.00 | 181.21 |
| Median | 194.69 | 129.26 | 105.33 | 149.21 | 158.79 | 197.09 | 56.65 | 152.73 | 170.76 | 166.77 | |
| Q1-Q3 | 168.36 241.77 | 104.53 177.14 | 90.17 145.22 | 116.50 178.74 | 141.23 196.29 | 174.75 217.04 | 31.92 100.54 | 132.38 201.40 | 153.00 192.70 | 150.81 194.10 | |
| K | Mean | 419.31 | 385.93 | 268.31 | 350.92 | 412.34 | 457.81 | 460.99 | 331.16 | 272.45 | 267.69 |
| Median | 411.19 | 376.33 | 235.33 | 378.89 | 425.55 | 456.31 | 467.59 | 331.47 | 270.45 | 275.07 | |
| Q1-Q3 | 362.49 510.66 | 355.31 405.55 | 172.27 340.95 | 292.76 421.96 | 378.38 459.90 | 392.74 500.40 | 431.19 515.27 | 294.81 351.08 | 259.43 310.45 | 217.52 307.11 | |
| Mg | Mean | 30.65 | 23.12 | 25.49 | 30.50 | 32.72 | 32.45 | 35.49 | 23.51 | 24.33 | 22.82 |
| Median | 31.73 | 22.74 | 24.85 | 29.08 | 32.79 | 32.26 | 34.37 | 24.33 | 24.85 | 22.21 | |
| Q1-Q3 | 23.27 35.43 | 14.28 31.73 | 21.15 30.14 | 26.44 33.32 | 28.03 34.90 | 28.56 35.43 | 29.61 40.19 | 20.09 27.10 | 20.23 26.84 | 21.15 24.85 | |
| Na | Mean | 125.56 | 127.97 | 138.55 | 92.84 | 69.34 | 71.51 | 108.41 | 99.99 | 86.64 | 93.25 |
| Median | 89.01 | 94.10 | 133.09 | 72.06 | 54.25 | 72.90 | 116.98 | 92.40 | 79.68 | 77.99 | |
| Q1-Q3 | 62.73 193.28 | 52.56 169.54 | 93.25 175.48 | 45.78 95.79 | 40.69 72.90 | 59.34 83.92 | 60.19 142.42 | 65.06 128.85 | 68.45 90.07 | 57.22 114.65 | |
| P | Mean | 275.15 | 220.79 | 235.51 | 260.34 | 277.53 | 286.80 | 286.99 | 214.89 | 224.26 | 225.99 |
| Median | 283.01 | 233.88 | 228.34 | 255.33 | 276.09 | 282.32 | 285.08 | 217.62 | 234.23 | 225.58 | |
| Q1-Q3 | 242.18 298.92 | 186.83 268.48 | 207.59 256.71 | 236.65 287.85 | 267.09 294.08 | 255.33 317.61 | 265.71 307.92 | 189.25 236.65 | 213.29 240.45 | 211.39 249.79 | |
| S | Mean | 50.14 | 50.05 | 57.59 | 54.17 | 45.19 | 71.00 | 56.26 | 66.16 | 57.55 | 59.19 |
| Median | 52.27 | 47.92 | 58.81 | 54.99 | 45.19 | 71.33 | 56.08 | 67.25 | 58.81 | 57.44 | |
| Q1-Q3 | 38.66 61.53 | 35.94 68.61 | 51.73 65.88 | 44.65 63.16 | 34.85 55.54 | 60.98 76.77 | 46.28 66.97 | 59.76 74.60 | 46.96 66.84 | 51.46 70.38 | |
| K/Na | 3.34 | 3.02 | 1.94 | 3.78 | 5.95 | 6.40 | 4.25 | 3.31 | 3.14 | 2.87 |
All concentrations are expressed in millimoles of each element per kilogram of cell dry weight. Mean and median values are shown, followed by the first and third quartiles (Q1–Q3). The bottom row corresponds to the K/Na ratio (based on mean K and Na concentrations at each cell passage).
FIG. 4.
Histograms representing the mean intracellular concentrations of potassium, sodium, and chlorine and the K/Na ratio for the ten consecutive HWJSC passages (P1 to P10). Statistically significant changes (increases or decreases) are indicated with asterisks. Green bars correspond to the control human fibroblasts, while red bars represent the same cells treated with 10 mM HEMA to induce apoptosis. All measures are expressed as millimoles per kilogram of cell dry weight. Color images available online at www.liebertonline.com/tec
Table 4.
Significance (P) Values for Differences in Intracellular Ionic Concentrations of Calcium, Chlorine, Potassium, Magnesium, Sodium, Phosphorous, and Sulfur Among Consecutive HWJSC Passages (P1 to P10)
| Cell passages compared | P1–P2 | P2–P3 | P3–P4 | P4–P5 | P5–P6 | P6–P7 | P7–P8 | P8–P9 | P9–P10 | All (Friedman) |
|---|---|---|---|---|---|---|---|---|---|---|
| Ca | 0.101 | 0.276 | 0.000a | 0.602 | 0.117 | 0.248 | 0.000a | 0.012 | 0.610 | 0.000a |
| Cl | 0.001a | 0.427 | 0.065 | 0.242 | 0.067 | 0.000a | 0.000a | 0.139 | 0.241 | 0.000a |
| K | 0.211 | 0.001a | 0.030 | 0.035 | 0.030 | 0.696 | 0.000a | 0.284 | 0.878 | 0.000a |
| Mg | 0.014 | 0.174 | 0.063 | 0.247 | 0.964 | 0.125 | 0.001a | 0.575 | 0.414 | 0.000a |
| Na | 0.830 | 0.545 | 0.006 | 0.607 | 0.192 | 0.012 | 0.970 | 0.386 | 0.047 | 0.000a |
| P | 0.000a | 0.253 | 0.062 | 0.135 | 0.304 | 0.840 | 0.000a | 0.386 | 0.721 | 0.000a |
| S | 0.788 | 0.187 | 0.326 | 0.022 | 0.000a | 0.000a | 0.035 | 0.169 | 0.959 | 0.000a |
The nonparametric Wilcoxon test was used for pair-wise comparisons. The nonparametric Friedman test was used for global comparisons.
Statistically significant p-values.
Based on the results of the trypan blue, LIVE/DEAD, WST-1, and electron-probe X-ray microanalysis assays, we calculated the ACVL for each cell passage. As shown in Figure 3, the highest ACVLs corresponded to the sixth and fifth cell passages and the lowest to the eighth and tenth passages.
Identification of genes associated with cell viability at different cell passages
Correlation analysis of all genes in the array revealed a significant positive correlation between ACVL and 630 probe sets and a significant negative correlation between ACVL and 801 probe sets (data available on request). Functional analysis of these genes (Table 5) showed an over-representation of 10 gene functions among the 630 probe sets positively correlated with cell viability, which were all related to the synthesis and maintenance of cell components and cell structures (Table 5). No gene functions were over-represented among the 801 probe sets negatively correlated with ACVL.
Table 5.
Cytoscape-BiNGO Functional Analysis of Genes Showing a Significant Positive Correlation (R>0.7) With Average Cell Viability Levels in Ten Consecutive HWJSC Passages, as Determined by Microarray
| GO-ID | p-value | Corrected p-value | Gene functions |
|---|---|---|---|
| 5739 | 3.13E-08 | 4.15E-05 | Mitochondrion |
| 5622 | 2.49E-05 | 1.65E-02 | Intracellular |
| 43231 | 5.26E-05 | 1.79E-02 | Intracellular membrane-bound organelle |
| 43227 | 5.41E-05 | 1.79E-02 | Membrane-bound organelle |
| 313 | 9.16E-05 | 2.02E-02 | Organellar ribosome |
| 5761 | 9.16E-05 | 2.02E-02 | Mitochondrial ribosome |
| 44429 | 2.19E-04 | 3.47E-02 | Mitochondrial part |
| 314 | 2.36E-04 | 3.47E-02 | Organellar small ribosomal subunit |
| 5763 | 2.36E-04 | 3.47E-02 | Mitochondrial small ribosomal subunit |
| 44424 | 3.68E-04 | 4.89E-02 | Intracellular part |
GO-ID, gene ontology reference for each specific term or gene function; p-value: statistical significance of the overrepresentation of each GO term or gene function using a hypergeometric distribution; Corrected p-value: statistical significance of the overrepresentation of each GO term or gene function using a hypergeometric distribution with Bonferroni correction for multiple testing; Gene functions: GO terms identified by Cytoscape-BiNGO with a significant p-value.
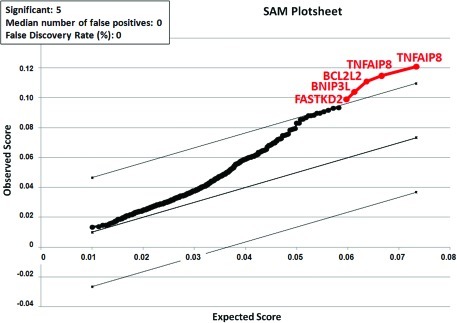
SAM analysis of apoptosis-related genes revealed five probe sets encoding for four different genes whose expression was significantly associated with ACVL (Fig. 5). Two of these genes (FASTKD2 and BNIP3L) have been associated with pro-apoptotic functions, and two (TNFAIP8 and BCL2L2) have been associated with anti-apoptotic functions.
FIG. 5.
Significance Analysis of Microarrays (SAM) results for all genes with a role in apoptotic cell death using multiclass analysis of genes associated with the average cell viability levels (ACVLs). All differentially expressed probe sets are represented in red color, and their names are shown in the plot. We used a δ value that permitted a false discovery rate of 0. Color images available online at www.liebertonline.com/tec
Discussion
MSCs derived from the human umbilical cord are being considered a potential alternative source to bone marrow-derived MSCs for cell therapy.4 HWJSCs are more readily accessible and have a greater expansion potential in comparison to bone marrow-derived MSCs,35 which can only be obtained after a painful and invasive procedure. Moreover, it has been suggested that HWJSCs are immune privileged and could, therefore, be tolerated in allogeneic transplantation.36 In this study, HWJSCs were found to fulfill all of the main criteria for the use of human MSCs in cell therapy and tissue engineering, including good proliferation rate in adherent cell culture; high differentiation capability to osteogenic, chondrogenic, and adipogenic cell lineages over multiple cell passages; and, as previously reported,1 the expression of several markers of undifferentiated MSC (CD105, CD103, and CD90) and the nonexpression of CD45, among other markers.32 These results confirm that the cultured cells were HWJSCs and indicate that our cultured HWJSCs may be clinically useful at the 10 cell passages analyzed.
A critical factor influencing the clinical outcomes of cell therapy/transplantation procedures and the success of tissue engineering is the viability and proliferative capacity of the cells used.37–40 However, no fully sensitive and specific method has yet been established that evaluates the cell viability of human MSCs before their application in these contexts. It should also be taken into account that cell cultures tend to age and lose viability after several passages in culture.10 Thus, evident morphological changes were recently described in corneal endothelial cell cultures from the fourth passage onward, with a trend for cell enlargement, cell growth alterations, and a higher percentage of cell death with increasing passage number.41 However, no data are available on specific intracellular changes in HWJSCs kept in culture for several passages, and little is known about the influence of sequential subculturing on the cell viability and proliferation capabilities of cultured HWJSCs.
In this study, we used classical (trypan blue exclusion) and novel highly sensitive techniques (electron-probe X-ray microanalysis, calcein AM and ethidium homodimer-1, WST-1, microarray, and RTCA) to determine cell viability and cell proliferation. The highest ACVL was observed at the sixth passage, although high cell viability levels were also found at the first, fifth, and seventh passages. No significant change in proliferative capability was observed at any of the 10 cell passages analyzed. A high correlation was found among the techniques used in this study, contrasting with the poor correlation previously observed between trypan blue and electron-probe X-ray microanalysis results for cell viability levels in corneal endothelial cells9 and vascular endothelium.10 The fact that HWJSCs are more undifferentiated than adult endothelial cells and have a higher stemness potential may account for this closer correlation.
Electron-probe X-ray microanalysis allows both qualitative and quantitative determinations of the ionic elements that play a role in cell viability. This approach permits the accurate determination of cell viability and the identification of mechanisms underlying the cell death found at given cell passages.13–18 Although it is clear that most physiological cell functions require a close control of intracellular levels of P, S, Cl, Ca, and other ions, no data have been published on the physiological concentrations of these elements in human native umbilical cords or cultured HWJSCs.
According to the K/Na ratios observed in our study, the highest cell viability levels were at the fifth and sixth passages, which also showed elevated cell proliferation rates. Intracellular concentrations of Na and K are known to be excellent markers of cell viability, and the K/Na ratio is one of the most powerful microanalytical indicators of cell damage.14,18,42 Decreased cell viability at the second, third, and seventh passages may be apoptosis related, given the significantly reduced intracellular levels of Cl in these subcultures, whereas the decrease at passages 8, 9, and 10 may be necrosis related, because elevated Cl concentrations were found in the corresponding subcultures. The high Cl concentration at passage 1 may be associated with a necrosis-related increase in mortality, which could result from the cell isolation process and cell adaptation to the culture conditions. A typical pattern of increased Na and depleted K and Cl is well documented in cells undergoing death by apoptosis.19 Various authors have found an early decrease in K and Cl to be an early predictor of apoptosis, reporting that the decrease in Cl may be the more important of the two and may be used to predict early cell damage.16 Hence, our findings are compatible with reports on ionic changes in preapoptotic cells and suggest that HWJSCs at the second, third, and seventh passages should not be used for clinical purposes because of the number undergoing cell death in the corresponding subcultures. HWJSCs at the eighth, ninth, and tenth passages also showed low viability levels and may not be suitable for human cell therapy and tissue engineering.
Our analysis of the other major intracellular elements revealed an initial decrease in Mg and P between the first and the second cell passage that was statistically significant for P, followed by a progressive increase in both elements until a significant decrease after the seventh passage. According to previous reports,9,43 these findings may be associated with an initial reduction in the intracellular concentration of ATP that would normalize over successive cell passages and then decrease at passages 8, 9, and 10, when cells may be entering senescence. Our observation of a significant increase in intracellular S concentration at the sixth cell passage is consistent with an increase in sulfated glycosaminoglycans at this passage, when the cell viability is higher, confirming the good functional status of HWJSCs in this subculture.44 Intracellular Ca levels were initially elevated but normalized after the fourth cell passage, when a significant decrease in Ca coincided with an increase in ACVL; Ca levels significantly increased between the seventh and tenth passages, when there was a decrease in ACVL. These results are consistent with the known increase in intracellular Ca levels in response to cell damage and apoptosis.45,46
We identified some of the genes that may be responsible for the differences that have just been mentioned with regard to cell viability among HWJSC passages. SAM analysis results demonstrated that the ACVL was significantly associated with four genes related to apoptotic activity. Two are apoptosis-inhibitor genes, BCL2L2 and TNFAIP8, and the latter has two different probe sets. BCL2L2 (GO:0006916: anti-apoptosis and GO:0042981: regulation of apoptosis) is a member of the BCL-2 apoptotic inhibitor gene family, whose anti-apoptotic effects have been demonstrated in various human cells and tumors.47 TNFAIP8 belongs to the tumor-necrosis-factor gene family, which plays a role in the control of cell viability and cell death (GO:0006916: anti-apoptosis, GO:0006915: apoptosis, GO:0019987: negative regulation of anti-apoptosis and GO:0043027: caspase inhibitor activity), and the protein can be induced by TNF-α and other apoptotic factors.48 The significant association of the expression of these anti-apoptotic genes with cell viability may explain some of the cell viability changes found in the first 10 consecutive passages of these cells. The combined action of these genes may be responsible for the high cell viability found at passages 5 and 6. The other two apoptosis-related genes associated with the ACVL were the pro-apoptotic genes BNIP3L and FASTKD2. The mitochondrial protein BNIP3L (GO:0006917: induction of apoptosis and GO:0043066: negative regulation of apoptosis) is known to induce apoptosis and cell growth arrest in transfected cells.49 Likewise, FASTKD2 (GO:0006915: apoptosis) may play a role in the regulation of mitochondrial apoptosis, given that FASTKD2 knockdown prevented apoptosis of breast cancer cells in an ex vivo model of apoptosis.50 Taken together, all of the results just mentioned suggest that a complex equilibrium of pro- and anti-apoptotic gene signals may drive cell death and senescence during sequential HWJSC subculture. These results are in agreement with electron-probe X-ray microanalysis findings on the preapoptotic distribution of intracellular elements in these cells.
Interestingly, all gene functions that were overrepresented among the genes positively correlated with ACVL corresponded to the synthesis and turnover of cellular components. It appears that cells with the highest cell viability levels (especially at fifth and sixth passages) activate multiple cell processes associated with protein synthesis, cell growth, and proliferation, although the differences in cell proliferation among the 10 cell passages did not reach statistical significance. The activation of these gene functions in cell passages with high cell viability confirms the good metabolic status of HWJSCs at these passages. It should be taken into account that the cell viability may have been influenced by our study conditions (e.g., trypsin concentration, cell confluence level, etc.), and results may vary under different conditions.
In summary, these results support the utilization of the highly sensitive methods described in this study for the analysis of all cells selected for clinical purposes in order to determine their vital status and ensure an adequate in vivo survival rate. In general, our data indicate that the highest cell viability levels correspond to the fifth and sixth HWJSC passages, suggesting that these passages should be preferentially used in cell therapy or tissue engineering protocols.
Acknowledgments
This study was supported by the Spanish Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I+D+I) from the National Ministry of Science and Innovation (Instituto de Salud Carlos III), grant FIS 08/615 and by Junta de Andalucia, Consejería de Economía, Innovación y Ciencia, grant P10-CTS-6060 (proyectos de excelencia). The authors are grateful to Richard Davies for assistance with the English version.
Disclosure Statement
There is no conflict of interest for any of the authors. No competing financial interests exist.
References
- 1.Alaminos M. Perez-Kohler B. Garzon I. Garcia-Honduvilla N. Romero B. Campos A. Bujan J. Transdifferentiation potentiality of human Wharton's jelly stem cells towards vascular endothelial cells. J Cell Physiol. 2010;223:640. doi: 10.1002/jcp.22062. [DOI] [PubMed] [Google Scholar]
- 2.Weiss M.L. Troyer D.L. Stem cells in the umbilical cord. Stem Cell Rev. 2006;2:155. doi: 10.1007/s12015-006-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu K.H. Zhou B. Lu S.H. Feng B. Yang S.G. Du W.T. Gu D.S. Han Z.C. Liu Y.L. In vitro and in vivo differentiation of human umbilical cord derived stem cells into endothelial cells. J Cell Biochem. 2007;100:608. doi: 10.1002/jcb.21078. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y.S. Oh W. Choi S.J. Sung D.K. Kim S.Y. Choi E.Y. Kang S. Jin H.J. Yang Y.S. Park W.S. Human umbilical cord blood-derived mesenchymal stem cells attenuate hyperoxia-induced lung injury in neonatal rats. Cell Transplant. 2009;18:869. doi: 10.3727/096368909X471189. [DOI] [PubMed] [Google Scholar]
- 5.Can A. Karahuseyinoglu S. Concise review: human umbilical cord stroma with regard to the source of fetus-derived stem cells. Stem Cells. 2007;25:2886. doi: 10.1634/stemcells.2007-0417. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell S.L. Niklason L.E. Requirements for growing tissue-engineered vascular grafts. Cardiovasc Pathol. 2003;12:59. doi: 10.1016/s1054-8807(02)00183-7. [DOI] [PubMed] [Google Scholar]
- 7.Lund R.D. Wang S. Lu B. Girman S. Holmes T. Sauve Y. Messina D.J. Harris I.R. Kihm A.J. Harmon A.M. Chin F.Y. Gosiewska A. Mistry S.K. Cells isolated from umbilical cord tissue rescue photoreceptors and visual functions in a rodent model of retinal disease. Stem Cells. 2007;25:602. doi: 10.1634/stemcells.2006-0308. [DOI] [PubMed] [Google Scholar]
- 8.Troyer D.L. Weiss M.L. Wharton's jelly-derived cells are a primitive stromal cell population. Stem Cells. 2008;26:591. doi: 10.1634/stemcells.2007-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alaminos M. Sanchez-Quevedo M.C. Muñoz-Avila J.I. Garcia J.M. Crespo P.V. Gonzalez-Andrades M. Campos A. Evaluation of the viability of cultured corneal endothelial cells by quantitative electron probe X-ray microanalysis. J Cell Physiol. 2007;211:692. doi: 10.1002/jcp.20976. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Morata A. Garzon I. Alaminos M. Garcia-Honduvilla N. Sanchez-Quevedo M.C. Bujan J. Campos A. Cell viability and prostacyclin release in cultured human umbilical vein endothelial cells. Ann Vasc Surg. 2008;22:440. doi: 10.1016/j.avsg.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Bilic G. Zeisberger S.M. Mallik A.S. Zimmermann R. Zisch A.H. Comparative characterization of cultured human term amnion epithelial and mesenchymal stromal cells for application in cell therapy. Cell Transplant. 2008;17:955. doi: 10.3727/096368908786576507. [DOI] [PubMed] [Google Scholar]
- 12.Solomon M. Wofford J. Johnson C. Regan D. Creer M.H. Factors influencing cord blood viability assessment before cryopreservation. Transfusion. 2010;50:820. doi: 10.1111/j.1537-2995.2009.02491.x. [DOI] [PubMed] [Google Scholar]
- 13.Roomans G.M. Application of X-ray microanalysis to the study of cell physiology in cells attached to biomaterials. Eur Cell Mater. 2002;3:1. doi: 10.22203/ecm.v003a01. [DOI] [PubMed] [Google Scholar]
- 14.Salido M. Vilches J. Roomans G.M. Changes in elemental concentrations in LNCaP cells are associated with a protective effect of neuropeptides on etoposide-induced apoptosis. Cell Biol Int. 2004;28:397. doi: 10.1016/j.cellbi.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Warley A. Potassium concentration is reduced in cultured rabbit tracheal smooth muscle cells after withdrawal of serum. Cell Biol Int. 2001;25:691. doi: 10.1006/cbir.2000.0711. [DOI] [PubMed] [Google Scholar]
- 16.Zierold K. Effects of cadmium on electrolyte ions in cultured rat hepatocytes studied by X-ray microanalysis of cryosections. Toxicol Appl Pharmacol. 1997;144:70. doi: 10.1006/taap.1996.8094. [DOI] [PubMed] [Google Scholar]
- 17.Vilches J. Salido M. Fernandez-Segura E. Roomans G.M. Neuropeptides, apoptosis and ion changes in prostate cancer. Methods of study and recent developments. Histol Histopathol. 2004;19:951. doi: 10.14670/HH-19.951. [DOI] [PubMed] [Google Scholar]
- 18.Warley A. Fernandez-Segura E. Lopez-Escamez J.A. Campos A. Changes in elemental concentrations in K562 target cells after conjugation with human lymphocytes studied by X-ray microanalysis. Cell Biol Int. 1994;18:915. doi: 10.1006/cbir.1994.1131. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Segura E. Canizares F.J. Cubero M.A. Warley A. Campos A. Changes in elemental content during apoptotic cell death studied by electron probe X-ray microanalysis. Exp Cell Res. 1999;253:454. doi: 10.1006/excr.1999.4657. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Segura E. Canizares F.J. Cubero M.A. Revelles F. Campos A. Electron probe X-ray microanalysis of cultured epithelial tumour cells with scanning electron microscopy. Cell Biol Int. 1997;21:665. doi: 10.1006/cbir.1997.0157. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Quevedo M.C. Alaminos M. Capitan L.M. Moreu G. Garzon I. Crespo P.V. Campos A. Histological and histochemical evaluation of human oral mucosa constructs developed by tissue engineering. Histol Histopathol. 2007;22:631. doi: 10.14670/HH-22.631. [DOI] [PubMed] [Google Scholar]
- 22.Gress T.M. Hoheisel J.D. Lennon G.G. Zehetner G. Lehrach H. Hybridization fingerprinting of high-density cDNA-library arrays with cDNA pools derived from whole tissues. Mamm Genome. 1992;3:609. doi: 10.1007/BF00352477. [DOI] [PubMed] [Google Scholar]
- 23.Iqbal J. Liu Z. Deffenbacher K. Chan W.C. Gene expression profiling in lymphoma diagnosis and management. Best Pract Res Clin Haematol. 2009;22:191. doi: 10.1016/j.beha.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Gill R.T. Enabling inverse metabolic engineering through genomics. Curr Opin Biotechnol. 2003;14:484. doi: 10.1016/s0958-1669(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 25.Jaluria P. Chu C. Betenbaugh M. Shiloach J. Cells by design: a mini-review of targeting cell engineering using DNA microarrays. Mol Biotechnol. 2008;39:105. doi: 10.1007/s12033-008-9048-5. [DOI] [PubMed] [Google Scholar]
- 26.Schena M. Shalon D. Davis R.W. Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 27.Alaminos M. Gonzalez-Andrades M. Muñoz-Avila J.I. Garzon I. Sanchez-Quevedo M.C. Campos A. Volumetric and ionic regulation during the in vitro development of a corneal endothelial barrier. Exp Eye Res. 2008;86:758. doi: 10.1016/j.exer.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Boekestein A. Kuijpers G.A. Stols A.L. Stadhouders A.M. Elemental analysis of individual rat blood platelets by electron probe X-ray microanalysis using a direct quantification method. Histochemistry. 1985;82:257. doi: 10.1007/BF00501402. [DOI] [PubMed] [Google Scholar]
- 29.Warley A. Standards for the application of X-ray microanalysis to biological specimens. J Microsc. 1990;157:135. doi: 10.1111/j.1365-2818.1990.tb02954.x. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez I.A. Fernández-Segura E. Ceballos G. Arrebola F. Sánchez-Quevedo M.C. Campos A. Hybrid cell death induced by exposure to 2-hydroxyethyl methacrylate (HEMA): an ultrastructural and X-ray microanalytical study. J Adhes Dent. 2008;10:105. [PubMed] [Google Scholar]
- 31.Nieto-Aguilar R. Serrato D. Garzon I. Campos A. Alaminos M. Pluripotential differentiation capability of human adipose-derived stem cells in a novel fibrin-agarose scaffold. J Biomater Appl. 2011;25:743. doi: 10.1177/0885328209360425. [DOI] [PubMed] [Google Scholar]
- 32.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F.C. Krause D.S. Deans R.J. Keating A. Prockop D.J. Horwitz E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 33.Maere S. Heymans K. Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 34.Boyer L.A. Plath K. Zeitlinger J. Brambrink T. Medeiros L.A. Lee T.I. Levine S.S. Wernig M. Tajonar A. Ray M.K. Bell G.W. Otte A.P. Vidal M. Gifford D.K. Young R.A. Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 35.Prasanna S.J. Gopalakrishnan D. Shankar S.R. Vasandan A.B. Pro-inflammatory cytokines, IFNgamma and TNFalpha, influence immune properties of human bone marrow and Wharton jelly mesenchymal stem cells differentially. PLoS One. 2010;5:9016. doi: 10.1371/journal.pone.0009016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss M.L. Anderson C. Medicetty S. Seshareddy K.B. Weiss R.J. VanderWerff I. Troyer D. McIntosh K.R. Immune properties of human umbilical cord Wharton's jelly-derived cells. Stem Cells. 2008;26:2865. doi: 10.1634/stemcells.2007-1028. [DOI] [PubMed] [Google Scholar]
- 37.Alaminos M. Del Carmen Sanchez-Quevedo M. Muñoz-Avila J.I. Serrano D. Medialdea S. Carreras I. Campos A. Construction of a complete rabbit cornea substitute using a fibrin-agarose scaffold. Invest Ophthalmol Vis Sci. 2006;47:3311. doi: 10.1167/iovs.05-1647. [DOI] [PubMed] [Google Scholar]
- 38.Garzon I. Sanchez-Quevedo M.C. Moreu G. Gonzalez-Jaranay M. Gonzalez-Andrades M. Montalvo A. Campos A. Alaminos M. In vitro and in vivo cytokeratin patterns of expression in bioengineered human periodontal mucosa. J Periodontal Res. 2009;44:588. doi: 10.1111/j.1600-0765.2008.01159.x. [DOI] [PubMed] [Google Scholar]
- 39.Garzon I. Serrato D. Roda O. Del Carmen Sanchez-Quevedo M. Gonzales-Jaranay M. Moreu G. Nieto-Aguilar R. Alaminos M. Campos A. In vitro cytokeratin expression profiling of human oral mucosa substitutes developed by tissue engineering. Int J Artif Organs. 2009;32:711. doi: 10.1177/039139880903201002. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Andrades M. Garzon I. Gascon M.I. Munoz-Avila J.I. Sanchez-Quevedo M.C. Campos A. Alaminos M. Sequential development of intercellular junctions in bioengineered human corneas. J Tissue Eng Regen Med. 2009;3:442. doi: 10.1002/term.178. [DOI] [PubMed] [Google Scholar]
- 41.Zhu C. Joyce N.C. Proliferative response of corneal endothelial cells from young and older donors. Invest Ophthalmol Vis Sci. 2004;45:1743. doi: 10.1167/iovs.03-0814. [DOI] [PubMed] [Google Scholar]
- 42.Roomans G.M. Pharmacological treatment of the ion transport defect in cystic fibrosis. Expert Opin Investig Drugs. 2001;10:1. doi: 10.1517/13543784.10.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Di Francesco A. Desnoyer R.W. Covacci V. Wolf F.I. Romani A. Cittadini A. Bond M. Changes in magnesium content and subcellular distribution during retinoic acid-induced differentiation of HL60 cells. Arch Biochem Biophys. 1998;360:149. doi: 10.1006/abbi.1998.0937. [DOI] [PubMed] [Google Scholar]
- 44.Roomans G.M. X-ray microanalysis of epithelial cells in culture. Methods Mol Biol. 2002;188:273. doi: 10.1385/1-59259-185-X:273. [DOI] [PubMed] [Google Scholar]
- 45.Berger M.L. Reynolds R.C. Hagler H.K. Bellotto D. Parsons D. Mulligan K.J. Buja L.M. Anoxic hepatocyte injury: role of reversible changes in elemental content and distribution. Hepatology. 1989;9:219. doi: 10.1002/hep.1840090210. [DOI] [PubMed] [Google Scholar]
- 46.Roomans G.M. Von Euler A. X-ray microanalysis in cell biology and cell pathology. Cell Biol Int. 1996;20:103. doi: 10.1006/cbir.1996.0014. [DOI] [PubMed] [Google Scholar]
- 47.Sasi N. Hwang M. Jaboin J. Csiki I. Lu B. Regulated cell death pathways: new twists in modulation of BCL2 family function. Mol Cancer Ther. 2009;8:1421. doi: 10.1158/1535-7163.MCT-08-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar D. Whiteside T.L. Kasid U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin- 1beta-converting enzyme-inhibitory protein. J Biol Chem. 2000;275:2973. doi: 10.1074/jbc.275.4.2973. [DOI] [PubMed] [Google Scholar]
- 49.Yasuda M. Han J.W. Dionne C.A. Boyd J.M. Chinnadurai G. BNIP3alpha: a human homolog of mitochondrial proapoptotic protein BNIP3. Cancer Res. 1999;1:533. [PubMed] [Google Scholar]
- 50.Yeung K.T. Das S. Zhang J. Lomniczi A. Ojeda S.R. Xu C.F. Neubert T.A. Samuels H.H. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol Cell Biol. 2011;31:2287. doi: 10.1128/MCB.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]