Abstract
HIV-1 transmitted drug resistance (TDR) could reverse the gains of antiretroviral rollout. To ensure that current first-line therapies remain effective, TDR levels in recently infected treatment-naive patients need to be monitored. A literature review and data mining exercise was carried out to determine the temporal trends in TDR in South Africa. In addition, 72 sequences from seroconvertors identified from Africa Centre's 2010 HIV surveillance round were also examined for TDR. Publicly available data on TDR were retrieved from GenBank, curated in RegaDB, and analyzed using the Calibrated Population Resistance Program. There was no evidence of TDR from the 2010 rural KwaZulu Natal samples. Ten datasets with a total of 1618 sequences collected between 2000 and 2010 were pooled to provide a temporal analysis of TDR. The year with the highest TDR rate was 2002 [6.67%, 95% confidence interval (CI): 3.09–13.79%; n=6/90]. After 2002, TDR levels returned to <5% (WHO low-level threshold) and showed no statistically significant increase in the interval between 2002 and 2010. The most common mutations were associated with NNRTI resistance, K103N, followed by Y181C and Y188C/L. Five sequences had multiple resistance mutations associated with NNRTI resistance. There is no evidence of TDR in rural KwaZulu-Natal. TDR levels in South Africa have remained low following a downward trend since 2003. Continuous vigilance in monitoring of TDR is needed as more patients are initiated and maintained onto antiretroviral therapy.
Introduction
Concern that a rapid massive scale-up of antiretroviral therapy (ART) would lead to the widespread emergence and transmission of drug-resistant virus1,2 was expressed early in the development of ART access programs in Africa. These were based largely on the challenges associated with early treatments (high pill burden, frequent dosing, toxicity) and financial constraints that would cause drug “stock-outs” and prohibit essential laboratory monitoring.3 Although some programs in sub-Saharan Africa have experienced difficulty maintaining a constant supply of drugs,4 pooled data obtained before the widespread roll-out of ART programs indicate that adherence levels in Africa have been as high, or higher, than as those in North America (77% vs. 55%, respectively, p≤0.001).5 Although high levels of adherence may be maintained following widespread roll-out of ART, missed drug pick-ups due to family obligations and travel costs have been leading causes of viremia and the selection of drug resistance in Africa.6–8
Transmitted drug resistance (TDR) levels vary widely, depending on factors such as the population studied, availability, and access to ART, duration of the treatment programs, quality of medical care, risk-taking behaviors, mode of transmission, and viral subtype.9,10 In developed countries, the use of mono therapy and dual therapy in the pre-highly active antiretroviral therapy (HAART) era (1980s and early 1990s) resulted in the rapid emergence of TDR with peak levels as high as 27%11–17 in some population groups. However, recent evidence suggests that TDR is decreasing with the current levels ranging between 5% and 15% in Europe (2007) and 10% and 18% in the United States.18 Since transmitted resistance can seriously limit future therapeutic options, North American and European treatment guidelines recommend resistance testing for all treatment-naive patients prior to the initiation of HAART. This provides an opportunity to avoid ineffective drug combinations and allows for individualized optimization of first-line HAART.
In most African countries, formalized ART became available in the public health system only after 2002/2003 with the introduction of first-line treatment with combination drug regimens consisting of two nucleoside combined with one nonnucleoside reverse transcriptase inhibitor (NRTIs, NNRTIs). These simple and affordable treatments have been effective and, until recently, levels of primary resistance have remained low (i.e., <5%). However, several recent studies in southern (Lusaka, Durban, Cape Town) and East (Entebbe, Kigali, Kilifi) Africa have reported TDR rates ≥5%.19, 20 All of these studies were conducted in large urban centers, either in young primagravidas attending antenatal clinics (ANC) or in treatment-naive individuals starting ART.
In resource-constrained countries where a substantial proportion of the population may be in need of treatment (i.e., have an AIDS-defining illness and/or a CD4+ T cell count <350 cells/μl), WHO recommends resistance genotyping of remnant specimens collected from recently infected treatment-naive individuals, either young (<25 years of age) primagravida women, individuals consecutively diagnosed with HIV-1 in seroprevalence surveys, or individuals who have laboratory confirmed evidence of recent infection using a test such as the cBED assay.21 With approximately 57 samples, a binomial sequential sampling method is recommended to estimate the potential prevalence of resistance as <5%, 5–15%, or >15%.22,23 To date, most WHO “threshold” surveys have been conducted in the antenatal setting. Whether the results can be extrapolated to the general population remains to be determined. Population-based estimates of TDR are rare but can be derived from surveillance, prevention, and vaccine studies including the identification of individuals with laboratory evidence of recent infection.
The current population-based study is nested within a large 7-year HIV-1 surveillance study conducted in the Hlabisa subdistrict of rural KwaZulu-Natal (KZN).24,25 Using samples collected in 2010, we estimated the level of transmitted drug resistance in a group of 72 recent seroconverters (defined by a confirmed positive serological test in 2010 preceded by a negative test in the previous surveillance round in which the person participated). To track temporal changes and assess demographic differences in TDR, we compared our population-based results to previous resistance studies conducted in diverse setting across South Africa.
Methods and Materials
Study participants
The Africa Centre hosts a large demographic and health surveillance in a circumscribed area of Hlabisa subdistrict in Northern, rural KwaZulu-Natal. HIV surveillance of eligible resident adults was initiated in 2003/2004 and has been followed annually thereafter.26 Furthermore, the Africa Centre partners with the Department of Health in the delivery of a large HIV treatment and care program.27 Plasma samples obtained from individuals with documented evidence of recent seroconversion were selected for resistance genotyping from the 2010 HIV surveillance round. Inclusion criteria for enrollment in studies of primary HIV-1 drug resistance included a confirmed seropositive test in 2010, preceded by an HIV-1 seronegative test in the previous surveillance round in which the person had participated. Ethics approval for the study was obtained from the University of KwaZulu-Natal Biomedical Research Ethics Committee (Ethics number: BE066107).
Specimen processing and laboratory testing
Blood samples for serological testing were collected in Beckton Dickinson (BD) EDTA microtainer tubes and transported to the Africa Centre laboratory in Durban for processing. An aliquot of the whole blood was sent to a commercial laboratory for CD4 testing. Plasma was prepared within 24 h of collection and tested for the presence/absence of HIV-1 antibodies using an initial screening (SD HIV ½ ELISA ver. 3.0, Standard Diagnostics, Inc., Kyonggi-do, Korea) followed by a confirmatory (Bio-Rad GS rLAV HIV-1 EIA, Bio-Rad Laboratories, Redmond, WA) ELISA. These tests were done as part of the annual HIV surveillance within the Africa Centre's main demographic study. Residual plasma was stored at −80°C until required for drug resistance testing. Genotyping of resistance mutations was performed retrospectively on individuals with confirmed HIV-1 seroconversion.
Sequencing of the HIV-1 pol gene
Sequencing of the HIV-1 protease and the first 300 codons of the reverse transcriptase (RT) gene was performed on a 1315-bp PCR product generated from the HIV-1 pol region. RNA was extracted from 140 μl of plasma using the QiaAmp viral RNA extraction kit (Qiagen) and reverse transcribed into cDNA with the Superscript III kit (Invitrogen Corporation, Carlsbad, CA) and a gene specific primer: RT21_MOD (5′-CTG TAT TTC AGC TAT CAA GTC CTT TGA TGG G-3′). The protease and RT gene regions were amplified from the cDNA using Platinum Taq polymerase (Invitrogen Corporation, Carlsbad, CA) and a nested PCR protocol. The following primers where used: first round PCR: RT21_MOD and MAW26 (5′-TTG GAA ATG TGG AAA GGA AGG AC-3′) and second round PCR: RT20_MOD (5′-CTG CCA ATT CTA ATT CTG CTT C-3′) and PRO-1_MOD (5’-TAG AGC CAA CAG CCC CAC CA-3′). The cycling conditions for both the first and second round PCR were 94°C for 2 min, 30 cycles of 95°C for 30 s, 58°C for 20 s, and 72°C for 2 min, followed by a final extension of 72°C for 10 min. To assess the success of the reaction, second round PCR products were stained with a fluorescent dye, Novel Juice (GeneDireX, Taipei Taiwan), subjected to agarose gel (1%) electrophoresis (45 min at 70 V and 400 mA), and visually compared to a 200-bp DNA ladder from Fermentas (Maryland, USA).
Successfully amplified samples were purified using the PureLink Invitrogen PCR purification kit (Invitrogen Corporation, Carlsbad, CA) according to the manufacturer's instruction. The concentration and quality of the DNA in each PCR product were assessed using a nanodrop scanning spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). Sequencing reactions were done using the Big Dye terminator chemistry (Applied Biosystems Inc., Foster City, CA) for each of the following primers: RTC1F (5’-ACC TAC ACC TGT CAA CAT AAT TG-3’), RTC2R (5’-TGT CAA TGG CCA TTG TTT AAC CTT TGG-3’), RTC3F (5’-ACC AGG GAT TAG ATA TCA ATA TAA TGT GC-3’), RTC4R(5’-CTA AAT CAG ATC CTA CAT ACA AGT CAT CC-3’), RTY (5’-CCT AGT ATA AAC AAT GAG ACA C-3’), AND MAW46 (5’-TCC CTC AGA TCA CTC TTT GGC AAC GAC-3′). Sequencing electrophoresis was done on a 3130xl Genetic Analyzer (Applied Biosystems Inc, Foster City, CA).
Analysis and interpretation of sequence results
Protease and reverse transcriptase nucleotide sequences were assembled using a Geneious Pro genetic analyzer.28 Quality assessment and HIV subtyping of these sequences were performed using the HIV-1 Quality Analysis Tool and REGA HIV-1 Subtyping Tool v. 2.0, respectively.29,30 To aid in quality assurance, neighbor-joining phylogenetic trees were created in Geneious using Clustal W for sequence alignments. Maximum likelihood (ML) trees were generated in PhyML v. 2.4.4,31 and 500 replicates were bootstrapped. Trees were viewed using FigTree. Sequence data were analyzed using the Calibrated Resistance Program (CPR).32 Resistance mutations were identified using the Stanford Drug Resistance Mutation List of 2009. The statistical program STATA version 10 (StataCorp LP, Texas, USA) was used to perform all the descriptive analysis used in this article.
Temporal studies of TDR
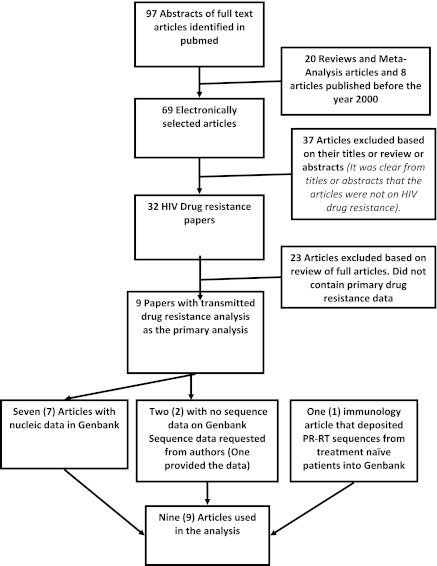
To better understand our KZN data in the context of other TDR studies conducted in South Africa, we performed a comprehensive review searching for previously published papers on primary drug resistance in treatment-naive individuals with sequences in GenBank. The key search terms used were “HIV-1 AND drug resistance AND South Africa.” HIV-1 pol sequences linked to these articles were then retrieved from GenBank and archived for further analysis (Fig. 1). For articles that did not have linked genotypes in GenBank, the data were requested from the authors. This was done as part of the curation of the Southern African mirror of the Stanford HIV Drug Resistance Database.33 Sequences published during the past 10 years were pooled and reanalyzed using the CPR tool at the Southern African Mirror of the Stanford HIV Drug Resistance Database. The Stanford Drug Resistance Mutation (SDRM) list (2009 version) was used for the analyses of transmitted resistance.
FIG. 1.
Literature search methodology and dataset construction.
Results
Current TDR rates as assessed in rural KZN
A total of 91 individuals participating in the Africa Centre's annual population-based HIV surveillance program met the inclusion criteria of having a confirmed HIV-1-seropositive HIV test in 2010, preceded by an HIV-1-seronegative test in the previous surveillance round in which the person had participated. The median time between the last seronegative and first seropositive result was 3.2 years (standard deviation: 1.76 years). The mean estimated time to seroconversion, as defined by the midpoint between the first positive test and the last negative test, was 1.8 years (standard deviation: 0.9 years). The median CD4+ T cell count for the samples genotyped was 413 cells/μl (interquartile range; 286–519 cells/μl).
Of the 91 samples selected for drug resistance testing, 72 (79%) were successfully genotyped. As would be expected, the majority (71/72, 99%) of recent seroconverters were infected with HIV-1 C subtype, the predominant subtype in KZN: the remaining seroconverter was infected with subtype A. There was no evidence of transmitted drug resistance in the samples of the 72 seroconverters tested.
Changes in prevalence of TDR mutations over the past 20 years
The PUBMED search identified 32 HIV-1 drug resistance studies. Eight of these published datasets34–41 contained sequences derived from HIV-1-infected, ART-naive individuals. Additional HIV-1 pol sequences42 amplified from untreated patients were retrieved from GenBank and included in the analysis. Two unpublished datasets were also included (Wilkinson et al., unpublished and the 2010 data from rural KZN). The total number of sequences analyzed was 1618 (Fig. 2). The samples for these studies were collected between 2000 and 2010, representing a period of almost 10 years. Figure 1 describes the selection process used to search for these articles. Table 1 summarizes the studies and publications analyzed.
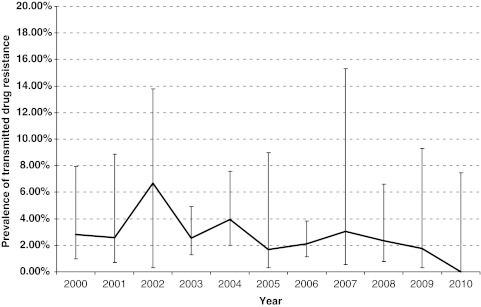
FIG. 2.
Trend in the prevalence of transmitted drug resistance between 2000 and 2010.
Table 1.
Data from Nine Published Studies on Primary Drug Resistance Using Inhouse Genotyping Methods on Plasma Samples
| First author | Sampling year | Publication year | Region | Number of patients | Number of sequences | Study participants | Study design |
|---|---|---|---|---|---|---|---|
| Pillay et al.34 | 2000 | 2002 | Gauteng | 52 | 37 | ANC women | pMTCT Clinical Trial Screening |
| Gordon et al.35 | 2001 | 2003 | KwaZulu-Natal | 72 | 72 | Patients | Cross-sectional |
| Bessong et al.37 | 2005 | 2005 | Limpopo | 14 | 13 | Patients | Cross-sectional |
| Bessong et al.36 | 2006 | 2006 | Limpopo and Gauteng | 53 | 35 | Patients | Cross-sectional |
| Seioghe et al.38 | 2004–2007 | 2007 | Johannesburg and Durban | 300 | 279 | ANC women | pMTCT baseline |
| Matthews et al.42 | 2008 | 2008 | KwaZulu-Natal | 438 | 475 | Patients | Cross-sectional |
| Pillay et al.40 | 2002 and 2004 | 2008 | Gauteng | 245 | 101 | ANC women | WHO protocol |
| Jacobs et al.39 | 2002–2004 | 2008 | Cape Town | 140 | 91 | Patients | Cross-sectional |
| Huang et al.41 | 2006 | 2009 | Free State | 390 | 354 | Patients | VCT |
ANC, antinatal clinic; pMTCT, prevention of mother-to-child transmission; VCT, voluntary HIV counseling and testing.
Analysis of the aggregated data indicated that the prevalence of TDR in South Africa has remained at low levels over the past 10 years. The year with the highest TDR rate was 2002 [6.67%, 95% confidence interval (CI): 3.09–13.79%; n=6/90]. After 2002, TDR levels returned to <5% (WHO low-level threshold) and showed no statistically significant increase in the interval between 2002 and 2010 (Fig. 2).
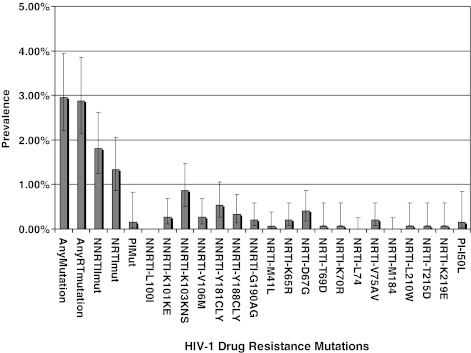
The most common mutations were associated with NNRTI resistance, K103N, followed by Y181C and Y188C/L. One sequence had drug resistance mutations associated with nucleosides and nonnucleoside RT inhibitors (M184V and Y181C, respectively). Another five sequences had multiple resistance mutations associated with NNRTI resistance35,41 (Fig. 3).
FIG. 3.
HIV‐1 drug resistance mutations identified in the 1650 sequences used in the pooled analysis.
Discussion
Studies examining the frequency of transmitted HIV-1 drug resistance in newly infected individuals are rare with only small numbers of participants being evaluated in each study. This is particularly true of Africa and other regions of the developing world where most HIV-1-infected individuals are diagnosed only when they present with advanced symptomatic disease. The current study takes advantage of a large long-standing (7 year) serosurvey to assess the frequency of TDR in rural KZN, a highly endemic region of South Africa where the prevalence of HIV-1 infection in the adult population is currently 22.3% for women and 11.5% for men.43 Using WHO criteria to select for eligible seroconverters (i.e., consecutively diagnosed, recently infected ART-naive individuals), we found no evidence of TDR in the general population. When analyzed according to the WHO threshold method, the level of resistance in rural KZN fell into the <5% category. This low level of TDR in KZN, 6 years after the introduction of HAART, is similar to resistance studies conducted in ART-naive populations of South Africa over the past 10 years.
As in North America and Europe, interstudy comparisons and TDR trend analyses in Africa are likely to be complicated by a number of confounding variables that include study location, local incidence rates, sample size, variation in study populations (heterosexual, homosexual, pediatric), changes in drug regimens and treatment practices, and differences in the definition (high vs. low level resistance, clinically relevant resistance) and methods used to assess resistance (phenotypic, genotypic assays of varying sensitivity ranging from population-based sequencing, allele-specific priming, and deep sequencing).9 These biases make it difficult to establish firm epidemiological generalizations based on the prevalence and trends of TDR in individual demographic and population groups.
Nevertheless, despite differences between the Africa Centre and previous TDR studies conducted in Gauteng, the Free State, the Western Cape, and KZN, we were able to create a temporal profile of primary resistance in South Africa over the past 10 years. A pooled analysis of available data (obtained from multiple sources including GenBank and eight published studies conducted at antenatal clinics and in treatment-naive patients initiating HAART) indicated that there has been no statistically significant increase in TDR rates in South Africa over time and that, with the exception of the year 2002, which showed a slight spike in TDR prevalence (6.7%), levels of transmitted resistance have remained below the <5% threshold. The spike in 2002 occurred just prior to the public sector rollout of ART and may be due to undisclosed participation in the prevention of mother-to-child transmission (pMTCT) programs or to the residual effects of mono therapies and dual therapies used in the private sector. Previous exposure to single-dose nevirapine (SD NVP) or short course AZT might have been the reasons for the higher TDR prevalence in 2002. Two recent publications on TDR from clinics in KwaZulu-Natal and Gauteng have also showed that TDR levels are still low (less than 5%) in South Africa.44,45
TDR surveillance in newly infected individuals with documented evidence of recent seroconversion, as described in this study, minimizes the possibility of including ART-experienced participants. The disadvantage of this approach is that it is limited to a relatively small number of early cases. Other studies have suggested that TDR may also be common in chronically infected individuals who never received ART, presumably due to the persistence of TDR following the resolution of acute infection.46 Resistance mutations in women exposed to SD NVP for pMCTC purposes have been shown to fade in the absence of ART, with most mutations becoming undetectable 18 months after NVP exposure.47–49 Much less is known about the waning of drug-resistant mutations acquired during acute infections. However, there are suggestions that transmitted drug resistance can persist for at least 4 years.50 At least two studies have suggested that such mutations may persist for prolonged periods of time due to massive fueling of cellular reservoirs with drug-resistant virus.51,52
The comprehensive nature of Africa Centre's Demographic Surveillance System provides an ideal platform for addressing these issues. Continued longitudinal monitoring of TDR-positive seroconverters who are not yet eligible for HAART provides an opportunity to examine the acquisition and evolution of drug resistance in individuals chronically infected with HIV-1. Studies of TDR-positive seroconverters will allow for an improved understanding of the reversion from drug-resistant to wild-type virus. As additional follow-up surveys are conducted, it should be possible to construct an in-depth understanding of the risk factors and temporal trends associated with TDR, similar to what has been accomplished in HIV-1 incidence studies.
In summary, the prevalence of TDR in rural KZN is still very low, similar to reports from other provinces of South Africa. Although widespread resistance testing in South Africa is not recommended at the present time, pretreatment genotypic testing has been shown to significantly improve the outcome of first-line drug regimens in resource-rich countries.53 In Southern Africa as ART access and treatment programs grow, it is critical to maintain surveillance among recent seroconverters and treatment-naive and treated populations. Programs covering well-defined geographic areas and populations minimize the biases inherent in TDR surveillance and provide guidance in implementing risk reduction and secondary prevention to maintain the effectiveness of first-line treatment.
Sequence data
GenBank accession numbers are JN664970 to JN665041.
Acknowledgments
The results generated in this paper were funded by the Wellcome Trust (082384/Z/07/Z), European Union (SANTE 2007 147–790), the US Centre for Diseases Control via CAPRISA (project title: Health Systems Strengthening and HIV Treatment Failure (HIV-TFC)) and the Swiss South African Joint Research Programme (SSJRP) research grant entitled “Swiss Prot/South Africa: Protein Bioinformatics Resource Development for Important Health-related Pathogens.” The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to acknowledge all the authors and SATuRN collaborators who contributed to the sequences data used in the temporal analysis through their GenBank sequence submissions and direct REGADB submissions as well as the submission of the ART exposure histories. We are also grateful to the Africa Centre Virology laboratory staff for their support in the genotyping of the 2010 Africa Centre samples.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Stevens W. Kaye S. Corrah T. Antiretroviral therapy in Africa. BMJ. 2004;328(7434):280–282. doi: 10.1136/bmj.328.7434.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popp D. Fisher JD. First, do no harm: A call for emphasizing adherence and HIV prevention interventions in active antiretroviral therapy programs in the developing world. AIDS. 2002;16(4):676–678. doi: 10.1097/00002030-200203080-00025. [DOI] [PubMed] [Google Scholar]
- 3.Trotta MP. Ammassari A. Melzi S, et al. Treatment-related factors and highly active antiretroviral therapy adherence. J Acquir Immune Defic Syndr. 2002;31(Suppl 3):S128–131. doi: 10.1097/00126334-200212153-00008. [DOI] [PubMed] [Google Scholar]
- 4.Uzochukwu BS. Onwujekwe OE. Onoka AC. Okoli C. Uguru NP. Chukwuogo OI. Determinants of non-adherence to subsidized anti-retroviral treatment in southeast Nigeria. Health Policy Plan. 2009;24(3):189–196. doi: 10.1093/heapol/czp006. [DOI] [PubMed] [Google Scholar]
- 5.Mills EJ. Nachega JB. Buchan I, et al. Adherence to antiretroviral therapy in sub-Saharan Africa and North America: A meta-analysis. JAMA. 2006;296(6):679–690. doi: 10.1001/jama.296.6.679. [DOI] [PubMed] [Google Scholar]
- 6.El-Khatib Z. Katzenstein D. Marrone G, et al. Adherence to drug-refill is a useful early warning indicator of virologic and immunologic failure among HIV patients on first-line ART in South Africa. PLoS ONE. 2011;6(3):e17518. doi: 10.1371/journal.pone.0017518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett DE. Bertagnolio S. Sutherland D. Gilks CF. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl 2):1–13. [PubMed] [Google Scholar]
- 8.Mills EJ. Nachega JB. Bangsberg DR, et al. Adherence to HAART: A systematic review of developed and developing nation patient-reported barriers and facilitators. PLoS Med. 2006;3(11):e438. doi: 10.1371/journal.pmed.0030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillay D. Current patterns in the epidemiology of primary HIV drug resistance in North America and Europe. Antivir Ther. 2004;9(5):695–702. [PubMed] [Google Scholar]
- 10.Sagir A. Oette M. Kaiser R, et al. Trends of prevalence of primary HIV drug resistance in Germany. J Antimicrob Chemother. 2007;60(4):843–848. doi: 10.1093/jac/dkm274. [DOI] [PubMed] [Google Scholar]
- 11.Weinstock H. Respess R. Heneine W, et al. Prevalence of mutations associated with reduced antiretroviral drug susceptibility among human immunodeficiency virus type 1 seroconverters in the United States, 1993–1998. J Infect Dis. 2000;182(1):330–333. doi: 10.1086/315686. [DOI] [PubMed] [Google Scholar]
- 12.UK Collaborative Group on Monitoring the Transmission of HIV Drug Resistance: Analysis of prevalence of HIV-1 drug resistance in primary infections in the United Kingdom. BMJ. 2001;322(7294):1087–1088. doi: 10.1136/bmj.322.7294.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant RM. Hecht FM. Warmerdam M, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288(2):181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 14.Little SJ. Holte S. Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 15.Weinstock HS. Zaidi I. Heneine W, et al. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1-infected persons in 10 US cities. J Infect Dis. 2004;189(12):2174–2180. doi: 10.1086/420789. [DOI] [PubMed] [Google Scholar]
- 16.Cane P. Chrystie I. Dunn D, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: Multicentre observational study. BMJ. 2005;331(7529):1368. doi: 10.1136/bmj.38665.534595.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wensing AM. van de Vijver DA. Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: Implications for clinical management. J Infect Dis. 2005;192(6):958–966. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar]
- 18.Hurt CB. McCoy SI. Kuruc J, et al. Transmitted antiretroviral drug resistance among acute and recent HIV infections in North Carolina from 1998 to 2007. Antivir Ther. 2009;14(5):673–678. [PMC free article] [PubMed] [Google Scholar]
- 19.Hamers RL. Siwale M. Wallis CL, et al. HIV-1 drug resistance mutations are present in six percent of persons initiating antiretroviral therapy in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2010;55(1):95–101. doi: 10.1097/QAI.0b013e3181e544e0. [DOI] [PubMed] [Google Scholar]
- 20.Price MA. Wallis CL. Lakhi S, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses. 2011;27(1):5–12. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parekh BS. Kennedy MS. Dobbs T, et al. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: A simple assay for detecting recent HIV infection and estimating incidence. AIDS Res Hum Retroviruses. 2002;18(4):295–307. doi: 10.1089/088922202753472874. [DOI] [PubMed] [Google Scholar]
- 22.Myatt M. Bennett DE. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross-sectional survey for use in low resource settings. Antivir Ther. 2008;13(Suppl 2):37–48. [PubMed] [Google Scholar]
- 23.Bennett DE. Myatt M. Bertagnolio S. Sutherland D. Gilks FC. Recommendation for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):12. [PubMed] [Google Scholar]
- 24.Barnighausen T. Tanser F. Newell ML. Lack of a decline in HIV incidence in a rural community with high HIV prevalence in South Africa, 2003–2007. AIDS Res Hum Retroviruses. 2009;25(4):405–409. doi: 10.1089/aid.2008.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanser F. Hosegood V. Barnighausen T, et al. Cohort Profile: Africa Centre Demographic Information System (ACDIS) and population-based HIV survey. Int J Epidemiol. 2008;37(5):956–962. doi: 10.1093/ije/dym211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanser F. Barnighausen T. Cooke GS. Newell ML. Localized spatial clustering of HIV infections in a widely disseminated rural South African epidemic. Int J Epidemiol. 2009;38(4):1008–1016. doi: 10.1093/ije/dyp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houlihan CF. Bland RM. Mutevedzi PC, et al. Cohort Profile: Hlabisa HIV Treatment and Care Programme. Int J Epidemiol. 2011;40(2):318–326. doi: 10.1093/ije/dyp402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond AJ. Ashton B. Buxton S, et al. Geneious. 2010;5.1 [Google Scholar]
- 29.de Oliveira T. Deforche K. Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21(19):3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 30.Alcantara LC. Cassol S. Libin P, et al. A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res. 2009;37(Web Server issue):W634–642. doi: 10.1093/nar/gkp455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guindon S. Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood systems biology. Syst Biol. 2003;52(5):9. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 32.Gifford RJ. Liu TF. Rhee SY, et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics. 2009;25(9):1197–1198. doi: 10.1093/bioinformatics/btp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira T. Shafer RW. Seebregts C. Public database for HIV drug resistance in southern Africa. Nature. 2010;464(7289):673. doi: 10.1038/464673c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillay C. Bredell H. McIntyre J. Gray G. Morris L. HIV-1 subtype C reverse transcriptase sequences from drug-naive pregnant women in South Africa. AIDS Res Hum Retroviruses. 2002;18(8):605–610. doi: 10.1089/088922202753747950. [DOI] [PubMed] [Google Scholar]
- 35.Gordon M. De Oliveira T. Bishop K, et al. Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: Implications for vaccine and antiretroviral control strategies. J Virol. 2003;77(4):2587–2599. doi: 10.1128/JVI.77.4.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bessong PO. Mphahlele J. Choge IA, et al. Resistance mutational analysis of HIV type 1 subtype C among rural South African drug-naive patients prior to large-scale availability of antiretrovirals. AIDS Res Hum Retroviruses. 2006;22(12):1306–1312. doi: 10.1089/aid.2006.22.1306. [DOI] [PubMed] [Google Scholar]
- 37.Bessong PO. Larry Obi C. Cilliers T, et al. Characterization of human immunodeficiency virus type 1 from a previously unexplored region of South Africa with a high HIV prevalence. AIDS Res Hum Retroviruses. 2005;21(1):103–109. doi: 10.1089/aid.2005.21.103. [DOI] [PubMed] [Google Scholar]
- 38.Seoighe C. Ketwaroo F. Pillay V, et al. A model of directional selection applied to the evolution of drug resistance in HIV-1. Mol Biol Evol. 2007;24(4):1025–1031. doi: 10.1093/molbev/msm021. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs GB. Laten A. van Rensburg EJ, et al. Phylogenetic diversity and low level antiretroviral resistance mutations in HIV type 1 treatment-naive patients from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008;24(7):1009–1012. doi: 10.1089/aid.2008.0028. [DOI] [PubMed] [Google Scholar]
- 40.Pillay V. Ledwaba J. Hunt G, et al. Antiretroviral drug resistance surveillance among drug-naive HIV-1-infected individuals in Gauteng Province, South Africa in 2002 and 2004. Antivir Ther. 2008;13(Suppl 2):101–107. [PubMed] [Google Scholar]
- 41.Huang KH. Goedhals D. Fryer H, et al. Prevalence of HIV type-1 drug-associated mutations in pre-therapy patients in the Free State, South Africa. Antivir Ther. 2009;14(7):975–984. doi: 10.3851/IMP1416. [DOI] [PubMed] [Google Scholar]
- 42.Matthews PC. Prendergast A. Leslie A, et al. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82(17):8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnighausen T. Tanser F. Gqwede Z. Mbizana C. Herbst K. Newell ML. High HIV incidence in a community with high HIV prevalence in rural South Africa: Findings from a prospective population-based study. AIDS. 2008;22(1):139–144. doi: 10.1097/QAD.0b013e3282f2ef43. [DOI] [PubMed] [Google Scholar]
- 44.Parboosing R. Naidoo A. Gordon M. Taylor M. Vella V. Resistance to antiretroviral drugs in newly diagnosed, young treatment-naive HIV-positive pregnant women in the province of KwaZulu-Natal, South Africa. J Med Virol. 2011;83(9):1508–1513. doi: 10.1002/jmv.22143. [DOI] [PubMed] [Google Scholar]
- 45.Hamers RL. Wallis CL. Kityo C, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: A multicentre observational study. Lancet Infect Dis. 2011;11(10):750–759. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 46.Little SJ. Koelsch KK. Ignacio C, et al. Persistence of transmitted drug-resistant virus among subjects with primary HIV infection deferring antiretroviral therapy; Paper presented at the 11th Conference on Retroviruses and Opportunistic Infections; San Francisco, CA. 2004. [Google Scholar]
- 47.Palmer S. Boltz V. Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci USA. 2006;103(18):7094–7099. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eshleman SH. Mracna M. Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) AIDS. 2001;15(15):1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 49.Loubser S. Balfe P. Sherman G. Hammer S. Kuhn L. Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006;20(7):995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Little SJ. Frost SD. Wong JK, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008;82(11):5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbour JD. Hecht FM. Wrin T, et al. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS. 2004;18(12):1683–1689. doi: 10.1097/01.aids.0000131391.91468.ff. [DOI] [PubMed] [Google Scholar]
- 52.Ghosn J. Pellegrin I. Goujard C, et al. HIV-1 resistant strains acquired at the time of primary infection massively fuel the cellular reservoir and persist for lengthy periods of time. AIDS. 2006;20(2):159–170. doi: 10.1097/01.aids.0000199820.47703.a0. [DOI] [PubMed] [Google Scholar]
- 53.Oette M. Kaiser R. Daumer M, et al. Primary HIV drug resistance and efficacy of first-line antiretroviral therapy guided by resistance testing. J Acquir Immune Defic Syndr. 2006;41(5):573–581. doi: 10.1097/01.qai.0000214805.52723.c1. [DOI] [PubMed] [Google Scholar]