Abstract
Severe pulmonary hypertension (PH) associated with vascular remodeling is a long-term complication of HIV infection (HIV-PH) affecting 1/200 infected individuals vs. 1/200,000 frequency in the uninfected population. Factors accounting for increased PH susceptibility in HIV-infected individuals are unknown. Rhesus macaques infected with chimeric SHIVnef virions but not with SIV display PH-like pulmonary vascular remodeling suggesting that HIV-Nef is associated with PH; these monkeys showed changes in nef sequences that correlated with pathogenesis after passage in vivo. We further examined whether HIV-nef alleles in HIV-PH subjects have signature sequences associated with the disease phenotype. We evaluated specimens from participants with and without HIV-PH from European Registries and validated results with samples collected as part of the Lung-HIV Studies in San Francisco. We found that 10 polymorphisms in nef were overrepresented in blood cells or lung tissue specimens from European HIV-PH individuals but significantly less frequent in HIV-infected individuals without PH. These polymorphisms mapped to known functional domains in Nef. In the validation cohort, 7/10 polymorphisms in the HIV-nef gene were confirmed; these polymorphisms arose independently from viral load, CD4+ T cell counts, length of infection, and antiretroviral therapy status. Two out of 10 polymorphisms were previously reported in macaques with PH-like pulmonary vascular remodeling. Cloned recombinant Nef proteins from clinical samples down-regulated CD4, suggesting that these primary isolates are functional. This study offers new insights into the association between Nef polymorphisms in functional domains and the HIV-PH phenotype. The utility of these polymorphisms as predictors of PH should be examined in a larger population.
Introduction
Pulmonary hypertension (PH) is characterized histologically by remodeling of pulmonary arteries and complex vascular lesions.1 PH is a significant noninfectious complication affecting individuals infected with the human immunodeficiency virus (HIV) at a higher frequency (1/200), compared to 1/200,000 in the HIV-negative population.2–6 An echocardiography-based study reported increased pulmonary artery systolic pressures (>30 mm Hg) in 35% of HIV+ pulmonary asymptomatic individuals, compared with only 7.7% of HIV-negative individuals,7 suggesting a higher risk of HIV-associated PH (HIV-PH) in this population. Whether the prevalence is higher in developing countries with limited access to screening tools remains unknown. Although mutations in the bone morphogenetic protein receptor-II have been identified in heritable PH, these mutations are not more prevalent in HIV-infected individuals.8
HIV can be recovered from the lungs of humans and experimentally infected animals.9–11 Hence, pulmonary vascular disease may be a consequence of the chronic exposure to viral proteins in the lung that may in turn drive the immune response. The lung cells infected with HIV or its simian counterpart, SIV, are sources of viral proteins, including Nef. HIV/SIV Nef (negative factor) is a 27- to 35-kDa cytoplasmic protein expressed early in HIV/SIV infection. Nef down-regulates CD4 expression12 and blocks traffic of major histocompatibility complex type-I (MHC-I) to the membrane allowing the infected cell to evade immune surveillance.13 Nef plays a pivotal role in pathogenesis: some HIV-infected long-term nonprogressors have deletions in the nef open reading frame,14 large deletions in nef attenuate viral pathogenicity in infected macaques,15 and sequence variations in Nef motifs are associated with different stages of pathogenesis.16 Altogether, these studies suggest that alterations in Nef sequence domains have pathophysiological consequences. Furthermore, Nef is associated with significant pulmonary vascular remodeling including obliterative vascular lesions indistinguishable from those in end-stage human PH.17,18 Recent studies from Sehgal's group revealed remarkable Golgi fragmentation in endothelial and smooth muscle cells in the PH-like vascular lesions in macaques and, strikingly, the same cells displayed endosomes bearing HIV-nef.
The present study translated these findings to humans and tested the hypothesis that Nef signature sequences are overrepresented in European HIV-infected individuals with PH but not in normotensive subjects. We validated the results in a separate group of research subjects from San Francisco, California. We found nef alleles associated with the pulmonary hypertensive phenotype, independent of virological and immunological parameters. Although most of the variant amino acids mapped to Nef functional domains, molecularly cloned Nef primary isolates retain the ability to down-regulate CD4 expression. These studies suggest a pathogenetic association between genotypic changes in Nef functional domains and higher prevalence of PH in HIV-infected individuals.
Materials and Methods
Study subjects
We analyzed peripheral blood mononuclear cells (PBMCs) DNA from HIV+ individuals with HIV-PH (n=10), enrolled in the French Registry of Pulmonary Hypertension,19 and lung tissues from a subject (It.13) enrolled in the Latium Registry of HIV-PH in Rome.3 Plasma samples from French HIV-infected individuals (n=7) with no evidence of pulmonary disease were used as controls. To validate the genetic association studies in the European cohort, we further examined banked PBMC DNA from HIV-PH subjects (n=11) and confirmed normotensive subjects (n=22) enrolled in the Lung-HIV studies at the University of California, San Francisco hospital (URL: http://www.lunghiv.com). Please refer to the Supplementary Data for inclusion/exclusion criteria and human subject study approvals.
HIV Nef sequence analyses
We analyzed 763 HIV-nef molecular clones from the 51 subjects (initial and validation groups) infected with HIV-subtype B strains. The full-length HIV-nef from the subject samples was cloned and nucleotide sequences were aligned and translated using Geneious.20 We also compared the HIV-nef alleles recovered from subjects in this study to nef sequences in SHIV-nef-infected macaques18 with PH-like pulmonary vascular remodeling.17
Statistical analyses
The characteristics of normotensive and hypertensive subjects were summarized with descriptive statistics and compared by geographic region. Amino acid residues at each position were tabulated per clone in each subject. Sensitivity and specificity for the identification of a HIV-PH phenotype based on number of Nef functional domains with polymorphisms were summarized with a receiver-operator characteristic (ROC) curve; overall discrimination was quantified with area under the curve (AUC). Statistical analyses were conducted using PyCogent, SAS 9.2 for Windows and GraphPad v.5.01. To study the similarities/differences between HIV-nef sequences from HIV-PH vs. controls, we reconstructed phylogenetic trees using Clustal W v1.83.21 Please refer to Supplementary Data for detailed statistical approaches.
Homology modeling of Nef
Nef protein structures were modeled by the web server PHYRE22 and visualized in Pymol23 (Delano Scientific LLC, San Carlos, CA). van der Waals overlaps were determined by Mol Probity.24
Functional characterization of HIV nef isolates associated with pulmonary hypertension
We subcloned HIV nef isolates into the HaloTag pFC14A vector (Promega) according to the manufacturer's instructions and tested Nef protein ability to down-regulate CD4 in HeLa-CD4+ cells (NIH-AIDS Reagents Program25). Expression of the nef-HaloTag fusion constructs was assessed by Western blots under denaturing/nonreducing and native conditions, using a polyclonal anti-HaloTag antibody. The expression of CD4 antigen receptor was determined by flow cytometry. Please see the Supplementary Data for details.
Results
Study subjects
HIV-infected European subjects with and without HIV-PH were not different regarding demographic characteristics (Table 1). While most of the European subjects with HIV-PH were receiving antiretroviral therapy (ART) (91%), compared to 43% of controls (p=0.0474), CD4+ T cell counts, HIV viral loads, or duration of HIV infection were not different between the groups (Table 1).
Table 1.
General Demographic, Virological, and Immunological Characteristics of HIV-Infected Individuals from the Initial and Validation Cohorts
| |
|
Initial cohort: France/Italy |
Validation cohort: San Francisco, California, USA |
||||
|---|---|---|---|---|---|---|---|
| Statistic | Without PH (plasma, n=7) | HIV-PH (PBMC, n=11) | p-valueTEST | Normotensive (PBMC, n=22) | HIV-PH (PBMC, n=11) | p-valueTEST | |
| Sex: male | n (%) | 4 (57%) | 9 (82%) | 0.3260FE | 18 (82%) | 8 (73%) | 0.661FE |
| Age (years) | mean (SD) | 41.4 (7.0) | 45.7 (8.7) | 0.2886TE | 52.5 (7.7) | 51.8 (8.5) | 0.8304TE |
| Vital status: deceased | n (%) | 0 (0%) | 5 (45%) | 0.1555LR | 0% | 0% | N/A |
| Duration of HIV (years) | Mean (SD) | 6.6 (2.0) | 8.0 (6.3) | 0.4945TU | 16 (5.7) | 14.1 (7.1) | 04509TE |
| HIV viral load (copies/ml) | Median (p75) | 31,200 (59,300) | 13,452 (38,657) | 0.2746W | <40 (1150) | <40 (75) | 1.000TE |
| CD4+ T cell counts (cells/mm3) | Mean (SD) | 496 (339) | 406 (329) | 0.5857TE | 512 (224) | 547 (396) | 0.7478TE |
| Receiving ART: yes | n (%) | 3 (43%) | 10 (91%) | 0.0474FE | 18 (82%) | 10 (91%) | 0.6431FE |
| mPAP (mm Hg) | Mean (SD) | Not determined | 46.8 (8.5) | N/A | 18.5 (3.4) | 39.3 (12) | <0.0001FE |
| Number of clones | Mean (SD) | 12.6 (4.1) | 18.8 (5.6) | 0.0223TE | 15 (3.0) | 12 (6.0) | 0.0649TE |
Banked HIV-infected samples as follows: PBMC DNA from subjects with confirmed diagnosis of HIV-PH from France and San Francisco, lung tissue from a subject from Italy, with echocardiograph-based indication of HIV-PH; controls consisted of plasma from French HIV-infected subjects with no evidence of PH and PBMC DNA for the validation cohort from San Francisco. The duration of HIV infection was calculated using the HIV diagnosis date and the date the specimen was collected. Controls and HIV-PH groups were compared using the appropriate tests in SAS. p values <0.05 were considered significant.
PBMC, peripheral blood mononuclear cells; SD, standard deviation; ART, antiretroviral therapy; p75, 75th percentile; N/A, not applicable; FE, Fisher's exact test; TE/TU, two-sample t-test assuming equal/unequal variance; LR, log rank test; W, Wilcoxon Mann–Whitney with continuity correction.
Hemodynamic data were not collected for the French controls because these individuals had no evidence of dyspnea and therefore echo Doppler was not indicated as per the French PH diagnostic algorithm.4 To date, control individuals have not developed symptoms (personal communication). Similar to the European cohort, there were no statistically significant differences in demographic or clinical parameters, including ART, CD4 counts, HIV viral loads, and duration of HIV infection between individuals with or without HIV-PH in the San Francisco group. Nevertheless, compared to the European cohort, the San Francisco validation group had a longer duration of HIV infection (∼15 and 7 years for San Francisco and European subjects, respectively (p<0.0001, t test), and a mean age of 44 and 52 years, respectively (p=0.0010, t test).
Nef sequences from HIV-PH individuals displayed polymorphisms in functional domains
Unifrac phylogenetic analyses26 of the full-length Nef protein showed that sequences from each individual clustered with a high degree of significance (p<10−40). However, there was no significant clustering by disease phenotype, suggesting that there is high variability of Nef among individuals and that the HIV-PH/normotensive phenotype is not due to a specific clade of Nef.
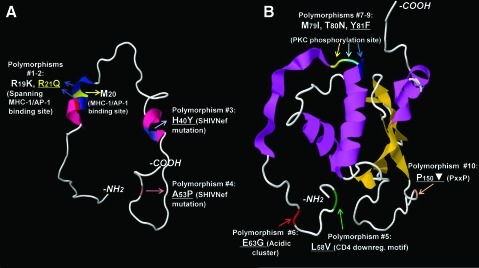
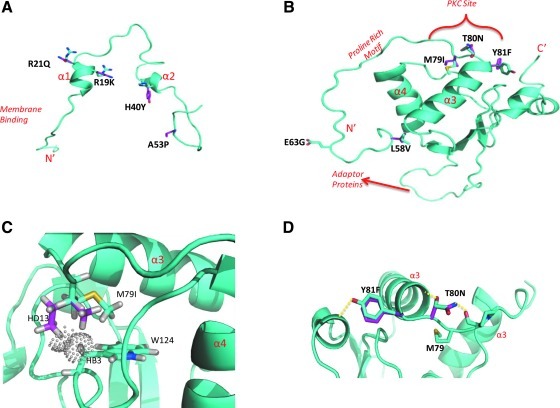
Interestingly, polymorphisms in nef mapped to known functional domains and these were overrepresented in European individuals with HIV-PH compared to controls, based on G statistic ranking data (Fig. 1, Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/aid); subsequent analyses focused on these functional domains. Specifically, we found 10 polymorphisms: the PxxP motif (proline-rich area essential for Nef interaction with SH3 domain-containing proteins,27 where x is any amino acid), the L58V CD4 down-regulation domain,28 the E63G acidic cluster mediating the sequestration of MHC-1 in the trans-Golgi network,29 and the M79I/T80N/Y81F phosphorylation site for protein kinase C.30 We also found changes near M20 whose functional role is to interact with the adaptor protein 1 (AP-1) and efficiently prevent MHC-1 trafficking to the membrane.31 Importantly, 7 of these 10 nef polymorphisms were validated in the San Francisco group with HIV-PH (Fig. 1, Supplementary Table S1).
FIG. 1.
HIV Nef polymorphisms overrepresented in subjects with HIV-associated PAH. Individual variations (using one-letter amino acid codes, numbered, colored, and indicated by arrows) are indicated in cartoons of Nef crystal structures. The Nef structure data are available as separate models.15 We used the models of (A) myristoylated Nef anchor domain, which includes Nef amino acids 2–57, and (B) Nef core to the C-terminus (amino acids 56–206). Alpha-helixes are colored in pink, beta-sheets are in orange. The N- and C-termini are indicated in each model. Nonconservative polymorphisms are indicated in yellow. Of the 10 polymorphisms identified in European subjects with HIV-associated PAH, seven were also found in the cohort from San Francisco (underlined). Refer to Supplementary Table S1 for frequency data. Symbol: ▾ indicates hydrophilic amino acid.
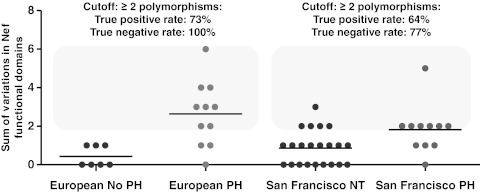
The number of variant functional domains was catalogued for each subject from each cohort (Europe and San Francisco) and presented as the sum of polymorphisms for each group (Fig. 2). Interestingly, the HIV-PH group in both cohorts was characterized by two or more polymorphisms in Nef domains: 5/22 normotensive controls from San Francisco showed >2 polymorphisms and 0/7 controls from Europe showed >2 polymorphisms.
FIG. 2.
Variations in functional domains in HIV-PH vs. HIV-infected subjects with no PH. The number of variations in HIV-1 Nef functional domains was determined for each subject and is shown as a sum of variations in Nef domains. Each symbol represents a subject. Note that the HIV-PH group is characterized by two or more variations in Nef functional domains (odds ratio 10.29, 95% confidence interval, 2.76 to 38.38; p=0.0004).
ROC curve analyses from the European cohort revealed that if a similar number of clones as obtained from the subjects in this study (i.e., 14±6) were obtained from HIV-infected individuals, using two variations in Nef as a cutoff, 73% (95% CI: 39%, 94%) of subjects with HIV-PH would be correctly identified, and 100% (95% CI: 59%,100%) without the disease would be correctly identified as negative. These data have an AUC of 0.8961 (95% CI: 0.75 to 1.05, p=0.005761, Supplementary Table S2). In the validation group, ROC analyses demonstrated that using two polymorphisms in Nef as a cutoff, 64% (95% CI: 31%, 89%) of subjects with HIV-PH would be correctly identified, and 77% (95% CI: 55%,92%) of normotensive subjects would be correctly identified as PH negative (AUC=0.7417, 95% CI:56%, 92%, p=0.02552). In addition, retrospective analyses showed that the survival of HIV-PH individuals in both cohorts was significantly lower (median survival of 18 years after the HIV diagnosis, p-value=0.0008) compared to normotensive controls (undefined because 100% are alive; Kaplan–Meier survival estimates).
Length of HIV infection or antiretroviral therapy is not associated with Nef polymorphisms
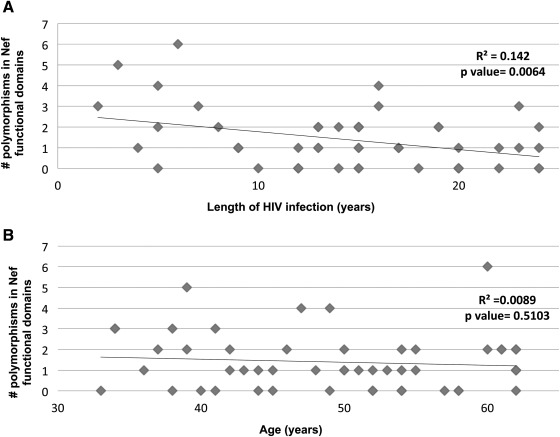
Using ≥2 polymorphisms in Nef functional domains as cutoff, we studied whether the length of HIV infection, age, or ART would have an impact on the selection of Nef variants. There is no correlation between the number of polymorphisms and length of HIV infection (R2=0.142) or age (R2=0.0089, Fig. 3). In addition, ART is not associated with the presence of >2 Nef variants in subjects with PH from Europe (p=1.000) or San Francisco (p=1.000). Although we identified variants that tended to cluster together: L58V-Y81F, PxxP-A53P, PxxP-H40, and A53P-H40Y in four subjects and Y81F-PxxP in five subjects, contingency analyses showed that there was no particular mutation associated with the presence of ART (Table 2).
FIG. 3.
Correlation between the number of polymorphisms in HIV Nef functional domains and length of HIV infection and age in the study group. The length of HIV infection and age (both in years) were plotted for each study subject (n=51). Linear regression analyses showed that there is no correlation between the number of polymorphisms in HIV-Nef functional domains and length of HIV infection (R2=0.142 (A) or age (R2=0.0089 (B).
Table 2.
Impact of Antiretroviral Therapy on Polymorphisms in HIV Nef Functional Domains
| Cohort | R19K | R21Q | H40Y | A53P | L58V | E63G | M79I | T80N | Y81F | Pxx_ |
|---|---|---|---|---|---|---|---|---|---|---|
| Europe | Not determined | p=1.000 | p=0.6078 | p=0.5221 | p=0.5221 | p=1.000 | p=0.2778 | p=0.4902 | p=0.5956 | p=0.2489 |
| San Francisco | p=0.5686 | p=0.3996 | p=0.4996 | p=0.0662 | p=1.000 | p=1.000 | p=0.1515 | p=0.3996 | p=0.1329 | p=0.1504 |
p values were calculated using contingency tables that included the number of individuals with the specific Nef polymorphisms in each cohort. Nef polymorphisms are shown using the one-letter amino acid code.
Lack of compartmentalization of Nef variants in PBMCs compared to plasma
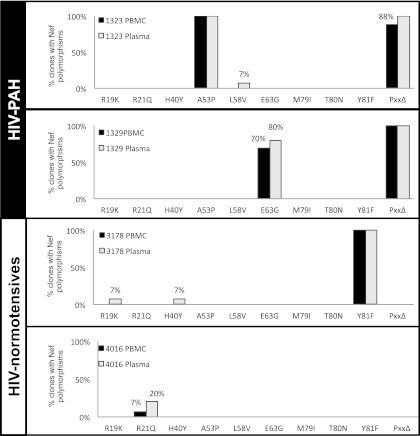
Different selective pressures on HIV nef have been reported when provirus from PBMCs is compared to virions from plasma.32 We genotyped nef from banked plasma samples and compared the sequences with PBMC proviral DNA from two HIV-PH and two HIV-normotensive subjects; these samples were contemporaneous. Figure 4 shows the frequencies of Nef polymorphisms in both compartments. Note that although there were slightly higher frequencies in plasma, these data suggest that there is no differential distribution of Nef variants in the two blood compartments.
FIG. 4.
HIV Nef sequence variations in blood compartments. HIV nef was genotyped from the peripheral blood mononuclear cells (PBMCs) and plasma from two HIV-infected individuals with PH and two normotensives of the San Francisco group. PBMCs and plasma samples analyzed were collected on the same day in all subjects except for Subject #4016, from whom the plasma sample was collected a month after the PBMCs. Sequences were aligned and translated; the frequency of polymorphisms in Nef functional domains was calculated for each case and plotted.
Individuals with HIV-PH and SHIV-nef-infected macaques with complex pulmonary lesions share substitutions in Nef residues
We compared Nef residues recovered from HIV+ with and without PH to Nef residues recovered from macaques infected with chimeric SHIVnef and with pulmonary vascular remodeling.17 Sequence analyses uncovered polymorphisms that were shared between HIV-PH individuals and the macaques: histidine to tyrosine at position 40 (H40Y) and alanine to proline at position 53 (A53P, Supplementary Table S1). The identification of common polymorphisms between Nef amino acid residues that arose spontaneously during passage in vivo in macaques and in the HIV-PH individuals supports a potential link between specific Nef sequences and the PH phenotype.
Comparison between consensus and HIV-PH Nef using three-dimensional homology models
We sought to analyze the potential impact of nef polymorphisms by comparing models of HIV-PH-Nef to the consensus Nef. None of the models suggested major global structural impact from the polymorphisms (Fig. 5); changes occurred in regions known to mediate intermolecular interactions.
FIG. 5.
Homology modeling of HIV Nef isolates from HIV-PH subjects. Models of Nef isolates from the HIV-PH subjects were generated by the server PHYRE. (A) Model of the N-terminus/anchor domain of the Nef consensus B Sequence (Los Alamos Databases) (cyan) based on model 1 of the 1QA5 NMR structure.33 Nef point mutants R19K, R21Q, H40Y, and A53P are indicated in magenta. (B) Model of the consensus B sequence (cyan) with Nef point polymorphisms L58V, E63G, M79I, T80N, and Y81F (magenta) superimposed. P150X was not modeled by PHYRE. (C) Model of Nef M79I (magenta) superimposed on the structure of Nef with the FYN SH3 domain (cyan/1EFN). In the wild-type structure, the methionine side chain fits within the hydrophobic pocket formed between helixes α3 and α4. The larger side chain of isoleucine is flipped 180° to the methionine because it does not fit within the hydrophobic pocket. Even removed from the pocket HD13 makes significant van der Waals (VdW) clashes (–0.414 Å determined by Mol Probity24) with HB3 of W124 (VdW radius indicated by spheres). The isoleucine is unfavorable for the observed positioning of the putative PKC site when Nef is bound to an SH3 domain-containing protein (Fyn). (D) Nef model of T80N and Y81F superimposed on the Consensus B 1EFN model34 (cyan) with possible T80 to G83 and Y81 to A190 hydrogen bonds shown (yellow dashed line). These hydrogen bonds potentially stabilize the positioning of the PKC site as well as the proline-rich helix for interaction with an SH3 domain. The T80N mutant not only is incapable of forming a hydrogen bonding to G83 within α3 but also could form a new hydrogen bond with the carbonyl of P78, which would increase the flexibility of the PKC site from α3.
Nef variants associated with PH down-regulate CD4
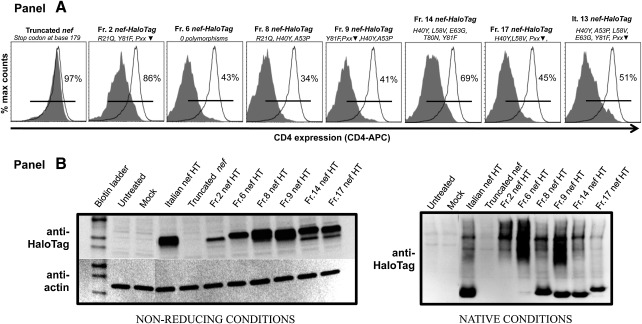
Down-regulation of CD4 receptor is one canonical function of Nef; we therefore explored whether the polymorphisms would interfere with Nef-dependent CD4 down-regulation. Nef-HaloTag fusion constructs were nucleofected into HeLa-CD4 cells and expression assessed by flow cytometry (see the Supplementary Methods for details). Baseline expression was 97–99%; the presence of Nef resulted in a 34–86% range of CD4 expression (Fig. 6A). Denaturing/non-reducing immunoblots of cell extracts showed that Nef from the various constructs were expressed at different levels. Given that Nef is a gregarious protein that interacts with numerous cellular proteins,33 we also analyzed the Nef-HaloTag constructs under native conditions. We observed the presence of higher molecular weight forms of Nef, suggesting that these cloned variants retained the ability to form putative intracellular oligomers/complexes (Fig. 6B).
FIG. 6.
Functional assay and immunoanalyses of Nef-HaloTag fusion constructs derived from HIV-PH individuals. Hela-CD4 cells were nucleofected with the indicated fusion constructs. Polymorphism information of each molecular construct (as per recovered from the subjects) is shown using one-letter amino acid codes; ▾ indicates hydrophilic amino acid. (A) The HaloTag portion of the construct was labeled with Oregon Green HaloTag ligand and CD4 receptor was labeled with an anti-CD4 antibody conjugated to allophycocyanin. Samples were acquired by flow cytometry; CD4 expression was assessed in mock-nucleofected and in cells nucleofected with truncated-Nef by gating in the whole cell population, while in the Nef-HaloTag-nucleofected cells, it was measured by gating in Nef-HaloTag positive cells. Baseline CD4 expression in percentage was determined using the mock-nucleofected cells. The histograms show the CD4 expression in cells nucleofected with each individual construct (tinted) compared to mock-nucleofected cells (open); percentages are shown. (B) Lysates from nucleofected cells were immunoblotted with anti-HaloTag polyclonal antibody and subsequently stripped and probed with antiactin monoclonal antibodies under denaturing/non-reducing conditions (left bottom panel). The panel at the lower right shows an immunoblot of the lysates with the anti-HaloTag polyclonal antibody under fully native conditions. Note the presence of multiple oligomeric forms in some Nef-HaloTag constructs.
Discussion
HIV-PH is a manifestation of HIV infection regardless of the unquestioned success of antiretroviral treatments. There is an increased mortality in HIV-PH subjects compared to normotensive HIV+ individuals.8 Unfortunately, many of the noninfectious complications of HIV infection, including PH, present with nonspecific symptoms and may be misdiagnosed.
Our group previously reported lung vascular pathologies characteristic of PH in macaques infected with chimeric SHIV-nef virions but not in monkeys infected with strains containing the native SIV nef alleles. Nef was also found in the lungs of HIV-infected patients with pulmonary hypertension.17 These findings suggested that HIV Nef protein, perhaps in conjunction with host genetic factors or persistent immune dysregulation, contributes to the development of pulmonary vascular changes leading to endothelial cell angioproliferative disease. The nef recovered from the SHIV-nef-infected monkeys (named nefSF33A) displayed four consistent amino acid changes acquired during passage in vivo.18 These substitutions (including H40Y and A53P) became the predominant residues in close to 100% of the molecular clones recovered 53 weeks postinfection, suggesting positive selection pressures.
We translated these results to humans and found polymorphisms in five of the known Nef functional domains; specifically, the MHC-1 binding site, PKC phosphorylation site, acidic cluster, CD4 down-regulation motif, and the proline-rich area (PxxP). Our studies were focused solely on HIV-Nef because of our previous findings in SHIV-nef-infected macaques, although we do not rule out additional viral/host factors in the pathogenesis of HIV-PH. For example, George et al.34 described pulmonary arteriopathy in monkeys infected with SHIVenv, suggesting that other HIV genes might also play a role in the development of PH-like pulmonary vascular lesions. Perhaps HIV-nef is a snapshot of the selective pressures driving HIV gene polymorphisms.
In light of the important pathogenic functions ascribed to Nef,35–37 the wild-type Nef protein would be expected to be more likely associated with a disease phenotype rather than a polymorphic Nef. Nonetheless, if the native Nef protein were the one mechanistically related to PH, then every HIV-infected subject would suffer PH, which is unlikely.
Functional analyses, based on down-regulation of the CD4 receptor (one canonical function of Nef), were intended to assess whether the “mutant” Nef protein retained function even if fused to a C-terminus tag. Our results demonstrate that Nef variant alleles associated with HIV-PH retain this ability, suggesting these are not functional dead-ends. Of note, some of the Nef polymorphisms identified in our study are localized in the CD4 down-regulation domain. These unique polymorphisms unlikely impair function because motifs located elsewhere on the polypeptide are also critical for Nef-mediated CD4 down-regulation.38
Nef is an adaptor molecule that interacts with cellular proteins; its structure plays a critical role in these interactions. None of the polymorphisms was predicted to have major structural changes on the core, but rather the impact would be on residues located on the surface; these would be predicted to disrupt solvent and intermolecular interactions. Our protein modeling studies suggest that in the context of Nef bound to an SH3 domain-containing protein, e.g., Fyn (Fig. 5), changes within the PKC phosphorylation site (M79, T80, and Y81) would alter hydrogen bonds that normally stabilize the phosphorylation site and the neighboring proline-rich motif. These may be sufficient to expose new motifs and favor new, yet to be studied interactions with cellular proteins (e.g., Src kinases, adaptor proteins).The substitution of T80N would theoretically not form hydrogen bonds with the consensus G83, but would instead form hydrogen bonds with P78, which would increase the flexibility of the PKC site from alpha-helix 3. However, the impact of these polymorphisms remains to be studied in crystal structures and in vitro.
Interestingly, we identified a series of Nef polymorphisms that were found in combination (L58V-Y81F, PxxP-A53P, PxxP-H40Y, and A53P-H40Y, and Y81F-PxxP). Although none of the polymorphisms was pathognomonic of PH, these warrant further mechanistic studies, given the significance of tyrosines and prolines in signaling pathways. For example, mutations in the PxxP motif favor cytotoxic T cell recognition by decreasing the activation of Hck in macrophages39; overexpression of PKC with Y to F mutations results in increased proliferative and tumorigenic phenotypes in fibroblasts.40 In addition, the substitution of H to Y may provide new substrates for Src-kinases, which may become newly accessible in polypeptides bent by prolines (A to P, for example). Our in vitro studies focused on full length Nef, which is structurally different than truncated versions of Nef41; the relevance of truncated/missense Nef sequences in HIV-PH remains currently unknown, which offers another fascinating aspect to explore in future studies.
The finding of significantly more polymorphisms in Nef functional domains in the HIV-PH groups may initiate vibrant discussions regarding selective pressures on Nef. Genetic divergence in Nef has been ascribed to the CD8+ T cell-mediated viral control.32,42,43 Accumulation of mutations in the HIV genome is conceivable, especially in chronically HIV-infected subjects exposed to ART, which was a feature of our validation cohort from San Francisco. We found no evidence that the Nef polymorphisms found in subjects with HIV-PH were affected by the presence of ART, length of HIV infection, or age, at least in the cohorts we analyzed.
Our studies are limited by the differences among the subjects analyzed retrospectively from both cohorts, including the use of ART (European controls were mostly naïve), HIV viral load (suppressed in the San Francisco) duration of HIV infection (more chronic in the San Francisco), age (younger in the European), and identification of normotensives (no cath in the European controls). In addition, our sample size may be too small to reach definitive conclusions; confirmation in larger, powered cohorts may confirm our results or detect polymorphisms in Nef due to ART, age, chronicity of HIV infection, and/or compartmentalization in plasma vs. PBMCs. Also, we acknowledge that at least 23% of the normotensive subjects recruited at San Francisco showed >2 polymorphisms in Nef. Whether these study subjects, although asymptomatic, are already at risk of HIV-PH will be elucidated prospectively because these subjects are currently enrolled in the longitudinal Lung-HIV studies focused on HIV-mediated lung complications, including PH.
Our findings offer new insights into the association between polymorphisms in Nef functional domains and an HIV-PH phenotype in two separate cohorts. We believe our data suggest the possibility that genetic testing is feasible and may be potentially useful in the identification of these individuals. Future longitudinal studies will help explore whether these polymorphisms are also associated with development and progression of HIV-PAH.
Supplementary Information
Study subjects
HIV-infected individuals were diagnosed with HIV-PH according to the standard diagnostic algorithm, which includes echo-Doppler (if dyspnea is present and unexplained) and right heart catheterization (RHC) if echocardiography suggested PH.1 Mean pulmonary artery pressures (mPAP) >25 mm Hg and pulmonary capillary wedge pressure ≤15 mm Hg via RHC defined the PH diagnosis. We also analyzed DNA isolated from paraffin-embedded, formalin-fixed fragments of lung tissues from a subject (It.13) enrolled in the Latium Registry of HIV-PH in Rome2; the subject had a systolic pulmonary artery pressure of 63 mm Hg by transthoracic echocardiography. Plasma samples from seven French HIV-infected individuals with no evidence of pulmonary disease (personal communication) were used as controls; these subjects did not have dyspnea or any other indication to undergo echo Doppler nor right heart catheterization as per the French diagnostic algorithm; we refer to these as “French with no PAH.”
Our validation group consisted of banked PBMC samples from HIV-PH subjects (n=11) and RHC-confirmed normotensives (n=22, with mPAP <25 mm Hg, referred to as “normotensives”) enrolled as pulmonary healthy controls for the Lung-HIV studies at the University of California, San Francisco (UCSF) hospital. The inclusion criteria included documented HIV infection longer than 6 months and the ability to provide anecdotal or medical evidence of HIV medication history. Subjects with PH-associated comorbidities such as left-heart disease, severe respiratory diseases, or chronic thromboembolic pulmonary hypertension, connective tissue disease, congenital heart disease, or portal hypertension were excluded. Information about coinfection with human herpesvirus-type 8 (HHV-8) was not available for all individuals; however, there is a similar prevalence of HHV8 seropositivity in HIV-PH subjects and HIV controls.3 Therefore, coinfection with HHV8 is less likely to be a confounder factor. This retrospective cross-sectional study was approved by the NIH/NHLBI Lung HIV Data Safety and Monitoring Board, CPP IIe-de-France VII, Le Kremlin-Bicetre, Committee for Human Research at UCSF, Colorado Multiple Institutional Review Board, and the Ethics Committee-Italian Institute of Infectious Diseases “Spallanzani.”
Nucleic acid extractions and amplification of HIV-nef via PCR
PBMC DNA from the French HIV-PH samples was used as template for nested PCR, using nef-specific primers4 and Maxime PCR Pre-Mix (Boca Scientific). DNA was extracted from paraffin-embedded lung tissues from It.13 after deparaffinization, extraction of DNA with phenol-chloroform, and ethanol precipitation of the extracted DNA. HIV viral RNA was extracted from plasma samples from HIV+ controls using the Viral RNA Mini Kit (Qiagen). HIV-nef alleles were amplified from viral RNA using the One-step RT-PCR kit (Qiagen), Nef9589R and Nef9595R as outer primers,5 and HIV-nef-specific inner primers.4 PCR was performed in triplicate for each subject (HIV-PH and controls), and the amplicons were pooled and ligated into a pCR-TOPO vector (Invitrogen) to minimize PCR resampling bias. By using this approach, each molecular clone would most likely contain sequence information from the quasispecies harbored by the host and less likely due to artifacts generated in vitro. Multiple molecular clones were purified and sequenced using ABI 3730XL sequencer (Applied Biosystems). All the molecular clones were generated in the same laboratory from samples obtained from the collaborating centers. Therefore, the differential distribution of polymorphisms due to center-specific technical biases is unlikely.
HIV-nef sequence analyses
A total of 763 HIV-nef molecular clones from archived material of 51 individuals infected with subtype B HIV strains were analyzed. The full-length HIV-nef sequences were aligned and translated using Geneoius,6 and realigned after visual inspection. Although the relevance of truncated/missense Nef sequences in HIV-PH remains currently unknown, we excluded molecular clones with premature stop codons, large deletions, and mutated myristoylation sequences from the analyses because their inclusion would hinder in vitro studies with the full-length protein.
We aligned the Nef sequences from the HIV-infected individuals with and without PH and compared these to reference sequences from the Los Alamos HIV databases at http://www.hiv.lanl.gov to determine the HIV subtype. The frequency of Nef amino acid residues at each position was determined for each subject by counting in an MS Excel spreadsheet and then compared by group (HIV-PH vs. control). The HIV-1 subtype B Nef sequences compiled by O'Neill et al.7 were used to determine the most frequent amino acid at each position of the full-length Nef polypeptide.
Phylogenetic analyses
Reference HIV-nef sequences (2007 dataset) were obtained from the Los Alamos HIV sequence database at http://www.hiv.lanl.gov. The reference sequences were used to build a hidden Markov model (HMM) using HMMER v2.3.2's hmmbuild with default parameters8 and included in the alignment of the Nef sequences recovered from the HIV-PH and HIV+ controls. From this alignment, duplicate sequences were removed, as were high-entropy positions (top 10% of positions by Shannon entropy scores3,9 and positions with >10% gaps). Bootstrapped trees were created using Clustal W v1.839 via the PyCogent application controller10 using 1000 bootstrap replicates. The trees were rooted using Nef sequences recovered from SHIV-nef-infected macaques (unpublished data). We used UniFrac11,12 to test whether specific phenotypes (e.g., HIV-PH vs. controls) or individual subjects were nonrandomly distributed over the tree and extended this method by randomizing the states of the sets of sequences corresponding to a given individual. We tested for associations between phylogenetic structure and specific polymorphisms by adapting the method developed by Lozupone et al.13 and by using the analysis-of-traits (AOT) package.
Statistical analyses
The characteristics of the sample were summarized with appropriate descriptive statistics. Sensitivity and specificity for an HIV-PH diagnosis based on the number of functional domains with variations were summarized with a receiver-operator characteristic (ROC) curve; the ability to use the sum of functional domains with variations to discriminate between individuals with and without HIV-PH was quantified in terms of the area under the curve (AUC).
The presence/absence of amino acid residues at each position was averaged for each subject, creating a proportion of variants for each position. Rank statistics were used to evaluate the distribution of proportions across subjects. For these purposes, the G test was performed on a 2×2 contingency in which the columns corresponded to study group (HIV-PH or controls), rows corresponded to whether a particular amino acid at a given position was different from the HIV sequence database, and the cells corresponded to the count of clones from the HIV-PH/control group with and without a deviation from the historical databases.
Statistical analyses were conducted using PyCogent, SAS 9.2 for Windows and Graph Pad Prism v.5.01. Differences were tested with statistical significance at the alpha=0.05 level.
Nef protein homology modeling
Nef sequences from primary isolates were modeled by the web server PHYRE.14 Briefly, we used the models of the myristoylated Nef anchor domain, which includes Nef amino acids 2–57 (PBD ID: 1QA5, from Geyer et al.,15 and the Nef core-C-terminus, which includes amino acids 56–206. Crystal structure data were downloaded from the RCSB Protein Data Bank16 and models were depicted using Geneious.6
Functional assay of the HIV-nef isolates associated with pulmonary hypertension
Specific amino acid residues essential for Nef dimerization and conservation of function in vivo have been reported.17 Hence, we used Geneious6 to examine the HIV-nef sequences from the French subjects with HIV-PH (with polymorphisms in Nef functional domains) for the presence of amino acid residues required for Nef oligomerization/function. All of the sequences identified in silico contained the required residues and were therefore predicted to be functional. Since there could be functional constraints in Nef when fused to molecules at the C-terminus, we subcloned the well-characterized NA7-nef from a GFP construct (kindly provided by Dr. J.V. Garcia18) to the HaloTag vector (Promega) and tested its capacity to down-regulate CD4 expression (not shown). We chose the HaloTag vector because of its flexibility for tagging with various ligands for flow cytometry and pull-down assays.19
After validating Nef function when fused to HaloTag, we then cloned the HIV-nef isolates from the French HIV-PH into the HaloTag pFC14A (Promega) following the manufacturer's instructions. We also cloned a truncated form of HIV-nef (stop codon at position 179), isolated from one of the French subjects, and used it as an internal control. We then tested Nef protein function by nucleofection of the molecular clones into HeLa-CD4+ cells (clone HT4-6C, NIH-AIDS Research and Reference Reagents Program20), engineered to express CD4 constitutively. Briefly, two million cells were nucleofected with 2 μg of plasmid DNA using the Nucleofector II and the Cell Line Nucleofection Kit V (Amaxa) and incubated for 16–18 h at 37°C, 5% CO2. The expression of the nef-HaloTag fusion constructs was assessed by Western blot under nonreducing and native conditions, using a Mini-PROTEAN Tetra Cell (Bio-Rad). HaloTag constructs were immunodetected using the rabbit polyclonal α-HaloTag antibody (Promega) at a 1:1000 dilution. The nef-HaloTag fusion constructs were also detected using an HIV-1 Nef monoclonal antibody (EH1, NIH AIDS Research and Reference Reagent Program, data not shown). Membranes under nondenaturing conditions were stripped using Western Re-Probe (G-Biosciences) and reprobed for beta-actin as a loading control using anti-β-Actin clone AC-15 (Sigma) at a 1:100,000 dilution. CD4 expression was determined by flow cytometry using an allophycocyanin-conjugated anti-CD4 antibody (Caltag) followed by acquisition in a FACSCalibur (BD Biosciences) with the CellQuest software. The HaloTag component of the Nef-HaloTag fusion protein was labeled with the Oregon Green HaloTag Ligand (Promega). Flow cytometry data were analyzed using FlowJo (Treestar, Inc.)
Sequence Data
The HIV-nef sequences recovered from the study subjects have been deposited in GenBank under the following accession numbers: JQ255608 to JQ256368 (to be submitted).
Supplementary Material
Acknowledgments
HeLa CD4 cells and anti-Nef antibodies were obtained through the NIH AIDS Research and Reference Reagents Program. This work was supported by NIH/NHLBI research Grants R01 HL90480, R01 HL90480-S1 (ARRA Supplement), R01 HL083491 (to S.C.F.), R01 HL901526 (to P.Y.H), and T32-HL07171 (Cardiovascular Pulmonary Postdoctoral Training Program at UC-Denver), and in part by grants from the Colorado Center for AIDS Research Grant P30-AI054907 (R.K.), the Ministẻre de l'Enseignement Supérieur et de la Recherché, and the Université Paris-Sud 11.
The authors thank Drs. Julio V. Garcia and John Foster for kindly providing the Nef-GFP plasmid, Drs. Rubin Tuder and Marc Moss for critical evaluation of this manuscript, and Tamara Miller, Jessica Gilman, Melanie Hakar, Natalied Rivera, and Daphne Velez (UC-Denver) for technical assistance. Drs. Azzedine Yaïci, David Montani (Service de Pneumologie et Reanimation, Hôpital Antoine Béclère, Universite Paris-Sud, Clamart, France), and Enrico M. Trecarichi (Institute of Infectious Diseases, Catholic University of the Sacred Heart, Rome, Italy) are also recognized.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tuder RM. Groves B. Badesch DB. Voelkel NF. Exuberant endothelial cell growth and elements of inflammation are present in plexiform lesions of pulmonary hypertension. Am J Pathol. 1994;144:275–285. [PMC free article] [PubMed] [Google Scholar]
- 2.Speich R. Jenni R. Opravil M. Pfab M. Russi EW. Primary pulmonary hypertension in HIV infection. Chest. 1991;100(5):1268–1271. doi: 10.1378/chest.100.5.1268. [DOI] [PubMed] [Google Scholar]
- 3.Petrosillo N. Chinello P. Cicalini S. Pulmonary hypertension in individuals with HIV infection. AIDS. 2006;20(16):2128–2129. doi: 10.1097/01.aids.0000247569.03504.8b. [DOI] [PubMed] [Google Scholar]
- 4.Sitbon O. Lascoux-Combe C. Delfraissy JF, et al. Prevalence of HIV-related pulmonary arterial hypertension in the current antiretroviral therapy era. Am J Respir Criti Care Med. 2008;177:108–113. doi: 10.1164/rccm.200704-541OC. [DOI] [PubMed] [Google Scholar]
- 5.Ntsekhe M. Hakim J. Impact of human immunodeficiency virus infection on cardiovascular disease in Africa. Circulation. 2005;112(23):3602–3607. doi: 10.1161/CIRCULATIONAHA.105.549220. [DOI] [PubMed] [Google Scholar]
- 6.Thenappan T. Shah SJ. Rich S. Gomberg-Maitland M. A USA-based registry for pulmonary arterial hypertension: 1982-2006. Eur Respir J. 2007;30(6):1103–1110. doi: 10.1183/09031936.00042107. [DOI] [PubMed] [Google Scholar]
- 7.Hsue PY. Deeks SG. Farah HH, et al. Role of HIV and human herpesvirus-8 infection in pulmonary arterial hypertension. AIDS. 2008;22(7):825–833. doi: 10.1097/QAD.0b013e3282f7cd42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes H. Humbert M. Sitbon O, et al. Prognostic factors for survival in human immunodeficiency virus-associated pulmonary arterial hypertension. Am J Respir Crit Care Med. 2003;167(10):1433–1439. doi: 10.1164/rccm.200204-330OC. [DOI] [PubMed] [Google Scholar]
- 9.Clarke JR. Krishnan V. Bennett J. Mitchell D. Jeffries DJ. Detection of HIV-1 in human lung macrophages using the polymerase chain reaction. AIDS. 1990;4(11):1133–1136. doi: 10.1097/00002030-199011000-00012. [DOI] [PubMed] [Google Scholar]
- 10.Twigg HL., III Weiden M. Valentine F, et al. Effect of highly active antiretroviral therapy on viral burden in the lungs of HIV-infected subjects. J Infect Dis. 2008;197(1):109–116. doi: 10.1086/523766. [DOI] [PubMed] [Google Scholar]
- 11.Barber SA. Gama L. Li M, et al. Longitudinal analysis of simian immunodeficiency virus (SIV) replication in the lungs: Compartmentalized regulation of SIV. J Infect Dis. 2006;194(7):931–938. doi: 10.1086/507429. [DOI] [PubMed] [Google Scholar]
- 12.Piguet V. Schwartz O. Le GS. Trono D. The downregulation of CD4 and MHC-I by primate lentiviruses: A paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168:51–63. doi: 10.1111/j.1600-065x.1999.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 13.Swann SA. Williams M. Story CM. Bobbitt KR. Fleis R. Collins KL. HIV-1 Nef blocks transport of MHC class I molecules to the cell surface via a PI 3-kinase-dependent pathway. Virology. 2001;282(2):267–277. doi: 10.1006/viro.2000.0816. [DOI] [PubMed] [Google Scholar]
- 14.Deacon NJ. Tsykin A. Solomon A, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270(5238):988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 15.Kestler HW., III Ringler DJ. Mori K, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 16.Kirchhoff F. Easterbrook PJ. Douglas N, et al. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J Virol. 1999;73(7):5497–5508. doi: 10.1128/jvi.73.7.5497-5508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marecki JC. Cool CD. Parr JE, et al. HIV-1 Nef is associated with complex pulmonary vascular lesions in SHIV-nef-infected macaques. Am J Respir Crit Care Med. 2006;174(4):437–445. doi: 10.1164/rccm.200601-005OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandell CP. Reyes RA. Cho K, et al. SIV/HIV Nef recombinant virus (SHIVnef) produces simian AIDS in rhesus macaques. Virology. 1999;265(2):235–251. doi: 10.1006/viro.1999.0051. [DOI] [PubMed] [Google Scholar]
- 19.Humbert M. Sitbon O. Chaouat A, et al. Pulmonary arterial hypertension in France: Results from a national registry. Am J Respir Crit Care Med. 2006;173(9):1023–1030. doi: 10.1164/rccm.200510-1668OC. [DOI] [PubMed] [Google Scholar]
- 20.Drummond AJ. Ashton B. Cheung M, et al. Geneious v5.0. http://www.geneious.com http://www.geneious.com
- 21.Thompson JD. Higgins DG. Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley LA. Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 23.Delano WL. The PyMol molecular graphics system: Delano scientific. 2002.
- 24.Lovell SC. Davis IW. Arendall WB, III, et al. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 25.Chesebro B. Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62(10):3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone CA. Hamady M. Cantarel BL, et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci USA. 2008;105(39):15076–15081. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saksela K. Cheng G. Baltimore D. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995;14(3):484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grzesiek S. Stahl SJ. Wingfield PT. Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35(32):10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 29.Piguet V. Wan L. Borel C, et al. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol. 2000;2(3):163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shugars DC. Smith MS. Glueck DH. Nantermet PV. Seillier-Moiseiwitsch F. Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol. 1993;67(8):4639–4650. doi: 10.1128/jvi.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roeth JF. Williams M. Kasper MR. Filzen TM. Collins KL. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J Cell Biol. 2004;167(5):903–913. doi: 10.1083/jcb.200407031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salgado M. Brennan TP. O'Connell KA, et al. Evolution of the HIV-1 nef gene in HLA-B*57 positive elite suppressors. Retrovirology. 2010;7:94. doi: 10.1186/1742-4690-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu W. Sanders-Beer BE. Katz KS. Maglott DR. Pruitt KD. Ptak RG. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37(Database issue):D417–D422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George MP. Brower A. Kling H, et al. Pulmonary vascular lesions are common in SIV- and SHIV-env-infected macaques. AIDS Res Human Retroviruses. 2011;27(2):103–111. doi: 10.1089/aid.2009.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster J. Garcia JV. HIV-1 Nef: At the crossroads. Retrovirology. 2008;5:84. doi: 10.1186/1742-4690-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laguette N. Bregnard C. Benichou S. Basmaciogullari S. Human immunodeficiency virus (HIV) type-1, HIV-2 and simian immunodeficiency virus Nef proteins. Molec Asp Med. 2010;31(5):418–433. doi: 10.1016/j.mam.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Arhel NJ. Kirchhoff F. Implications of Nef: Host cell interactions in viral persistence and progression to AIDS. Curr Topics Microbiol Immunol. 2009;339:147–175. doi: 10.1007/978-3-642-02175-6_8. [DOI] [PubMed] [Google Scholar]
- 38.Geyer M. Fackler OT. Peterlin BM. Structure–function relationships in HIV-1 Nef. EMBO Rep. 2001;2(7):580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mwimanzi P. Hasan Z. Hassan R. Suzu S. Takiguchi M. Ueno T. Effects of naturally-arising HIV Nef mutations on cytotoxic T lymphocyte recognition and Nef's functionality in primary macrophages. Retrovirology. 2011;8:50. doi: 10.1186/1742-4690-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acs P. Beheshti M. Szallasi Z. Li L. Yuspa SH. Blumberg PM. Effect of a tyrosine 155 to phenylalanine mutation of protein kinase cdelta on the proliferative and tumorigenic properties of NIH 3T3 fibroblasts. Carcinogenesis. 2000;21(5):887–891. doi: 10.1093/carcin/21.5.887. [DOI] [PubMed] [Google Scholar]
- 41.Jung J. Byeon IJ. Ahn J. Gronenborn AM. Structure, dynamics, and Hck interaction of full-length HIV-1 Nef. Proteins. 2011;79(5):1609–1622. doi: 10.1002/prot.22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong T. Zhang Y. Xu KY, et al. Extensive HLA-driven viral diversity following a narrow-source HIV-1 outbreak in rural China. Blood. 2011;118(1):98–106. doi: 10.1182/blood-2010-06-291963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Culmann B. Gomard E. Kieny MP, et al. An antigenic peptide of the HIV-1 NEF protein recognized by cytotoxic T lymphocytes of seropositive individuals in association with different HLA-B molecules. Eur J Immunol. 1989;19(12):2383–2386. doi: 10.1002/eji.1830191231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.