Abstract
Reliable methods for measuring human immunodeficiency virus (HIV) incidence are a high priority for HIV prevention. They are particularly important to assess the population-level effectiveness of new prevention strategies, to evaluate the community-wide impact of ongoing prevention programs, and to assess whether a proposed prevention trial can be performed in a timely and cost-efficient manner in a particular population and setting. New incidence assays and algorithms that are accurate, rapid, cost-efficient, and can be performed on easily-obtained specimens are urgently needed. On May 4, 2011, the Division of AIDS (DAIDS), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), sponsored a 1-day workshop to examine strategies for developing new assays to distinguish recent from chronic HIV infections. Participants included leading investigators, clinicians, public health experts, industry, regulatory specialists, and other stakeholders. Immune-based parameters, markers of viral sequence diversity, and other biomarkers such as telomere length were evaluated. Emerging nanotechnology and chip-based diagnostics, including algorithms for performing diverse assays on a single platform, were also reviewed. This report summarizes the presentations, panel discussions, and the consensus reached for pursuing the development of a new generation of HIV incidence assays.
Introduction
Accurate determination of human immunodeficiency virus (HIV) incidence is critical for monitoring the HIV epidemic, evaluating ongoing prevention programs, and designing and implementing prevention trials. Unfortunately, current methods for assessing HIV incidence have proven to be inadequate. Techniques for estimating incidence from HIV prevalence, longitudinal cohort studies, and “first-generation” incidence assays all have significant limitations.1–7 Multiple expert consultations including the World Health Organization (WHO) Technical Working Group on HIV Incidence Assays have concluded that rapid, reliable, and cost-efficient incidence assays or algorithms are urgently needed.3–8
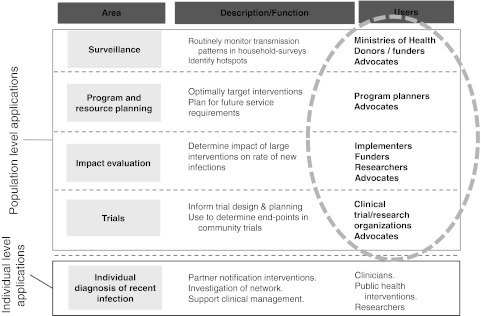
To the extent that we must depend on relatively crude incidence measures, our ability to target interventions, assess their impact, and devise more effective prevention strategies is weakened. When an incidence estimate is based on data collected over a prolonged period of time, as with cohort studies or repeated age-structured prevalence estimates, the results may already be outdated when they become available. Improved assays that can be applied in cross-sectional studies would undoubtedly be welcomed by program planners, ministries of health, international funding agencies, advocates, and researchers (Fig. 1).
FIG. 1.
Who would routinely use a reliable HIV incidence assay?
To facilitate a dialogue regarding the design, implementation, and optimization of new assays to detect recent HIV infections, the Division of AIDS (DAIDS), National Institutes of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), sponsored the “Novel Biomarkers for HIV Incidence Assay Development” workshop on May 4, 2011, in Bethesda, Maryland. Participants included leading investigators, regulatory specialists, clinicians, public health experts, industry, and other stakeholders. The meeting elicited valuable discussions in a number of key areas, including criteria to be met by “next- generation” incidence assays, host and viral biomarkers that might be exploited to develop a novel incidence assay, and the pathway from novel biomarker to a marketable assay.
Workshop Summary
The urgent need for new incidence assays and/or algorithms
Alex Welte [Director, South African Centre for Epidemiological Modeling and Analysis (SACEMA), Stellenbosch University, South Africa] outlined the urgent need for new incidence assays and/or algorithms, the criteria that they should fulfill, and the emerging theoretical framework in which these requirements and estimates can be made precise.9–12 Current incidence estimates based on cohort studies and mathematical modeling are not sufficient. Improved incidence assays are required to identify population groups that are at high risk of HIV infection (in as close to real time as possible) and to measure the impact of interventions that are tailored to reduce HIV incidence in the same population and setting. Currently, large investments are being made in HIV prevention programs by the U.S. President's Emergency Plan for AIDS Relief (PEPFAR), the Global Fund to Fight AIDS, Tuberculosis and Malaria, and national HIV program planners––even though there is limited ability to evaluate the impact of these programs.
Ideally, the assay would be able to distinguish recent and long-term HIV infections based on specimens collected during a single cross-sectional survey, circumventing the need for expensive longitudinal studies.1,9 This information could be used to identify transmission hotspots, target interventions, and allocate resources.
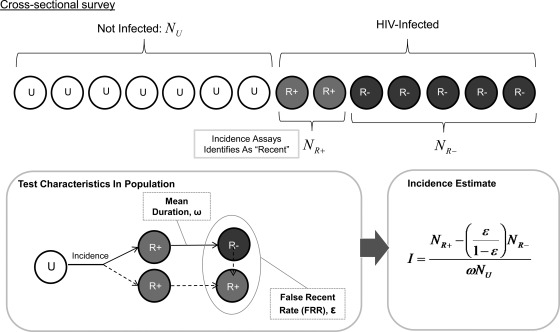
Welte emphasized that applications for estimating incidence, in contrast to clinical diagnostic assays, do not need to have a high predictive value at the individual level; however, they must meet other less-familiar but well-defined performance standards. The two crucial characteristics of an incidence assay are its false-recent rate (FRR) and mean duration of recent infection (MDRI) or “window period” (Fig. 2).
FIG. 2.
Key parameters of an HIV incidence assay.
An ideal HIV incidence assay should (1) provide a real measurement based on specimens collected in a cross-sectional manner, not an approximation based on a fitted model; (2) provide up-to-date information about the current rate of HIV infection; (3) have minimal biases introduced by observation; and (4) cost less to implement than large cohort studies. The assay should yield tightly reproducible incidence estimates when applied to realistically attainable sample sizes. The FRR––the probability that a chronically infected individual (e.g., one infected for ≥12 months) will be falsely categorized as recent––must be low (definitely <5%, but preferably <2%), and the MDRI must be sufficiently long (at least 4 months but preferably 6–12 months).
The challenges to HIV incidence assay development should not be underestimated. In light of this reality, developers will need to address the following:
Technical challenges: Small variations in test performance may result in much larger errors in the incidence estimate if certain context-dependent parameters are not taken into account [e.g., different HIV subtypes, varying antiretroviral therapy (ART) coverage rates, and other population factors].
Practical challenges: Larger sample sizes will be needed to estimate HIV incidence than are required for prevalence. For example, to document that an intervention/prevention program has reduced HIV incidence by 50%, two cross-sectional surveys on the order of 10,000 persons would be needed to obtain reasonable power to detect this change under realistic assumptions about test performance (FRR ≤5%), and other factors (e.g., 10% prevalence, 1% incidence).13–15 In addition, if sampling among the general population does not adequately capture hard-to-reach high-risk populations, it may underestimate HIV incidence.
Current lack of performance standards: No normative agency has established performance standards for HIV incidence assays. Since the unit for evaluation is the entire population, guidance that pertains to diagnostic (individual level) assays––with its emphasis on the tradeoff sensitivity and specificity––does not apply. No “gold-standard” method has been agreed upon, but it is clear that the appropriate metric of test performance is the variance of the incidence estimate; furthermore, this involves a tradeoff between lengthening the mean duration of recent infection (MDRI) and keeping the FRR acceptably low. A WHO working group has drafted a guidance document to estimate HIV incidence at a population level.13
Lack of well-characterized specimens to evaluate new assays: A centralized bank of well-characterized pedigreed specimens for feasibility/evaluation is currently being developed through a project led by the United Kingdom (UK) Health Protection Agency Centre for Infections.
Need for coherent user guidance: Until recently, the Centers for Disease Control and Prevention (CDC), the Joint United Nations Programme on HIV/AIDS (UNAIDS), and the package inserts accompanying BED assay materials all seemed to differ and their application in the field was often inconsistent.12 A recent WHO guidance document has addressed these issues for users of incidence assays.13
Lack of strong market incentive: It is unclear whether the global demand will reach a few hundred thousand or several million tests per year. This uncertainty tends to limit investment, which is required to achieve higher performance levels.2,8
Biomarkers for novel incidence assays
The focus of this workshop was biomarkers that may serve as the basis for novel incidence assays that can meet key requirements. Potential host biomarkers were discussed first, followed by viral biomarkers.
Potential host biomarkers
Innate immunity: Plasma proteins and other markers
Persephone Borrow (Weatherall Institute of Molecular Medicine, University of Oxford, England) discussed innate immunity and related plasma proteins. A growing body of research suggests the outcome of HIV infection may largely depend on innate responses that impact mucosal entry and viral replication and dissemination.16 In addition to contributing to host resistance to initial infection in the face of repeated exposures, the team proposed that innate responses may be a factor in determining the viral load set point.
To determine whether changes in soluble innate factors might serve as biomarkers of the passage of time after HIV infection, Borrow's group quantified these molecules using mass spectrometry. During the initial phase of infection, prior to acquired immunity, a surge of host innate antiviral defense factors was found. The initial peak of viremia was also associated with changes in the levels of a large number of cytokines and chemokines.16 However, these levels varied widely between subjects and HIV-seronegative persons may have similar levels in response to other infections and “stresses.” Furthermore, these levels quickly return to normal as viremia is contained and subsequently undergo little perturbation. Hence, elevation in acute-phase proteins associated with early HIV infection do not appear to be good candidates for the development of an HIV incidence assay.17
Changes in cellular markers
Andrew McMichael (Director, Weatherall Institute of Molecular Medicine, University of Oxford, England) presented an overview of the changes in cellular markers that occur during HIV infection, focusing on cellular immune responses that are critical to determining the outcome of infection. HIV infection results in activation of most immune cells, including NK cells, macrophages, B cells, and T cells, and is associated with increased levels of proinflammatory cytokines.18 CD8+ T cell responses, which vary according to HLA genetic polymorphisms, may be particularly important in suppressing the initial infection.19 Other key factors determining the course of HIV infection are the nature of the transmitted virus and the rate of virus mutation.18
McMichael showed how the earliest innate and adaptive immune responses detected after HIV-1 transmission can be aligned with Fiebig staging20 and viral load. For example, CD8+ T cell responses to only 1–3 epitopes are typically detectable during Fiebig stages 1–3, but they broaden to 5–10 or more epitopes in Fiebig stage 5 and beyond. In addition, neutralizing antibodies are not detected until more than 80 days after infection when they begin to select envelope (Env) escape mutations.
McMichael also presented data showing that the level of CD8+ T cell suppression of HIV-1 replication/infection in autologous CD4+ T cells correlates inversely with the rate of CD4+ T cell decline in blood.18 Thus, it may be possible to use the measurement of CD8+ suppression in conjunction with CD4+ T cell count to extrapolate back and estimate the time elapsed from initiation of infection. However, rather than serve as a stand-alone incidence marker, this measure might be useful as one component of an HIV incidence algorithm.
Immunoglobulin type and the pattern of change of anti-HIV antibodies
Georgia Tomaras (Duke University Medical Center, North Carolina) discussed the use of nonneutralizing antibody responses as potential biomarkers of recent infection. She emphasized that certain time-dependent variables must be considered including (1) the absence/presence of specific HIV-1 antigens/epitopes, (2) the affinity/avidity of specific antibody and antigen binding, and (3) the changes in these levels during early infection.18 The timing of the initial anti-Env response relative to HIV viral load may be particularly important.21
Mucosal and systemic Env-specific IgA antibodies usually decline rapidly during acute HIV infection and are therefore unlikely to serve as a stand-alone biomarker of HIV incidence. IgG antibodies tend to persist for longer periods of time. During acute HIV-1 infection, IgG antibodies directed against various envelope proteins arise sequentially with anti-gp41 appearing first, followed by anti-gp120 V3, anti-CD4 binding site, and nonneutralizing anti-MPER cluster II antibodies. Tomaras's laboratory found that IgG3 antibodies to p55 Gag, gp41 Env, p66 RT, gp120 Env, and p31 integrase consistently peaked during acute infection and then significantly declined. Peak levels were not directly associated with viral load, ART, or viral clade. The persistence of IgG3 antibodies appears to depend on antigenic stimulation and may decline after HIV viral load set point is reached. Tomaras suggested that the time-dependent pattern of change in specific antibodies may be useful as a component of an HIV incidence test.22
Telomere length due to HIV infection as a function of time
Beth Jamieson (David Geffen School of Medicine University of California at Los Angeles) described how telomere length is affected by HIV infection over time. Telomeres are a region of repetitive DNA sequences that protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes. These regions deter the degradation of genes near the ends of chromosomes by allowing chromosome ends to shorten during chromosome replication. The protection that telomeres provide to the chromosomal ends appears to be necessary to maintain the viability of cells and telomere shortening ultimately leads to programmed cell death.23 The enzyme telomerase, which is present in germ cells, stem cells, and activated lymphocytes, counteracts these effects. Researchers have shown that genetic and nongenetic factors, including age, influence telomere length. 24 On average, mean telomere length decreases by 20 to 60 base pairs every year throughout life. Strong correlations exist between age and telomere length in peripheral blood cells; however, extensive variation in individual telomere length limits our ability to use absolute telomere length to estimate age.
Jamieson's laboratory examined the relationship between HIV infection, aging, and impact of ART on de novo immune responses. In HIV-1-infected men, these cells have shortened telomeres compared to age-matched seronegative controls. The team then asked, “Can telomere length be used to distinguish recent from chronic HIV infection and thus be used as a biomarker of HIV incidence assay?” While the individual variability in telomere length was too great to make the absolute telomere length a useful measurement, they reasoned that telomere length may shorten at different rates in different cellular subsets as a consequence of HIV-1 infection.
Since neutrophils are short lived and lack telomerase, Jamieson compared telomere length between neutrophils and naïve CD4+ T cells. However, the ratio of telomere length in the two cell populations did not change as a result of HIV-1 infection, suggesting that telomere shortening also occurs in neutrophil progenitor cells during HIV-1 infection. In conclusion, research thus far has not demonstrated that telomere length is a useful biomarker of recent versus chronic HIV-1 infection.
Potential viral biomarkers
Mutations detected by clinical assays
Frank Maldarelli (HIV Drug Resistance Program National Cancer Institute, NIH) described a method for using HIV pol gene sequence data generated by routine HIV drug resistance assays to distinguish recent from chronic HIV infection. Viral sequence diversity in the population of HIV sequences in a given sample is detected when more than one nucleotide is present at the same position. An ambiguity index (AI) can be calculated as the number of ambiguous positions divided by the total number of positions in the sequence. A previous study demonstrated that this measure of genetic diversity increases with time after infection and could therefore be used to estimate the duration of infection.25
As per current recommendations in the United States and Europe, the pol sequence of the virus is examined in all newly diagnosed HIV infections in order to guide the selection of antiretroviral drugs for treatment. Therefore, sequencing of this region of the virus has been standardized and assays are available from commercial laboratories.
When Maldarelli used this approach to evaluate 100 recently infected patients and 97 patients chronically infected [Western blot (WB) positive for >1 year] with clade B virus, it gave 74.5% sensitivity and 87.2% specificity when the AI cutoff was set at 0.43.25 The method appears to be more accurate when CD4+ counts are also taken into consideration. Using the same AI cutoff, less than 5% of those with CD4+ count >250 had chronic infection and a similarly low proportion of those with CD4 counts <250 were recently infected. However, some samples could not be categorized as early or chronic—suggesting that additional parameters need to be assessed before this technology can be optimized for use as an HIV incidence assay.
A viral sequence-based HIV incidence assay
Ha Youn Lee (Department of Biostatistics and Computational Biology, University of Rochester, New York) discussed another potential genome sequence diversification-based strategy for developing an HIV incidence assay.26 This approach is based on the analysis of multiple envelope sequences generated using a single genome sequencing method.1,27,28 Differences among approximately 20 aligned sequences from each subject are used to determine the extent of HIV genetic diversity in the original sample. Lee's analysis involves calculating the mean Hamming Distance (HD: a measure of the number of nucleotide differences between sequence pairs) between all possible pairs of sequences from each patient sample.26 This mean HD was designated as the diversity of the sample.
Plots of envelope diversity versus stage of infection demonstrated that incident infections resulting from a single founder virus did not overlap in diversity with chronic infections, but that incident infections resulting from multiple founder viruses could overlap in diversity with chronic infections. Thus, individuals whose infections resulted from multiple founder viruses might be misclassified as chronically infected.
To overcome this potential problem, Lee examined the 10% quantile of the HD distribution, or Q10. These Q10 values were much lower in incident infection compared to chronic infection, regardless of whether multiple founder viruses established infection. Using a cutoff of Q10=7, a specificity of 100% and a sensitivity of 97.3% was found in a group of specimens from 182 incident and 43 chronic cases. As the length of the gene segment analyzed decreased, the mean of the chronic Q10 decreased as well. However, significant differences were still seen with gene segments as small as 500 base pairs, with both sensitivity and specificity remaining above 97% (Table 1).
Table 1.
Q10 Cut-Off Values, Sensitivity, and Specificity for the Full Envelope Sequence and for Envelope Subregions (from Park et al.26)
| Full envelope | 2000 long (HXB2 6360) | 1000 long (HXB2 6860) | 500 long (HXB2 7110) | 500 long (HXB2 7125) | 500 long (HXB2 7625) | |
|---|---|---|---|---|---|---|
| Q10 cut-off | 7 | 2 | 1 | 0 | 0 | 0 |
| Sensitivity | 97.3% | 95.1% | 97.8% | 98.9% | 97.8% | 98.4% |
| Specificity | 100% | 100% | 100% | 100% | 97.7% | 97.7% |
Reference sequence and starting nucleotide position are in parentheses.
The approximately 20 single genome sequences required from each subject make this approach complicated and expensive to perform with current single genome amplification technology.26 Lee is therefore exploring the use of pooled 454 deep-sequencing technology to develop a high through-put (and ultimately less expensive) process. While this method appears promising at this early stage of development, further evaluation with a full range of specimen panels will be required to determine the FRR and MDRI.
Analysis of HIV diversity using a high-resolution melting assay
Susan Eshleman (Johns Hopkins University School of Medicine, Maryland) described the use of a high-resolution melting (HRM) assay to analyze HIV diversity in individuals with recent versus nonrecent HIV infection. The HRM diversity assay is relatively simple assay that does not require sequencing. The assay provides a single numeric HRM score that reflects the level of genetic diversity in a specific region of the HIV genome.29
The HRM diversity assay is performed using a LightScanner System for HRM analysis; this system was developed for detection of specific point mutations in deoxyribonucleic acid (DNA). Eshleman has adapted this system for analysis of HIV diversity.30 Validation studies showed that reproducibility is maintained over a wide range of sample volumes and viral loads. HRM scores are significantly associated with sequence-based measures of genetic diversity obtained from the analysis of HIV clones; these measures include genetic diversity, genetic complexity, and Shannon entropy.31
The HRM diversity assay was used to analyze HIV in samples obtained at the time of HIV seroconversion (recent); median, 189 days since the last negative HIV test, range 15–540 days and in samples obtained from adults who were infected for at least 2 years (nonrecent); individuals on ART, and those with AIDS. These samples were obtained from diverse populations (including men who have sex with men, women, and injecting drug users). HRM scores were measured for six different regions of the HIV genome (2 in gag, 1 in pol, 3 in env). High HRM scores in all six regions were significantly associated with nonrecent infection.31 HRM scores in three of those regions (one in gag and two in env) were independently associated with nonrecent infection in a multivariate model. Analysis of HIV diversity in different regions of the HIV genome may provide “diversity signatures” that are more reliable for determining recency than any single measurement. The group is also examining whether the HRM diversity assay can be used in conjunction with other assays to improve the precision of HIV incidence estimates.
Critical Pathway to Assay Development and Implementation
Nanotechnology, chip technology, and other possible platforms for converting biomarkers into assays
Shixing Tang [U.S. Food and Drug Administration (FDA), Maryland] reported on the development of a gold nanoparticle-based biobarcode amplification (BCA) assay for early and sensitive detection of HIV-1 capsid (p24) antigen.32 The assay consists of anti-p24 antibody-coated microplates for the capture of the p24 antigen, streptavidin-coated gold nanoparticles, and biotinylated biobarcode DNA for signal amplification.33 The signal is read by a chip-based scanometric method that can detect 100-fold less HIV-1 p24 antigen to conventional HIV p24 enzyme-linked immunosorbent assay (ELISA).
A similar test platform using europium nanoparticle-based immunoassay (ENIA) can also be used to detect HIV-1 p24 antigen. The ENIA employs nanospheres covered with thousands of europium atoms that emit light in the presence of the antigen. Tang's team enhanced the sensitivity of this test to ∼0.5 pg/ml of HIV p24 so that it is also capable of detecting HIV-1 p24 approximately 3-4 days earlier than conventional p24 ELISA assays. It is possible that the same platform could be applied to other biomarkers with a longer MDRI, making it useful for an HIV incidence assay.
Tang also presented data on anti-HIV-1 p24 host responses to 10 different peptide fragments that span the entire protein. These anti-p24 antibodies could be characterized as either polyclonal-like (multiple peptide specificities), monoclonal-like (specificity to 1–3 peptides), or nonreactive. Of particular interest was the appearance of a switch in the immune response from a polyclonal-like pattern during acute HIV infection to a monoclonal-like pattern or no response during chronic infection (unpublished data).
Michael Pollack (Advanced Liquid Logic, Inc., North Carolina) presented the digital microfluidics platform being developed at Advanced Liquid Logics. This technology uses their proprietary electrowetting-on-dielectric (EWOD) lab-on-a-chip system in which liquids are processed as droplets that can be moved together or apart between sites on demand.34 These devices use arrays of surface electrodes to modify the shape and position of droplets through the electrowetting effect. This technology can be used for sample preparation and for interrogating the sample at the protein35 (e.g., enzymatic, luminex, and ELISA-based assays) or nucleic acid36 [e.g., real-time reverse transcription, polymerase chain reaction (RT-PCR), pyrosequencing] level. In addition, Pollack's team is evaluating the possibility of using digital microfluidics to measure cell types, including CD4+ counts.37
The digital microfluidics platform can also be designed to perform sequential tests on the same specimen. If it is determined that two measurements (for example, a novel biomarker estimation and CD4 cell count) are needed for an HIV incidence assay/algorithm to achieve a sufficiently low FRR (for example, novel biomarker estimation and CD4 cell count), it is conceivable that a single compact digital microfluidics card could perform both tests. The platform also has the advantage of rapid turn-around times and low-power requirements.
Assessing the performance of incidence assays
Gary Murphy (United Kingdom Health Protection Agency, London) discussed the validation of HIV incidence assays. He defined validation as “the collection and evaluation of data, from the process design stage throughout production, which establishes scientific evidence that a process is capable of consistently delivering quality products.” Murphy noted that evaluation was the comparison of devices developed for the same purpose or an attempt to independently verify manufacturers' claims.
External validation has three primary purposes: (1) to provide reassurance, (2) to demonstrate transferability, and (3) to provide scientific evidence to support advice and guidance. Murphy noted that a partial validation of the current incidence assays requires the evaluation of a limited but relevant set of parameters using a broad range of well-characterized samples. To meet these goals, he and his collaborators have established the Consortium for the Evaluation and Performance of HIV Incidence Assays (CEPHIA) through funding provided by the Bill and Melinda Gates Foundation (BMGF), with the following four objectives:
To create a specimen repository composed of large panels of specimens from HIV positive persons for evaluating/performing a Recent Infection Testing Algorithm (RITA).
To evaluate candidate assays for their ability to correctly identify recent HIV Infection.
To derive and deploy improved methods for estimating an incident infection using RITA and to determine algorithms with low FRR that may be constructed from available components/tests.
To build collaborations and consensus around the characterization and deployment of RITAs in the field.
The proposed repository will house three types of specimen panels and incorporate a diverse set of HIV clades. This would include a long-term seroconversion panel that follows infected individuals for up to 2 years in order to distinguish recent from chronic HIV infections, specimens from chronically infected persons (beyond 2 years) to measure the false recent rate, and a false recent challenge panel containing samples from individuals who typically misclassify (i.e., elite controllers, ART-treated patients, and those with advanced disease). One major issue raised was that the repository may not be useful for all biomarker assays under investigation since the samples collected are limited to plasma and serum.
Alex Welte provided an overview of the statistical techniques used to evaluate an HIV incidence assay. He emphasized that although it is important for the assay to be accurate, precise, sensitive, and specific, the key performance characteristics are the MDRI and the FRR.6,38
Discussion
Although most HIV incidence assays currently undergoing external validation are based on host immune responses, novel assays based on viral markers have considerable promise. Further development and evaluation of these assays remain a priority. The CEPHIA group indicated that it would entertain requests for the use of repository samples for these purposes if there are sufficient data indicating that a novel assay is accurate, reproducible, and robust. Such data should be published in peer-reviewed journals.
To support the identification of novel biomarkers for HIV incidence assays, the Division of AIDS, NIAID, has released a Program Announcement [PA-12-012: HIV incidence assays with improved specificity (R01)], open through Sept. 2014. In addition, the Small Business Innovation Research (SBIR) mechanism is available for supporting the assay development process itself. The development of improved HIV incidence assays and algorithms remains a high priority for NIAID.
Key requirements of an HIV incidence assay are high reproducibility and a low FRR when applied across different populations and viral clades. The MDRI should ideally be between 6 and 12 months. For broad applicability, the assay should be capable of using currently available repository materials, typically serum, plasma and possibly dried blood spots. This may change in the future if other specimens [e.g., peripheral blood mononuclear cells (PBMCs), stimulated plasma, noninvasive samples] are collected more commonly.
A critical specimen requirement for HIV incidence assays based on viral biomarkers is the minimum amount of virus needed to amplify, rather than a minimum specimen volume per se. This will be affected by disease stage, treatment status, viral resistance, and occasionally host response (e.g., elite controller status).
Conclusions
Accurate determination of HIV incidence will continue to be critical for monitoring the HIV epidemic, evaluating ongoing prevention programs, and designing and evaluating new prevention trials. Novel assays must be developed that provide reproducible results when applied to diverse populations and large sample sizes. The two crucial characteristics of an incidence assay are its FRR and MDRI.
Many challenges to HIV incidence assay development must be overcome. A repository of well-characterized specimens to evaluate new assays is being assembled, but procedures that enable all qualified investigators to utilize these specimens are still being formulated. A consensus is emerging regarding essential performance standards but these have yet to be reviewed and endorsed by normative agencies.
Innovative investigators are beginning to address the need for novel biomarkers for new HIV incidence assays. Others must be encouraged to join them. Biomarkers based on the evolution of viral diversity appear to be particularly attractive candidates, but host biomarkers may yet prove to be equally valuable.
Once novel biomarkers have been identified, they must be incorporated into a highly reproducible assay. The assay should use easy-to-obtain specimens, be unaffected by clade, and, ideally, be capable of being performed in the field by persons with limited training and equipment. Fortunately, a number of innovative new platforms are available, including those based on nanotechnology and “lab-on-a-chip” technologies. Continuing improvements (and cost reductions) in viral sequencing technology may also contribute to the development of future assays.
Despite these many requirements and challenges, the participants in this workshop were optimistic that novel HIV incidence assays will become available in the foreseeable future. If achieved, progress toward controlling the HIV pandemic will be accelerated.
Acknowledgments
This project has been funded in part with Federal funds from the NIAID, NIH, Department of Health and Human Services (DHHS), under Contract No. HHSN272200800014C. The authors wish to extend special thanks to Michael Gilbreath, Michael Ussery, and Cynthia Soriano from DAIDS, NIAID, NIH; Jennifer Frasier and Jean Morrow, Henry M. Jackson Foundation (HJF)–DAIDS Contractors; and LaVietra Shannon from BL Seamon for their invaluable assistance in planning and/or implementing this workshop. Also, the authors would like to thank Brenda Collins, HJF-DAIDS Contractor (Science Writer), who helped enormously in putting this manuscript together; and Lester Freeman, HJF-DAIDS Contractor, for his technical assistance in preparing this manuscript for submission.
The opinions expressed herein are those of the authors and should not be construed as representing the official views of the DHHS. The views do not necessarily reflect the official policies of the DHHS; nor does mention of trade names, commercial practices, or organizations imply endorsement by the U.S. Government.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bar KJ. Li H. Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84(12):6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busch MP. Pilcher CD. Mastro TD, et al. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS. 2010;24(18):2763–2771. doi: 10.1097/QAD.0b013e32833f1142. [DOI] [PubMed] [Google Scholar]
- 3.Welte A. McWalter TA. Laeyendecker O. Hallett TB. Using tests for recent infection to estimate incidence: Problems and prospects for HIV. Euro Surveill. 2010;15:19589. [PMC free article] [PubMed] [Google Scholar]
- 4.Hallett TB. Estimating the HIV incidence rate: Recent and future developments. Curr Opin HIV AIDS. 2011;6(2):102–107. doi: 10.1097/COH.0b013e328343bfdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marinda ET. Hargrove J. Preiser W, et al. Significantly diminished long-term specificity of the BED capture enzyme immunoassay among patients with HIV-1 with very low CD4 counts and those on antiretroviral therapy. J Acquir Immune Defic Syndr. 2010;53(4):496–499. doi: 10.1097/qai.0b013e3181b61938. [DOI] [PubMed] [Google Scholar]
- 6.Mastro TD. Kim AA. Hallett T, et al. Estimating HIV incidence in populations using tests for recent infection: Issues, challenges and the way forward. J HIV/AIDS Surveill Epidemiol. 2010;2(7):1–14. [PMC free article] [PubMed] [Google Scholar]
- 7.Incidence Assay Critical Path Working Group: More and better information to tackle HIV epidemics: Towards improved HIV incidence assays. PLoS Med. 2011;8(6):e1001045. doi: 10.1371/journal.pmed.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proceedings of the Meeting on the Development Assays to Estimate HIV Incidence: May 13–14, 2009; Chapel Hill, North Carolina. [Google Scholar]
- 9.McWalter TA. Welte A. Relating recent infection prevalence to incidence with a sub-population of assay non-progressors. J Math Biol. 2010;60:687–710. doi: 10.1007/s00285-009-0282-7. [DOI] [PubMed] [Google Scholar]
- 10.McWalter TA. Welte A. A comparison of biomarker based incidence estimators. PLoS ONE. 2009;4:e7368. doi: 10.1371/journal.pone.0007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassanjee R. Welte A. McWalter TA. Viljoen J. Bärnighausen T. Newell ML. Fatti P. Calibration of BED assay for use in incidence estimation 5th IAS Conference, 2009, on HIV pathogenesis, treatment and prevention; Abstract CDB018. [Google Scholar]
- 12.Bärnighausen T. McWalter T. Rosner Z. Newell M-L. Welte A. HIV incidence estimation using the BED capture enzyme immunoassay: Systematic review and sensitivity analysis. Epidemiology. 2010;21:685–697. doi: 10.1097/EDE.0b013e3181e9e978. [DOI] [PubMed] [Google Scholar]
- 13.A UNAIDS/WHO publication: When and how to use assays for recent infection to estimate HIV infection at a population level. 2011. www.who.int/diagnostics_laboratory/110906_guidance_hiv_incidence.pdf www.who.int/diagnostics_laboratory/110906_guidance_hiv_incidence.pdf
- 14.Welte A. McWalter TA. Laeyendecker O. Hallett TB. Using tests for recent infection to estimate incidence: Problems and prospects for HIV. EuroSurveillance. 2010;15:19589. [PMC free article] [PubMed] [Google Scholar]
- 15.Web-based tool for assay based incidence estimation. www.incidence-estimation.com/page/tools www.incidence-estimation.com/page/tools
- 16.Borrow P, et al. Innate immune factors associated with HIV-1 transmission. Curr Opin HIV AIDS. 2011;6(5):341–347. doi: 10.1097/COH.0b013e3283499e11. [DOI] [PubMed] [Google Scholar]
- 17.Borrow P. Shattock RJ. Vyakarnam A. Innate immunity against HIV: A priority target for HIV prevention research. Retrovirology. 2010;7:84. doi: 10.1186/1742-4690-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCMichael A. Borrow P. Tomaras G, et al. The immune response during acute HIV-1 infection: Clues for vaccine development. Nat Rev Immunol. 2010;10(1):11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford H, et al. Compensatory mutation partially restores fitness and delays reversion of mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic HIV-1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feibig EW, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors; Implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 21.Tomaras GD. Haynes BF. HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr Opin HIV AIDS. 2009;4(5):373–379. doi: 10.1097/COH.0b013e32832f00c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yates NL. Lucas JT. Nolen T, et al. Multiple HIV-1 specific IgG3 responses decline during acute HIV-1: Implications for detection of incident HIV Infection. AIDS. 2011;25(17):2089–2097. doi: 10.1097/QAD.0b013e32834b348e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenberg DT. An evolutionary review of human telomere biology: The thrifty telomere hypothesis and notes on potential adaptive paternal effects. Am J Hum Biol. 2011;23(2):149–167. doi: 10.1002/ajhb.21127. [DOI] [PubMed] [Google Scholar]
- 24.Rickabaugh TM. Kilpatrick RD. Hultin LE, et al. The dual impact of HIV-1 infection and aging on naive CD4 T-cells: Additive and distinct patterns of impairment. PLoS One. 2011;6(1):e16459. doi: 10.1371/journal.pone.0016459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouyos RD. von Wyl V. Yerly S, et al. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis. 2011;52(4):532–539. doi: 10.1093/cid/ciq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY. Love TM. Nelson J. Thurston SW. Perelson AS. Lee HY. Designing a genome-based HIV incidence assay with high sensitivity and specificity. AIDS. 2011;25(16):F13–F19. doi: 10.1097/QAD.0b013e328349f089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keele BF. Giorgi EE. Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrahams MR. Anderson JA. Giorgi EE, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towler WI. James MM. Ray SC, et al. Analysis of HIV diversity using a high-resolution melting assay. AIDS Res Hum Retroviruses. 2010;26(8):913–918. doi: 10.1089/aid.2009.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James M. Lei W. Musoke P, et al. Association of HIV diversity and survival in HIV-infected Ugandan infants. PLoS One. 2011;6(4):e18642. doi: 10.1371/journal.pone.0018642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cousins MM. Laeyendecker O. Beauchamp G, et al. Use of a high resolution melting (HRM) assay to compare gag, pol, and env diversity in adults with different stages of HIV infection. PLoS One. 2011;6(11):e27211. doi: 10.1371/journal.pone.0027211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang S. Hewlett I. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J Infect Dis. 2010;201(Suppl 1):S59–S64. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang S. Zhao J. Wang A, et al. Characterization of immune responses to capsid protein p24 of human immunodeficiency virus type 1 and implications for detection. Clin Vaccine Immunol. 2010;17(8):1244–1251. doi: 10.1128/CVI.00066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack MG. Shenderov AD. Fair RB. Electrowetting-based actuation of droplets for integrated microfluidics. Lab Chip. 2002;2(2):96–101. doi: 10.1039/b110474h. [DOI] [PubMed] [Google Scholar]
- 35.Sista R. Hua Z. Thwar P, et al. Development of a digital microfluidic platform for point of care testing. Lab Chip. 2008;8(12):2091–2104. doi: 10.1039/b814922d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua Z. Rouse JL. Eckhardt AE, et al. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal Chem. 2010;82(6):2310–2316. doi: 10.1021/ac902510u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollack MG. Pamula VK. Srinivasan V. Eckhardt AE. Applications of electrowetting-based digital microfluidics in clinical diagnostics. Expert Rev Mol Diagn. 2011;11(4):393–407. doi: 10.1586/erm.11.22. [DOI] [PubMed] [Google Scholar]
- 38.Kassanjee R. Welte A. McWalter TA, et al. Seroconverting blood donors as a resource for characterising and optimising recent infection testing algorithms for incidence estimation. PLoS One. 2011;6(6):e20027. doi: 10.1371/journal.pone.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]