Abstract
COP1 is a p53-targeting E3 ubiquitin ligase that is downregulated by DNA damage through mechanisms that remain obscure. Here we report that COP1 is not downregulated following DNA damage in 14-3-3σ null cells, implicating 14-3-3σ as a critical regulator in the response of COP1 to DNA damage. We also identified that 14-3-3σ, a p53 target gene product, interacted with COP1 and controlled COP1 protein stability after DNA damage. Mechanistic studies revealed that 14-3-3σ enhanced COP1 self-ubiquitination, thereby preventing COP1-mediated p53 ubiquitination, degradation, and transcriptional repression. Additionally, we found that COP1 expression promoted cell proliferation, cell transformation, and tumor progression, manifesting its role in cancer promotion, whereas 14-3-3σ negatively regulated COP1 function and prevented tumor growth in a mouse xenograft model of human cancer. Immunohistochemical analysis of clinical breast and pancreatic cancer specimens demonstrated that COP1 protein levels were inversely correlated with 14-3-3σ protein levels. Together, our findings define a mechanism for posttranslational regulation of COP1 after DNA damage that can explain the correlation between COP1 overexpression and 14-3-3σ downregulation during tumorigenesis.
Keywords: 14-3-3σ, COP1, p53, ubiquitination
Introduction
Constitutive photomorphogenic 1 (COP1) is an evolutionarily conserved E3 ubiquitin ligase containing RING-finger, coiled-coil, and WD40-repeat domains. COP1 has been identified as a crucial mediator to block photomorphogenesis in the dark through the ubiquitinated proteasomal degradation of light-induced transcription factor HY5 in Arabidopsis (1). In mammalian cells, COP1 regulates various cellular functions, such as proliferation and survival, by facilitating the degradation of physiological substrates through ubiquitin-mediated protein degradation. The ubiquitinated targets of COP1 include stress-responsive transcription factors p53 tumor suppressor (2) and c-JUN (3–5), transducer of regulated CREB activity 2 (TORC2, a glucose metabolite regulator) (6), FOXO1 (7), and nucleosome remodeling factor MTA1 (8). COP1 plays an important role in DNA damage response (2), but COP1 is quickly downregulated following DNA damage (9). However, the mechanism involved in downregulation of COP1 after DNA damage remains elusive.
The 14-3-3 proteins are a family of regulatory chaperone molecules involved in many diverse physiological functions, including signaling transduction, stress response, apoptosis, and cell cycle checkpoint regulation (10, 11). Binding by 14-3-3 proteins mediates stability and/or subcellular localization of target proteins (10, 12, 13). 14-3-3σ is characterized as a human mammary epithelial-specific marker (HME1) (14), and is later found to be an essential regulator of apoptosis, cell migration, cell cycle, and DNA damage response (15–18). In contrast to the other 14-3-3 family members (β, ε, γ, ζ, η, and τ), which are able to form both homo- and heterodimers with other members, 14-3-3σ can form only homodimers (19). This unique characteristic implies that 14-3-3σ has exclusive functions and behaviors. 14-3-3σ, but not other family members, has been found to be frequently lost or decreased in various human cancers (20) and functions as a potential tumor suppressor. In response to DNA damage response, 14-3-3σ is known to be a p53 downstream target and may serve as a regulator to prevent oxidative and DNA-damaging stress-induced mitotic checkpoint dysfunction (21). Although 14-3-3σ may play an important role in protecting cells from DNA damage, the detailed mechanism by which 14-3-3σ modulates the DNA damage response remains not well characterized.
Given that both COP1 and 14-3-3σ are involved in DNA damage response, we examined the role of 14-3-3σ in DNA damage-mediated COP1 downregulation. We found that 14-3-3σ physically interacted with COP1 and accelerated COP1-mediated self-ubiquitination, thereby destabilizing COP1 protein, which in turn reduced COP1-mediated p53 ubiquitination. Our studies also indicate the applicability of employing 14-3-3σ-COP1-p53 axis as a therapeutic intervention in COP1-overexpressing cancers.
Materials and methods
Cell culture and reagents
HCT116 14-3-3σ−/− and 14-3-3σ+/+ cells (provided by Dr. Bert Vogelstein) were authenticated with western blot routinely. Human 293T, A549, H1299, HCT116, and U2OS were from ATCC (authenticated by short tandem repeat analysis). Cells were transfected with DNA using FuGENE HD (Roche) reagents. Antibodies used: Flag (M2 monoclonal antibody, Sigma), tubulin (Sigma), COP1 (Bethyl Laboratories), 14-3-3σ (RDI), p53 (FL393 and DO1, Santa Cruz Biotechnology), Myc (mouse monoclonal 9E10, Santa Cruz Biotechnology; rabbit polyclonal, Sigma), HA (12CA5, Roche), His (Cell Signaling), and ubiquitin (Zymed Laboratories, Inc.).
Plasmids
pCMV5-Flag-COP1 was provided by E. Bianchi. pCDNA6-Myc-COP1 was constructed by our lab. pCMV5-Flag-14-3-3σ, pCMV5-His-p53, pCMV5-p53, Ad-β-gal, and Ad-14-3-3σ were previously described (15). COP1-C136S/C139S mutant was generated using PCR-directed mutagenesis. The pET15b plasmids expressing Flag-tagged COP1 (1–226 aa, 216–420 aa, 392–731 aa) were generated using PCR.
Immunoprecipitation, immunoblotting, FACS Cell cycle analysis
Cell lysates were immunoprecipitated with indicated antibodies, followed by immunoblotting for detecting proteien-protein association or regulation. Samples were analyzed using a BD Facscanto II flow cytometer (BD Biosciences) for analyzing the distribution in different cell cycle phases (FACS). These experiments were performed as previously described (18).
In vitro binding assay
35 Slabeled COP1 was prepared by in vitro transcription and translation using TNT system as previously described (10). TNT products of COP1 were incubated with GST or GST-14-3-3σ. Also, the Flag-COP1 (aa 1–226, 216–420 or 392–731) was expressed using pET15b vector and purified for binding assay.
In vivo ubiquitination assay
293T cells were transiently cotransfected with the indicated plasmids to detect exogenous COP1 and p53 ubiquitination. Cells were treated with 5 μg/mL MG132 (Sigma) for 6 hr and lysed. The ubiquitinated COP1 was immunoprecipitated by anti-Myc (9E10) or anti-COP1, and immunoblotted by anti-HA or anti-ubiquitin. The ubiquitinated p53 was immunoprecipitated with anti-p53 (DO-1) and immunoblotted with anti-ubiquitin or anti-HA.
In vitro ubiquitination assay
Flag-COP1 and the Flag-COP1 RING mutant (C136S, C139S) proteins were prepared using TNT system and were incubated with a combination of 200 pmol ubiquitin, 2 pmol E1, 10 pmol E2 (UbcH5a/5b), 2 mM ATP and purified GST-14-3-3σ in a total volume of 50 μL for 1 hr at 37°C. The ubiquitinated COP1 was immunoprecipitated with anti-Flag as described (18) and immunoblotted with anti-ubiquitin. E1 and E2 were purchased from Biomol International.
Quantitative PCR
Primers for real-time quantitative PCR of p53 target genes—CDKN1A, SFN, BAX, and PUMA were as referenced in Primer Bank (http://pga.mgh.harvard.edu/primerbank/). 1 μg RNA was used for producing cDNA by iScript cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR analyses were performed using iQ SYBR Green Super mix (Bio-Rad, 170-8882) and the iCycler iQ Real-time PCR detection system.
Luciferase assay
The BDS2-3X-luc reporter containing a p53 responsive element (15) was co-transfected with the indicated expressing vectors into H1299 cells. Luciferase activity was assayed with the dual luciferase assay system (Promega).
Generation of stable transfectants
HCT116 cells were infected by Mission lentiviral shRNA transduction particles (Sigma) containing either control shRNA, or two specific 14-3-3σ shRNA. After infection, cells were selected with 2 μg/mL puromycin for 2 weeks. HCT116 and U2OS cells were transfected with indicated plasmids for the generation of overexpression stable transfectants.
Soft agar colony formation, Foci formation, MTT assay
These experiments were used to analyze anchorage-independent growth, transformation phenotype, and cell growth in COP1-overexpressing cells respectively. They were performed as previously described (10).
Nude mice experiment
nu/nu mice were xenografted with vector control stable cell lines, COP1 expressing cells left uninfected, and COP1-expressing cells infected with Ad-β-gal (MOI = 100) or Ad-14-3-3σ (MOI =100) for 48 hr. Cells were harvested and injected into the flank of each mouse. Tumor volumes were measured and recorded and collected.
Immunohistochemistry
Immunohistochemistry was performed on a tissue microarray consisting of 121 cases of primary pancreatic carcinoma specimens from patients who underwent initial pancreaticoduodenectomy at The University of Texas M. D. Anderson Cancer Center between 1990 and 2004. Immunohistochemical staining for 14-3-3σ and COP1 was performed on 5-μm unstained sections from the tissue microarray blocks. Sections were incubated for 90 min at 37°C with primary antibodies against 14-3-3σ (RDI) or COP1 (Bethyl Laboratories) at a 1:100 dilution. Standard avidin-biotin immunohistochemical analysis of the sections was done (Vector Laboratories). The staining results were evaluated by a board-certified pathologist (H.W.). The COP1 staining in the tumor cells was scored as 0 (no nuclear staining), 1 (< 10% nuclear staining), 2 (10%–50% nuclear staining), and 3 (> 50% nuclear staining).
Results
14-3-3σ is required for DNA damage-mediated COP1 downregulation
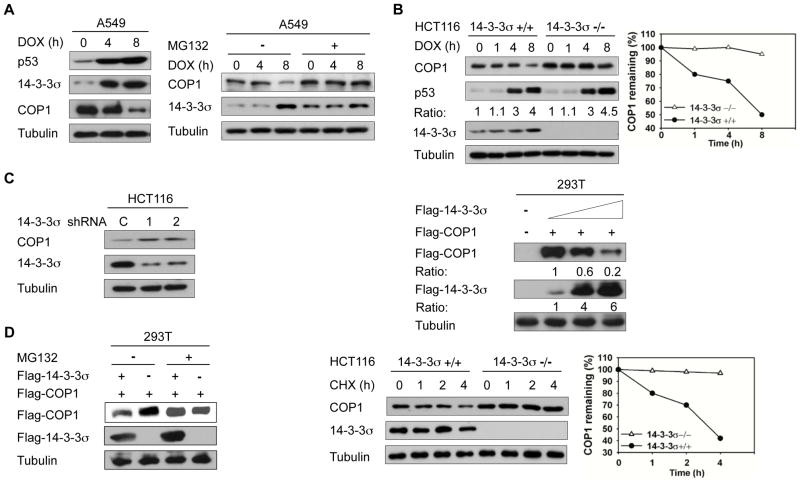
DNA damage induces p53 stabilization, which in turn induces the 14-3-3σ expression to execute cell cycle arrest (21). In a DNA damage response study using lung adenocarcinoma A549 cells expressing wild-type p53, we found that DNA damage caused by doxorubicin treatment led to the induction of 14-3-3σ. At the same time, COP1 expression was downregulated by the DNA damage (Fig. 1A, left panel). Moreover, MG132 rescued doxorubicin-induced COP1 downregulation (Fig. 1A, right panel), implying that ubiquitin-mediated proteasome degradation was involved in doxorubicin-mediated COP1 downregulation. We also found that the downregulation of COP1 upon doxorubicin-induced DNA damage observed in HCT116 14-3-3σ+/+ cells was absent in HCT116 14-3-3σ−/− cells (Fig. 1B). Consistently, DNA damage-mediated COP1 downregulation was also compromised when 14-3-3σ was knocked down (Fig. S1). These results suggest that 14-3-3σ plays a role in DNA-damage-mediated COP1 downregulation. To investigate the impact of 14-3-3σ on the expression of COP1, we investigated the steady state expression of COP1 when 14-3-3σ was depleted by shRNA, or overexpressed. 14-3-3σ depletion by shRNA led to accumulation of COP1 (Fig. 1C), while 14-3-3σ overexpression led to COP1 downregulation (Fig. 1C). Like DNA damage-mediated COP1 downregulation, 14-3-3σ-mediated COP1 downregulation was rescued by MG132 treatment (Fig. 1D, left panel). In the absence of de novo protein synthesis (cycloheximide treatment), the protein turnover rate of endogenous COP1 over time was reduced more in HCT116 14-3-3σ−/− cells than in the HCT116 14-3-3σ+/+ cells (Fig. 1D, right panel), implying that 14-3-3σ controls COP1 protein stability. Together, we conclude that 14-3-3σ is involved in DNA damage-mediated COP1 downregulation.
Figure 1. 14-3-3σ negatively regulates COP1 stability.
A, A549 cells were treated with 1 μg/mL doxorubicin (DOX) for the indicated hours. Lysates were analyzed by immunoblotting with indicated antiboies (Left). A549 cells were treated with 1 μg/mL doxorubicin (DOX) for the indicated hours and treated with or without MG132 for 3 hr. Lysates were immunoblotted with the indicated antibodies (Right).
B, Indicated cells were treated with 1 μg/mL doxorubicin (DOX) for the indicated hours. Cell lysates were immunoblotted with the indicated antibodies. The protein levels of COP1 in response to DNA damage at the different time points were determined (Right).
C, HCT116 cells were knocked down with two specific 14-3-3σ shRNA (1 & 2) or control Luciferase shRNA (C). Lysates were immunoblotted with anti-COP1, and anti-14-3-3σ (Left). 293T cells were transfected with the indicated expression vectors. Equal amounts of cell lysates were immunoblotted with the indicated antibodies (Right).
D, 293T cells were transfected with the indicated plasmids. Cells were treated with or without proteasome inhibitor MG132 before lysates were collected. Lysates were immunoblotted with the indicated antibodies (Left). Indicated cells were treated with cycloheximide (CHX) for the indicated hours. Cell lysates were immunoblotted with indicated antibodies. The protein levels of COP1 over time are shown (Right).
14-3-3σ directly interacts with COP1 and promotes COP1 ubiquitination
To further clarify the relationship between 14-3-3σ and COP1, we showed that DNA damage-induced 14-3-3σ interacts with COP1 (Fig. 2A). We also demonstrated the direct binding between 14-3-3σ and COP1 in vitro (Fig. 2B). We define the region on COP1 required for binding to 14-3-3σ by performing GST pull-down assays. The results showed that 14-3-3σ bound to the N-terminus of COP1 (aa 1–226 and 216–420) but not the C-terminus of COP1 (aa 392–731 containing WD40 repeats) (Fig. 2C). These data indicate that 14-3-3σ binds to the COP1 N-terminal RING and coiled-coil domain.
Figure 2. 14-3-3σ directly interacts with COP1.
A, A549 cells were treated with 1 μg/mL doxorubicin overnight. Before harvesting, cells were treated with MG132. Equal amounts of cell lysates were immunoprecipitated with either rabbit IgG (as control) or anti-COP1, and immunoblotted with anti-14-3-3σ (upper panel). Lysates were also immunoprecipitated with the indicated antibodies and immunoblotted with anti-COP1 (bottom panel). WCL: whole cell lysate.
B, 35S-labeled COP1 proteins were translated in vitro and incubated with either GST- or GST-14-3-3σ for binding assay. The bottom panel shows the input of GST proteins analyzed by Coomassie Blue staining. PD: pull down.
C, Bacterially purified Flag-COP1 domains were incubated with GST-14-3-3σ, and then was subjected to GST pull-down (PD) followed by immunoblotting using anti-Flag. Specific interaction of COP1 domains with 14-3-3σ is indicated by arrows. The asterisks indicate non-specific bands.
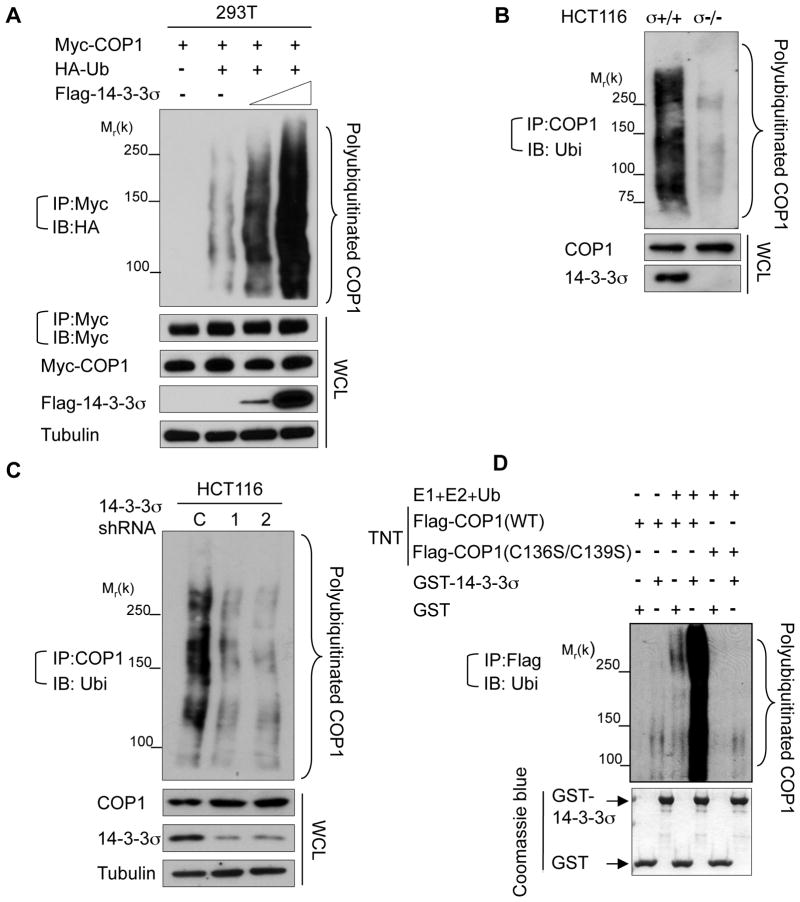
In the presence of MG132, increased expression of 14-3-3σ significantly accelerated COP1 polyubiquitination (Fig. 3A). Consistently, more polyubiquitinated COP1 was detected in HCT116 14-3-3σ+/+ cells than in HCT116 14-3-3σ−/− cells in the presence of MG132 (Fig. 3B), while stable knockdown of 14-3-3σ decreased endogenous COP1 ubiquitination (Fig. 3C). In vitro ubiquitination assay also revealed that COP1 has the self-ubiquitination activity and that the self-ubiquitination of COP1 was enhanced by the presence of purified GST-14-3-3σ but abolished by mutations in the RING domain of COP1 (C136S, C139S) (Fig. 3D). We conclude that 14-3-3σ interacts with COP1 directly and facilitates COP1 self-ubiquitination, a process that requires the RING domain of COP1.
Figure 3. 14-3-3σ promotes COP1 ubiquitination.
A, 293T cells were cotransfected with the indicated plasmids and increasing amounts of Flag-14-3-3σ. The cells were treated with MG132 for 6 hr before harvesting, and the ubiquitinated COP1 was immunoprecipitated with anti-Myc and immunobltted with anti-HA. Equal amounts of whole cell lysate (WCL) were immunoblotted with the indicated antibodies.
B, Indicated cells were treated with MG132 for 6 hr before harvesting. Polyubiquitinated COP1 was detected by immunoprecipitating with anti-COP1, followed by immunoblotting with anti-ubiquitin.
C, HCT116 cells were knocked down with two specific 14-3-3σ shRNA (1 & 2) or control Luciferase shRNA (C). Lysates were immunoprecipitated with anti-COP1 and immunoblotted with anti-ubiquitin.
D, Flag-COP1 and Flag-COP1 RING mutant (C136S, C139S) proteins were made using an in vitro Transcription/Translation system (TNT) and then incubated with the indicated purified GST proteins. Polyubiquitinated COP1 was examined by immunoprecipitating with anti-Flag, followed by immunoblotting with anti-ubiquitin (middle panel). The bottom panel shows the input of GST proteins (Coomassie Blue staining).
14-3-3σ antagonizes COP1-mediated p53 degradation
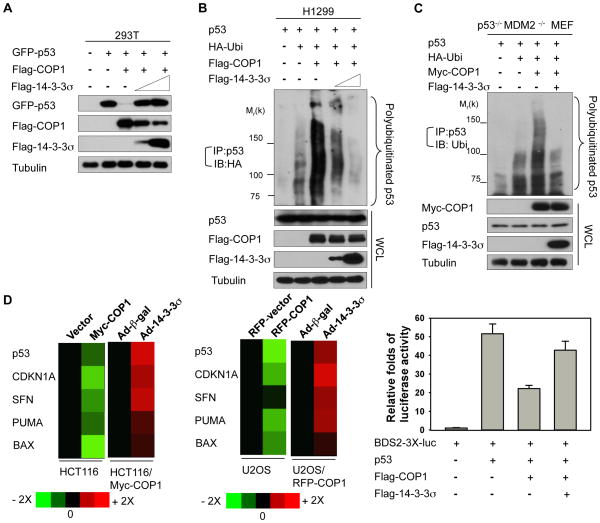
Because p53 tumor suppressor is an ubiquitination target of COP1 (22), we further evaluated whether 14-3-3σ–mediated COP1 downregulation has an impact on p53 stability and ubiquitination. We first showed that COP1 was able to reduce steady-state levels of p53 and that 14-3-3σ antagonized this process in a dose-dependent manner (Fig. 4A). Next, we found that increasing amounts of 14-3-3σ antagonized COP1-mediated polyubiquitination of p53 in a dose-dependent manner (Fig. 4B). To exclude the contribution of MDM2 in the polyubiquitination of p53 in this setting since 14-3-3σ is able to block MDM2-mediated p53 ubiquitination, we performed similar experiments by transfecting p53−/− MDM2−/− MEF cells and achieved similar results: 14-3-3σ reduced COP1-mediated polyubiquitination of p53 even in the absence of MDM2 (Fig. 4C). Subsequently, 14-3-3σ can accelerate the turnover of COP1, which translates into decelerating p53 turnover in p53−/− MDM2−/− MEF cells (Fig. S2). These data suggest that 14-3-3σ-COP1 axis is independent of 14-3-3σ-MDM2 axis in terms of p53 ubiquitination.
Figure 4. 14-3-3σ antagonizes COP1-mediated p53 degradation.
A, 293T cells were cotransfected with the indicated plasmids. Lysates were immunoblotted with the indicated antibodies.
B, H1299 cells were cotransfected with the indicated plasmids. Cells were treated with MG132 before harvesting. The polyubiquitinated p53 were immunoprecipitated with anti-p53 and immunoblotted with anti-HA. Lysates were also immunoblotted with the indicated antibodies.
C, p53−/−MDM2−/− MEF cells were cotransfected with the indicated plasmids. Cells were treated with MG132 before harvesting. Polyubiquitination of p53 was detected by immunoprecipitation with anti-p53 and immunoblotting with anti-ubiquitin. Lysates were also immunoblotted with the indicated antibodies.
D, The mRNA levels of the indicated p53 target genes, including CDKN1A, SFN, PUMA, and BAX, were detected by real-time PCR in indicated cells stably expressing the control vector, Myc-COP1, or RFP-COP1 and normalized to GAPDH mRNA levels. Cells were also infected with the indicated Ad-β-gal (control) or Ad-14-3-3σ. The expression levels of the indicated p53 target genes were measured, and the data are presented as a heat map (Left). Statistic analysis for the heap map using Student’s t-test, p<0.05. The BDS2-3X-luc reporter containing a p53-responsive element was transfected with the indicated plasmids. Relative luciferase activity was shown (Right). Error bars represent 95% confidence intervals.
To address the biological consequence of 14-3-3σ’s impact on COP1-mediated p53 degradation/ubiquitination, we sought to analyze the gene expression of p53 targets. We demonstrated that overexpressing Myc-COP1 or RFP-COP1 decreased the gene expression of p53 transcriptional targets, including CDKN1A, SFN, BAX, and PUMA, as measured by quantitative PCR in two p53-positive cell lines (colon carcinoma HCT116 and osteosarcoma U2OS), while adenoviral overexpression of 14-3-3σ in these COP1-overexpressing cells increased the p53 target gene expression when compared with adenoviral overexpression of β-galactosidase in controls (Fig. 4D, left panel). To verify the role of COP1 axis in 14-3-3σ’s positive impact on p53 target genes expression, we demonstrated that 14-3-3σ’s activity in upregulating p53 transcriptional targets was compromised in COP1-depleted cells (Fig. S3). Also, in a luciferase reporter gene assay 14-3-3σ antagonized COP1-mediated p53 transcriptional repression (Fig. 4D, right panel). Therefore, 14-3-3σ’s negative impact on COP1 can be translated into reducing p53 ubiquitination/degradation as well as increasing p53 transcriptional activity.
14-3-3σ suppresses COP1-mediated proliferation, survival, and anchorage-independent growth
Since COP1 can antagonize the activity of p53, COP1 may have a role in cell cycle regulation. We showed that stably expressing Myc-COP1 promoted cell growth of HCT116 cells as determined by MTT assay (Fig. 5A). The infection of Ad-14-3-3σ in these COP1-overexpressing HCT116 cells inhibited the increase in the number of live cells when compared with Ad-β-gal controls (Fig. 5A). COP1’s impact on p53 has led to COP1-expressing cells resistant to IR-inducing cell death (2); therefore, we investigated whether 14-3-3σ can sensitize COP1-expressing HCT116 cells to IR-mediated cell death. Importantly, Myc-COP1-expressing HCT116 cells infected with Ad-14-3-3σ had a greater reduction in the number of live cells than did Ad-β-gal-infected cells when cells were treated with IR (Fig. 5B, right two bars). The result indicates that 14-3-3σ can potentiate IR-mediated cell death in COP1-expressing HCT116 cells.
Figure 5. 14-3-3σ suppresses COP1-mediated proliferation, survival, and anchorage-independent growth.
A, The number of live HCT116/Myc-COP1 cells, HCT116/vector control cells, COP1-expressing cells infected with Ad-β-gal or Ad-14-3-3σ, was estimated by MTT assay every day for 7 days. Optical density at 570 nm (OD570) absorbance of each group is shown. This experiment was done in triplicate. Error bars represent 95% confidence intervals.
B, HCT116/Myc-COP1 cells were infected with Ad-β-gal or Ad-14-3-3σ on day 1. Infected cells were treated with or without 10 Gy IR on day 2. Live cells were detected as dye-excluding cells and counted after trypan blue staining on day 4. The bar graph shows the percentage of live cells. Error bars represent 95% confidence intervals.
C, Indicated cells were analyzed for cell cycle distribution using fluorescence-activated cell sorting (left panel). HCT116/Myc-COP1 cells were left uninfected (-) or infected with Ad-β-gal or Ad-14-3-3σ, followed by fluorescence-activated cell sorting for cell cycle analysis. The percentages of representative cells in each phase of the cell cycle are shown for each treatment group (right panel). This experiment was done in triplicate.
D, Indicated cells were analyzed for foci formation. HCT116/Myc-COP1 cells infected with Ad-β-gal or Ad-14-3-3σ were subjected to foci formation (left panel). The bar graphs show the average of the foci numbers in these cells. Error bars represent 95% confidence intervals. HCT116/Myc-COP1 and HCT116/vector cells were analyzed in a soft agar assay, as were HCT116/Myc-COP1 cells infected with Ad-β-gal or Ad-14-3-3σ (right panel). The average number of colonies per plate is represented. Error bars represent 95% confidence intervals.
Fluorescence-activated cell sorting analysis showed an average of 28% of Myc-COP1-overexpressing HCT116 cells were in the S phase, while only 18% of HCT116/vector control cells were in the S phase. This indicates that the overexpression of COP1 promotes progression to S phase (Fig. 5C, left panels). To investigate whether 14-3-3σ affects the cell cycle distribution of COP1-expressing HCT116 cells, we found that the average percentage of cells in the G2 phase was higher for Ad-14-3-3σ-infected COP1-expressing HCT116 cells than for Ad-β-gal-infected control cells and untreated control cells (Fig. 5C, right panels). These data suggest that 14-3-3σ can inhibit COP1-mediated cell proliferation by causing G2 arrest.
More numbers of foci were observed in Myc-COP1-overexpressing cells than in control cells (Fig. 5D, left panel). Infecting these Myc-COP1-overexpressing cells with Ad-14-3-3σ reduced the numbers of foci in these cells compared with the numbers in Ad-β-gal-infected control cells (Fig. 5D, left panel). Similar results were obtained for anchorage-independent growth as assessed by a soft agar colony formation assay (Fig. 5D, right panel). Therefore, these functional analyses demonstrate the important roles of the 14-3-3σ-COP1 axis in cancer cell proliferation, IR-mediated cell death, and cell transformation.
14-3-3σ suppresses COP1-promoted tumorigenicity
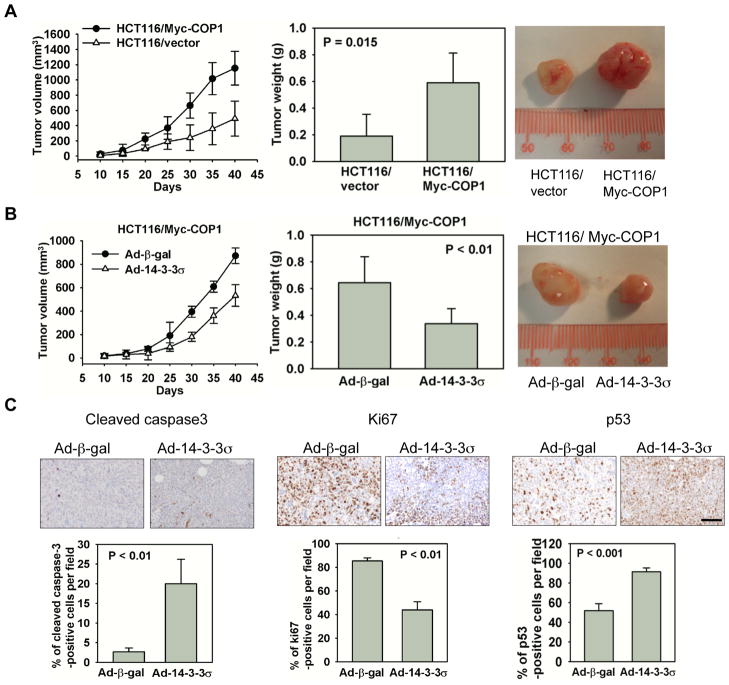
To confirm the above results in vivo, we examined the impact of COP1 overexpression on tumorigenicity in a nude mouse xenograft model. HCT116 cells stably transfected with either control plasmid (empty vector) or plasmid expressing Myc-COP1 were subcutaneously injected into the flanks of nude mice. COP1-overexpressing HCT116 cells have a faster tumor growth rate than HCT116 control cells (Fig. 6A, left panel). At the end of the experiments, the mice were euthanized and the xenografts were removed and measured. Myc-COP1-expressing HCT116 xenografts grew to an average weight of 0.590 g, while control xenografts grew to an average weight of only 0.190 g (Fig. 6A, middle panel), indicating that Myc-COP1 HCT116 cells formed larger tumors than HCT116/vector control cells (Fig. 6A, right panel).
Figure 6. 14-3-3σ suppresses COP1-promoted tumorigenicity.
A, Myc-COP1-overexpressing HCT116 stable transfectants and HCT116/vector control transfectants were injected subcutaneously into the flank regions of female nude mice. Tumor volumes were monitored for 40 days. Tumor growth curves are shown (left panel). Error bars represent 95% confidence intervals. Tumors were dissected and weighed 40 days after cell injection (middle panel). Bar graph shows tumor weights. Error bars represent 95% confidence intervals. The photograph shows size of representative tumors (right panel).
B, Myc-COP1-overexpressing HCT116 stable transfectants were infected with Ad-14-3-3σ and Ad-β-gal. Cells were injected subcutaneously into the flanks of nude mice. Tumor growth curves were plotted and tumor weights and sizes were measured in the same manner as in (A). Error bars represent 95% confidence intervals. Tumors were dissected and weighed (bar graph) 40 days after injection. Error bars represent 95% confidence intervals. The photograph shows size of representative tumors obtained.
C, Serial tumor sections from (B) experiment were immunostained with anti-cleaved caspase-3 (left panel), anti-Ki67 (middle panel), and anti-p53 (right panel). Bar graphs show the percentage of cells with positive signals from each staining. Error bars represent 95% confidence intervals. Scale bar, 50 μm.
Since 14-3-3σ had affected the growth of COP1-expressing cells (Fig. 5A), we sought to determine whether 14-3-3σ expression can antagonize COP1-promoted tumor growth. The infection of Ad-14-3-3σ in these COP1-overexpressing HCT116 cells decelerated the xenograft tumor growth when compared with infection of Ad-β-gal controls (Fig. 6B, left panel). The Ad-14-3-3σ infected tumors later removed from the mice were smaller than the ones formed by Ad-β-gal-infected cells (Figure 6B, middle and right panels), with average tumor weights of 0.337 g and 0.618 g, respectively. Xenografts were fixed and processed for immunohistochemistry. Levels of the proliferation marker Ki67 were lower and levels of p53 and the apoptotic marker cleaved caspase-3 were higher in Myc-COP1 HCT116 cells infected with Ad-14-3-3σ than in the control Ad-β-gal-infected cells (Fig. 6C). Thus, COP1 overexpression accelerates tumor formation and increases tumor size in vivo, but 14-3-3σ antagonizes COP1-promoted tumorigenicity.
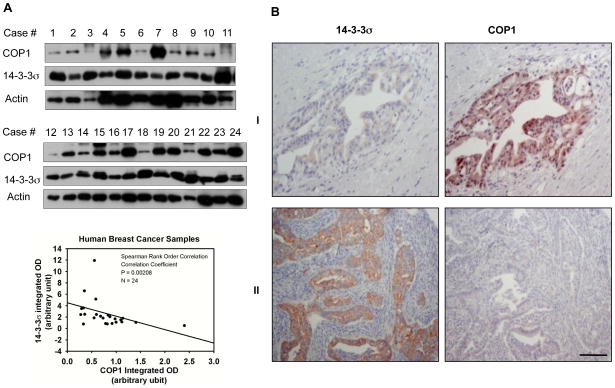
COP1 and 14-3-3σ are inversely correlated in primary breast cancer and pancreatic cancer specimens
We analyzed 14-3-3σ and COP1 protein expressions in 24 human primary breast cancer specimens by immunoblot analysis. 14-3-3σ expression was inversely correlated with COP1 expression (p < 0.001; Fig. 7A). We further studied this inverse relationship between 14-3-3σ and COP1 by examining 121 human pancreatic cancer specimens. High COP1 expression was detected in 40 (72.2%) of the 55 pancreatic cancer specimens with low 14-3-3σ expression, and COP1 expression was inversely correlated with 14-3-3σ expression (p < 0.005; Fig. 7B, Supplementary Table S1). Therefore, the inverse relationship between COP1 and 14-3-3σ was detected in at least two types of human cancer, suggesting that the negative regulation of COP1 by 14-3-3σ may be a clinically relevant regulatory pathway in human cancers.
Figure 7. COP1 and 14-3-3σ are inversely correlated in primary breast cancer and pancreatic cancer specimens.
A, Protein levels of COP1 and 14-3-3σ were analyzed by immunoblotting in malignant human breast cancer specimens. The integrated optical intensity of COP1 was plotted against that of 14-3-3σ. Spearman rank order correlation was used to demonstrate the inverse correlation between COP1 and 14-3-3σ protein levels (p < 0.001). Spearman correlation=−0.61.
B, Pancreatic cancer specimens were immunostained with anti-14-3-3σ and anti-COP1. Two representative cases of the 121 examined are shown. Consecutive tissue sections from tumor case I showed a low level of 14-3-3σ but a high level of COP1. In contrast, consecutive sections from tumor case II showed abundant 14-3-3σ but scanty COP1 (original magnification, 200×). Scale bar, 50 μm.
Discussion
COP1 drives the ubiquitination and proteasomal degradation of p53, thereby maintaining low steady-state levels of p53 in unstressed cells (22). COP1 is downregulated in response to DNA damage (9), but detailed mechanistic regulation remains to be characterized. In this study, our results indicate that 14-3-3σ is a negative regulator for COP1 in response to DNA damage.
At issue is how p53 protein accumulates via regulation of COP1 after DNA damage. It is conceivable that the inhibition of p53 E3 ligases is an important step for p53 stabilization after DNA damage, but it is not clear how COP1 behaves in response to DNA damage. Here we are able to provide critical insights. First, our results show that COP1 downregulation correlates with the induction of 14-3-3σ, suggesting that 14-3-3σ may have an impact on the expression of COP1. The impact of 14-3-3σ on COP1 is here documented by showing that 14-3-3σ deficiency leads to compromised downregulation of COP1 in response to DNA damage (Fig. 1B) and reduced turnover rate of COP1 (Fig. 1D). Second, 14-3-3σ associates with COP1 directly and the interaction between 14-3-3σ and COP1 is mapped to the coiled-coil domain (Fig. 2C), which is different from other COP1-associated proteins such as c-JUN (3–5) and FOXO1 (7). JUN and FOXO1 bind to COP1 at the WD40 repeats and are substrates degraded by COP1. The major function of WD40 motifs is to mediate the interaction between the E3 ligase and its substrates (23) to facilitate the degradation of substrates. In our study, 14-3-3σ does not bind to WD40 motif. One may speculate that 14-3-3σ binds to the coiled-coil region of COP1 to regulate the activity of COP1 as opposed to act as a substrate of COP1. Because the coiled-coil region is known to be a protein-protein interaction domain and is involved in dimerization of COP1 (24), it is possible that 14-3-3σ affects the self-association of COP1 or interferes with the binding of other COP1-associated proteins, thus potentially affecting COP1’s activity. Such a possibility remains to be investigated. We recently identify that 14-3-3σ preferentially binds to S387 of COP1 (25). This site (RTAS387QL) is indeed located within the fragment containing coiled-coil region (Fig. 2C). Third, 14-3-3σ is essential for the ubiquitination of COP1 as 14-3-3σ deficiency compromises ubiquitination of COP1 (Fig. 3), thereby decelerating turnover rate of COP1 (Fig. 1). In vitro ubiquitination study indicates that COP1 is self-ubiquitinated and 14-3-3σ facilitates such a process (Fig. 3D). These mechanistic studies provide insights into how 14-3-3σ inversely correlates with the expression of COP1 in response to DNA damage (Fig. 1A) and how loss of 14-3-3σ attenuates the DNA damage-mediated downregulation of COP1 (Fig. 1B). Consequently, 14-3-3σ impedes the COP1’s important biological function—p53 ubiquitination (Fig. 4B). Importantly, COP1 ubiquitinates p53 in a MDM2-independent manner (Fig. 4C), and this process can still be inhibited by the expression of 14-3-3σ (Fig. 4C). These data suggest that the COP1-p53 axis is independent of the MDM2-p53 axis in terms of regulating p53 ubiquitination. Together, it is important to point out that 14-3-3σ regulates both the MDM2-p53 axis (our previous study) (15) and the COP1-p53 axis (this study). These findings highlight the complexity of the p53 ubiquitination process and demonstrate that 14-3-3σ exerts its negative impacts on two p53 ubiquitin ligases to stabilize p53. Lastly, in addition to previous observation that COP1 is overexpressed in a high percentage of ovarian adenocarcinoma (2), we have now shown that COP1 is also deregulated in pancreatic cancer and breast cancer samples (Fig. 7). Together, these data suggest that COP1 may have a role in carcinogenesis. The deregulated expression of COP1 could be a potential pathological effect in other types of cancer, which remains to be investigated. However, how COP1 executes its oncogenic activity has not been demonstrated. For the first time, we have shown that COP1 overexpression leads to increased cell proliferation, cell transformation, cell survival, and promoting tumor formation (Fig. 5 and 6), attesting to its oncogenic role. Significantly, 14-3-3σ is able to antagonize these activities of COP1 (Fig. 5 and 6). Consistently, our clinical sample studies indicate that COP1 is overexpressed in a high percentage of the pancreatic cancer samples in which the expression of 14-3-3σ is low. Also, our set of breast cancer samples shows an inverse correlation between the protein levels of COP1 and 14-3-3σ. Together, our results demonstrate that COP1 promotes tumorigenesis. This report fills this gap in knowledge by demonstrating the inverse relationship between COP1 and 14-3-3σ in response to DNA damage, the molecular mechanism of 14-3-3σ in regulating COP1 ubiquitination and protein stability, and the biological significance of this 14-3-3σ-COP1 regulatory process in tumorigenesis. Therefore, our research paves the path to future investigation of the 14-3-3σ-COP1 axis as a target for novel anticancer therapies.
Supplementary Material
Acknowledgments
We are grateful to Drs. Y.Y. Wen, E. Bianchi, and R. Pardi for material support. We thank Dr. Zhenbo Han and Bill Spohn for microscopy support. This work was supported by grants from the National Institutes of Health (NIH) (R01CA089266), Directed Medical Research Programs (DOD SIDA BC062166 to S.J.Y. and M.H.L.) and Susan G. Komen Breast Cancer Foundation (KG081048). The University of Texas M. D. Anderson Cancer Center is supported by NIH core grant CA16672.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. Embo J. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dornan D, Bheddah S, Newton K, et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64:7226–30. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 3.Wertz IE, O’Rourke KM, Zhang Z, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–4. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 4.Savio MG, Rotondo G, Maglie S, Rossetti G, Bender JR, Pardi R. COP1D, an alternatively spliced constitutive photomorphogenic-1 (COP1) product, stabilizes UV stress-induced c-Jun through inhibition of full-length COP1. Oncogene. 2008;27:2401–11. doi: 10.1038/sj.onc.1210892. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi E, Denti S, Catena R, et al. Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J Biol Chem. 2003;278:19682–90. [Google Scholar]
- 6.Dentin R, Liu Y, Koo SH, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–9. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 7.Kato S, Ding J, Pisck E, Jhala US, Du K. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J Biol Chem. 2008;283:35464–73. doi: 10.1074/jbc.M801011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li DQ, Ohshiro K, Reddy SD, et al. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc Natl Acad Sci U S A. 2009;106:17493–8. doi: 10.1073/pnas.0908027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dornan D, Shimizu H, Mah A, et al. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science. 2006;313:1122–6. doi: 10.1126/science.1127335. [DOI] [PubMed] [Google Scholar]
- 10.Laronga C, Yang HY, Neal C, Lee MH. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J Biol Chem. 2000;275:23106–12. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- 11.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 12.Michaud NR, Fabian JR, Mathes KD, Morrison DK. 14-3-3 is not essential for Raf-1 function: identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–7. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–71. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 14.Prasad GL, Valverius EM, McDuffie E, Cooper HL. Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 1992;3:507–13. [PubMed] [Google Scholar]
- 15.Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol. 2003;23:7096–107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang HY, Wen YY, Lin YI, et al. Roles for negative cell regulator 14-3-3sigma in control of MDM2 activities. Oncogene. 2007;26:7355–62. doi: 10.1038/sj.onc.1210540. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Zhang Y, Zhao R, et al. Negative cell cycle regulator 14-3-3sigma stabilizes p27 Kip1 by inhibiting the activity of PKB/Akt. Oncogene. 2006;25:4585–94. doi: 10.1038/sj.onc.1209481. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Wen YY, Zhao R, et al. DNA damage-induced protein 14-3-3 sigma inhibits protein kinase B/Akt activation and suppresses Akt-activated cancer. Cancer Res. 2006;66:3096–105. doi: 10.1158/0008-5472.CAN-05-3620. [DOI] [PubMed] [Google Scholar]
- 19.Verdoodt B, Benzinger A, Popowicz GM, Holak TA, Hermeking H. Characterization of 14-3-3sigma dimerization determinants: requirement of homodimerization for inhibition of cell proliferation. Cell Cycle. 2006;5:2920–6. doi: 10.4161/cc.5.24.3571. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Liu JY, Zhang JT. 14-3-3sigma, the double-edged sword of human cancers. Am J Transl Res. 2009;1:326–40. [PMC free article] [PubMed] [Google Scholar]
- 21.Hermeking H, Lengauer C, Polyak K, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 22.Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 23.Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. Embo J. 2001;20:118–27. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torii KU, McNellis TW, Deng XW. Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. Embo J. 1998;17:5577–87. doi: 10.1093/emboj/17.19.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su C, Zhao R, Velazquez-Torres G, et al. Nuclear export regulation of COP1 by 14-3-3 sigma in response to DNA damage. Molecular Cancer. 2010;9:243. doi: 10.1186/1476-4598-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.