Abstract
In February 2006, a patient colonized with a multidrug-resistant sequence type 56 Acinetobacter baumannii strain was admitted to a hospital in Madrid, Spain. This strain spread rapidly and caused a large outbreak in the hospital. Clinicians should be alert for this strain because its spread would have serious health consequences.
Keywords: Acinetobacter baumannii, bacteria, bacteremia, multidrug resistance, OXA-24 carbapenemase, plasmid, septicolysin, Spain, dispatch
The increasing resistance of Acinetobacter baumannii to antimicrobial drugs, including carbapenems (1–3), and resistance to desiccation and disinfectants (4) contribute to its persistence in hospital environments and propensity to cause outbreaks (5,6). In February 2006, a patient colonized with a multidrug-resistant A. baumannii strain was admitted to the medical–surgical intensive care unit (ICU) of a hospital in Madrid, Spain. This strain then spread rapidly, persisted for >30 months, and caused a large outbreak in the hospital. We report details of this outbreak.
The Study
We conducted a retrospective longitudinal study at 12 de Octubre University Hospital, Madrid, Spain, of patients colonized/infected with A. baumannii during January 2006–May 2008. We also conducted a cohort study of patients with A. baumannii bacteremia during January 2002–May 2008.
MICs of drugs were confirmed by using Etest strips (AB Biodisk, Solna, Sweden) according to the manufacturer’s criteria. Multidrug-resistant (MDR) phenotypes were defined as resistance to 5 classes of drugs: antipseudomonal cephalosporins (ceftazidime, cefepime), carbapenems (imipenem, meropenem), piperacillin/tazobactam, fluoroquinolones, and aminoglycosides (gentamicin, tobramycin, amikacin). Isolates were classified on the basis of antimicrobial susceptibility patterns: antibiotype 1, MDR isolates; antibiotype 2, isolates resistant to carbapenems but not MDR; and antibiotype 3, isolates susceptible to carbapenems. Colonization was defined as isolation of A. baumannii from >1 clinical specimen in the absence of clinical symptoms consistent with infection. Bacteremia was determined by application of criteria proposed by the Centers for Disease Control and Prevention (Atlanta, GA, USA) (7).
Clonal relatedness between clinical isolates was determined by using pulsed-field gel electrophoresis (PFGE) and the CHEF DRIII system (Bio-Rad Laboratories, Hercules, CA, USA) according to reported techniques (8). Migration of DNA fragments was normalized, and computer-assisted analysis of PFGE patterns was conducted by using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium). Multilocus sequence typing (MLST) was performed according to published protocols (9). Isolates were assigned to a sequence type according to the allelic profiles database (http://pubmlst.org/abaumannii/). Univariate analysis was performed by using the t test for continuous variables and the χ2 or Fisher exact tests for categorical variables. Adjusted odds ratios (ORs) were calculated by using logistic regression analysis. Data were analyzed by using SPSS software (SPSS Inc., Chicago, IL, USA). A p value <0.05 was considered significant.
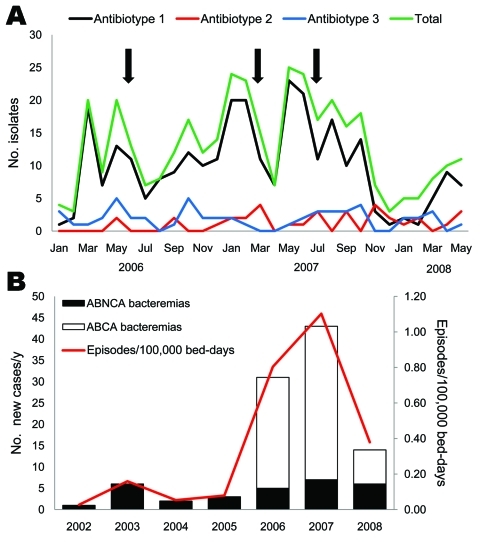
During January 2006–May 2008, a total of 377 patients were colonized/infected with A. baumannii. Mean age of the patients was 57 years and 63.4% were men. Patients were hospitalized mostly in ICUs (184, 48.8%), and in surgical (100, 26.5%), medical (85, 22.5%), and pediatric (8, 2.1%) wards. A total of 76.9% (290/377) of the isolates were antibiotype 1, 9.0% (34/377) were antibiotype 2, and 14.1% (53/377) were antibiotype 3. Temporal distribution of cases is shown in Figure 1, panel A. Bacterial isolates of antibiotype 1 were assigned to the major clonal type (clone AbH12O-A2) by PFGE. Of 290 patients with A. baumannii antibiotype 1 isolates (clone AbH12O-A2), 165 patients were infected (57%) and 125 (43%) were colonized.
Figure 1.
Temporal distribution of patients with Acinetobacter baumannii infections, Spain. A) Patients colonized/infected with A. baumannii classified by antibiotype. Arrows indicate times of intensification of infection control measures. The medical–surgical intensive care unit at Octubre University Hospital, Madrid, Spain, was refurbished in July 2007. B) Annual incidence of A. baumannii bacteremia. ABCA, A. baumannii clone A or AbH12O-A2; ABNCA: A. baumannii nonclone A.
MLST analysis of 3 isolates belonging to clone AbH12O-A2 was performed to determine the relationship between these isolates and other described strains. The 3 isolates showed the same allelic profile of 7 housekeeping genes (allele no. in brackets; gltA [1], gyrB [18], gdhB [18], recA [10], cpn60 [14], gpi [29], and rpoD [18]) and were identified as sequence type 56 according to the MLST database (http://pubmlst.org/abaumannii/).
A. baumannii clone AbH12O-A2, which showed a broad antimicrobial drug-resistance profile, resistance to carbapenems, and susceptibility only to tigecycline and colistin, was present throughout the entire 30-month study and peaked several times until the medical–surgical ICU was refurbished in July 2007. The number of new case-patients with clone AbH12O-A2 then decreased; <3 cases/month were observed during October 2007–February 2008 (Figure 1, panel A).
Annual incidence of A. baumannii bacteremia increased from 0.03 episodes/100,000 bed days in 2002 to 1.1/100,000 bed days in 2007 (Figure 1, panel B), which coincided with the outbreak peak caused by clone AbH12O-A2. Clinical features of patients with A. baumannii bacteremia are shown in Table 1. Multivariate analysis of bacteremia caused by clone AbH12O-A2 and nonclone AbH12O-A2 showed that variables independently associated with AbH12O-A2 bacteremia were hospitalization in ICUs (OR 3.48, 95% confidence interval [CI] 1.23–9.54), exposure to >3 antimicrobial drugs (OR 3.13, 95% CI 1.12–8.76), and ventilator-associated pneumonia as the source of bacteremia (OR 8.35, 95% CI 1.12–8.76).
Table 1. Clinical characteristic of patients with Acinetobacter baumannii bacteremia, Spain*.
| Characteristic | Clone ABCA, n = 65 | Clone ABNCA, n = 29 | p value | OR (95% CI) |
|---|---|---|---|---|
| Age, y | 57.5 ± 14.2 | 58.7 ± 19.6 | 0.730 | NA |
| Male sex |
50 (76.9) |
21 (72.4) |
0.639 |
1.27 (0.47–3.45) |
| Concurrent conditions | ||||
| Immunosuppression | 12 (18.5) | 6 (20.7) | 0.800 | 0.87 (0.29–2.60) |
| Solid tumor | 16 (24.6) | 6 (20.7) | 0.678 | 1.25 (0.43–3.62) |
| Hematologic malignancy | 1 (1.5) | 1 (3.4) | 0.553 | 0.44 (0.03–7.25) |
| Diabetes mellitus | 9 (13.8) | 9 (31.0) | 0.050 | 0.36 (0.12–1.03) |
| Liver cirrhosis | 11 (16.9) | 3 (10.3) | 0.408 | 1.76 (0.45–6.88) |
| Heart failure | 4 (6.2) | 3 (10.3) | 0.475 | 0.57 (0.12–2.72) |
| Chronic obstructive pulmonary disease | 7 (10.8) | 3 (10.3) | 0.951 | 1.05 (0.25–4.37) |
| Liver transplant |
15 (23.1) |
7 (24.1) |
0.911 |
0.94 (0.34–2.64) |
| Duration of hospitalization before A. baumannii bacteremia, d |
34.8 ± 36.1 |
23.9 ± 27.5 |
0.150 |
NA |
| Hospital location | ||||
| Intensive care unit | 41 (63.1) | 9 (31.0) | 0.004 | 3.80 (1.50–9.66) |
| Medical ward | 6 (9.2) | 12 (41.4) | 0.001 | 0.14 (0.05–0.44) |
| Surgical ward |
18 (27.7) |
8 (27.6) |
0.992 |
1.00 (0.38–2.68) |
| Source of bacteremia | ||||
| Catheter-related infection | 25 (38.5) | 9 (31.0) | 0.489 | 1.39 (0.54–3.52) |
| Pneumonia associated with mechanical ventilation | 18 (27.7) | 1 (3.4) | 0.006 | 10.72 (1.36–84.8) |
| None (primary bacteremia) | 12 (18.5) | 14 (48.3) | 0.003 | 0.24 (0.09–0.63) |
| Intraabdominal infection | 7 (10.8) | 2 (6.9) | 0.716 | 1.62 (0.32–8.37) |
| Urinary tract infection | 3 (4.6) | 2 (6.9) | 0.642 | 0.65 (0.10–4.13) |
| Other |
0 |
1 (3.4) |
0.309 |
3.32 (2.43–4.52) |
| Carbapenem resistance |
65 (100.0) |
7 (24.1) |
0.001 |
0.09 (0.50–0.20) |
| Prior colonization with A. baumannii |
43/62 (69.4) |
1/17 (5.9) |
0.001 |
36.21(4.47–293.1) |
| Antimicrobial drugs used | ||||
| Cephalosporin | 7/62 (11.3) | 3/29 (10.3) | 0.893 | 1.10 (0.26–4.61) |
| Piperacillin/tazobactam | 21/62 (33.9) | 4/29 (13.8) | 0.046 | 3.20 (0.98–10.41) |
| Fluorquinolone | 24/62 (38.7) | 9/29 (31.0) | 0.478 | 1.40 (0.54–3.59) |
| Glycopeptide | 44/62 (71.0) | 12/29 (41.4) | 0.007 | 3.46 (1.38–8.69) |
| Aminoglycoside | 17/62 (27.4) | 8/29 (27.6) | 0.987 | 0.99 (0.37–2.66) |
| Carbapenem | 41/62 (66.1) | 11/29 (37.9) | 0.011 | 3.20 (1.28–7.99) |
|
>3 drugs |
36/62 (58.1) |
8/29 (27.6) |
0.007 |
3.63 (1.40–9.47) |
| Invasive procedure or device | ||||
| Central venous catheter† | 51/64 (79.7) | 15/29 (51.7) | 0.006 | 3.66 (1.42–9.46) |
| Surgical procedure‡ | 33/64 (51.6) | 11/29 (37.9) | 0.223 | 1.74 (0.71–4.27) |
| Mechanical ventilation† |
49/64 (76.6) |
14/29 (48.3) |
0.007 |
3.50 (1.38–8.87) |
| Duration of hospitalization after A. baumannii bacteremia, d | 46.6 ± 72.9 | 20.5 ± 21.2 | 0.050 | NA |
| Died during hospitalization | 35 (53.8) | 9 (31.0) | 0.041 | 2.59 (1.03–6.54) |
*Values are mean ± SD or no. (%) except as indicated. Clone ABCA, A. baumannii clone A (AbH12O-A2); ABNCA, A. baumannii nonclone A; OR, odds ratio; CI, confidence interval; NA, not applicable. †Week before bacteremia. ‡Month before bacteremia.
Plasmid pMMA2 (GenBank accession no. GQ377752), which was isolated from the clone causing the outbreak (AbH12O-A2), harbored a blaOXA-24 gene (10) coding for carbapenemase OXA-24 (also called OXA-40) as described (11). Four additional clones were detected during the outbreak (AbH12O-D, AbH12O-CU1, AbH12O-CU2, and AbH12O -CU3), which harbored plasmids pMMD, pMMCU1, pMMCU2, and pMMCU3, respectively (GenBank accession nos. GQ904226, GQ342610, GQ476987, and GQ904227). Carbapenem resistance in all clones was linked to a plasmid harboring the blaOXA-24 gene flanked by XerC/XerD-like recombination sites (11). Comparative analysis among plasmid sequences showed different patterns and coding regions. All plasmids, including pMMA2, harbored the blaOXA-24 gene as part of a DNA module flanked by XerC/XerD-like sites, which suggested that these sites are involved in mobilization of DNA containing the bla OXA-24 gene by site-specific recombination (11).
Two genes with a putative role in virulence were detected in plasmids from clones AbH12O-A2 and AbH12O-CU3 upstream of blaOXA-24: a septicolysin-like gene coding for a pore-forming toxin (12), and a TonB-dependent receptor gene coding for an outer membrane protein involved in iron uptake and virulence (13–15). Insertion sequence 4, which provided an additional promoter sequence, was detected upstream from the septicolysin gene in plasmid pMMA2; this sequence was absent in plasmid pMMCU3 (Figure 2). Two nucleotide changes detected in promoter regions provided an additional promoter region for the TonB-dependent receptor gene in plasmid pMMA2.
Figure 2.
Nucleotide sequence of the region between the septicolysin and Ton-B dependent receptor genes of Acinetobacter baumannii in plasmids pMMA2 and pMMCU3 from clone AbH12O-A2 (upper panel) and AbH12O-CU3 (lower panel), respectively. Integrated insertion sequence 4 (IS4) (red letters) provided a new promoter sequence for septicolysin in plasmid pMMA2 from clone AbH12O-A2. Upper case letters indicate amino acids. IRL, inverted repeated left sequence; IRR, inverted repeated right sequence from IS4; Stop, stop (termination) codon.
Real-time PCR (Table 2) was performed to analyze expression of septicolysin and TonB-dependent receptor genes in clones AbH12O-A2 and AbH12O-CU3. Expression of septicolysin in clone AbH12O-A2 was 2.1× times higher than that of clone AbH12O-CU3. Conversely, the TonB-dependent receptor was also overexpressed in clone AbH12O-A2 (1.8× higher than in clone AbH12O-CU3).
Table 2. Oligonucleotides used in real-time reverse transcription PCRs for Acinetobacter baumannii, Spain*.
| Primer | Gene | Sequence, 5′ → 3′ |
|---|---|---|
| TonB-Forw | TonB-dependent receptor | GGACTGGTGATAAAGCACTAT |
| TonB-Rev | TonB-dependent receptor | GCCGCATAGAGTTATCACATC |
| Septicolysin-Forw | Septicolysin | CACCATCTTGTACCAATACATTT |
| Septicolysin-Rev | Septicolysin | GAAATTAGCAGAAGCTCTCTTAC |
| rpoB-Forw | RNA polymerase subunit B | CAGCCGCGAYCAGGTTGACTACA |
| rpoB-Rev | RNA polymerase subunit B | GACGCACCGCAGGATACCACCTG |
| gyrB-Forw | DNA gyrase subunit B | AAGTGAGGTAAAACCAGCGGTA |
| gyrB-Rev | DNA gyrase subunit B | AATCTTGCCTGCAATTGATTTT |
*Forw, forward; rev, reverse.
Conclusions
Outbreaks of MDR A. baumannii have been demonstrated in many studies (1,2,5). We report a large outbreak during 2006–2008 that persisted for >30 months. The AbH12O-A2 strain was pathogenic and caused 65 cases of bacteremia.
Clone AbH12O-A2 had unique characteristics. First, it was an MDR (including carbapenems) clone (ST56), susceptible only to tigecycline and colistin. Second, it harbored a carbapenemase blaOXA-24 gene, flanked by XerC/XerD binding sites located on a plasmid, which probably spread to other Acinetobacter clones by a Xer recombination system (11). Third, this clone overexpressed 2 putative virulence factors, septicolysin and TonB-dependent receptor.
The septicolysin gene showed 2× overexpression caused by insertion of IS4, which provided an additional promoter. Although the exact role of septicolysin is unknown, it has been designated a cholesterol-dependent cytolysin, which has been reported to be produced by pathogenic bacteria such as Clostridium perfringens, Bacillus anthracis, and Streptococcus pneumoniae to aid invasion of tissues or cells (12).
The protein produced by the TonB-dependent receptor gene has been associated with virulence and iron uptake in A. baumannii (13) and may be involved in survival of bacteria in the lungs and blood. This characteristic may explain the large rate of bacteremia caused by clone AbH12O-A2. Thus, clinicians should be alert for the MDR ST56 A. baumannii clone because its spread would have serious health consequences.
Acknowledgments
This study was supported by the Spanish Network for Research in Infectious Diseases (grants RD06/0008/0011, RD06/0008/0025, PI081613, and PS09/00687) for the Instituto de Salud Carlos III.
Biography
Dr Acosta is a clinical microbiologist at Hospital 12 de Octubre, Madrid, Spain. Her primary research interests are epidemiology of nosocomial infections and mechanisms of antimicrobial drug resistance.
Footnotes
Suggested citation for this article: Acosta J, Merino M, Viedma E, Poza M, Sanz F, Otero JR, et al. Multidrug-resistant Acinetobacter baumannii harboring OXA-24 carbapenemase, Spain. Emerg Infect Dis [serial on the Internet]. 2011 Jun [date cited]. http://dx.doi.org/10.3201/eid1706.091866
1These authors contributed equally to this article.
References
- 1.del Mar Tomas M, Cartelle M, Pertega S, Beceiro A, Llinares P, Canle D, et al. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin Microbiol Infect. 2005;11:540–6. 10.1111/j.1469-0691.2005.01184.x [DOI] [PubMed] [Google Scholar]
- 2.Corbella X, Montero A, Pujol M, Dominguez MA, Ayats J, Argerich MJ, et al. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii. J Clin Microbiol. 2000;38:4086–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee NY, Lee HC, Ko NY, Chang CM, Shih HI, Wu CJ, et al. Clinical and economic impact of multidrug resistance in nosocomial Acinetobacter baumannii bacteremia. Infect Control Hosp Epidemiol. 2007;28:713–9. 10.1086/517954 [DOI] [PubMed] [Google Scholar]
- 4.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villegas MV, Hartstein AI. Acinetobacter outbreaks, 1977–2000. Infect Control Hosp Epidemiol. 2003;24:284–95. 10.1086/502205 [DOI] [PubMed] [Google Scholar]
- 6.Naas T, Coignard B, Carbonne A, Blanckaert K, Bajolet O, Bernet C, et al. VEB-1 extended-spectrum β-lactamase–producing Acinetobacter baumannii, France. Emerg Infect Dis. 2006;12:1214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–32. 10.1016/j.ajic.2008.03.002 [DOI] [PubMed] [Google Scholar]
- 8.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis–generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4328–35. 10.1128/JCM.43.9.4328-4335.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382–90. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bou G, Oliver A, Martinez-Beltran J. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000;44:1556–61. 10.1128/AAC.44.6.1556-1561.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, et al. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob Agents Chemother. 2010;54:2724–7. 10.1128/AAC.01674-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, et al. The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 2008;10:1765–74. 10.1111/j.1462-5822.2008.01191.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorsey CW, Tolmasky ME, Crosa JH, Actis LA. Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology. 2003;149:1227–38. 10.1099/mic.0.26204-0 [DOI] [PubMed] [Google Scholar]
- 14.Torres AG, Redford P, Welch RA, Payne SM. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun. 2001;69:6179–85. 10.1128/IAI.69.10.6179-6185.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves SA, Torres AG, Payne SM. TonB is required for intracellular growth and virulence of Shigella dysenteriae. Infect Immun. 2000;68:6329–36. 10.1128/IAI.68.11.6329-6336.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]