Abstract
Transgenics using bacterial artificial chromosomes (BACs) offers a great opportunity to look at gene regulation in a developing embryo. The modified BAC containing a reporter inserted just before the translational start site of the gene of interest allows for the visualization of spatio-temporal gene expression. Though this method has been used in the mouse model extensively, its utility in zebrafish studies is relatively new. This review aims to look at the utility of making BAC transgenics in zebrafish and its applications in functional genomics. We look at the various methods to modify the BAC, some limitations and what the future holds.
Introduction
Transgenic assays in animal model systems have been the gold standard for functional testing of various regulatory elements over the years.1–5 The ability of DNA fragments to drive reporter gene expression in a correct spatio-temporal manner and recapitulating endogenous gene expression has led to the discovery of numerous promoters and enhancers.6–9 With the advent of chromatin immunoprecipitation followed by high throughput sequencing (ChIP-Seq) the need for functional validation of binding sites has increased10,11manifold. Functional validation helps to segregate binding sites (DNA fragments) which direct spatio-temporal expression of neighbouring genes from binding sites which might not have a direct role in gene regulation and this is critical in our understanding of transcriptional control and goes a long way in helping to build gene regulatory networks. Though some studies have also utilized the more rapid luciferase assay in cell lines to validate the functionality of a binding site,12–14 in vivo transgenics is still a more powerful and convincing method for such validations especially if working with developmental control genes.
Genome analysis of humans and closely related as well as divergent vertebrates like zebrafish has revealed the presence of highly conserved sequences that do not code for proteins.15–18 Part of these conserved noncoding elements (CNEs) play a role in regulating gene expression, and are believed to be essential to all vertebrate development.6,9,19 Simultaneously it has also been shown that DNA elements in the absence of any sequence similarity can function as developmental enhancers in zebrafish.20,21 In spite of these findings the approaches to test for regulatory activity of these elements have remained “targeted”, where the PCR amplified DNA is either ligated to a reporter vector4 or in the case of zebrafish is co-injected with the reporter construct,9,22,23 and analysed for transient expression of green fluorescent protein (GFP) or lacZ. Another approach uses the Tol2 transposon system that allowed CNE–reporter gene fusions to be integrated into the germline more efficiently thus achieving much more robust expression of the transgene and solved the problem of mosaicism associated with transient transgenics in zebrafish.24–27 Though such methods have increased our understanding of gene regulation and control of transcription, they encounter hurdles when multiple regulatory domains from non-contiguous DNA act in concert to regulate expression of a gene. Difficulties also arise when the non-coding regulatory DNA is not conserved across species and thus not recognizable prior to testing.
Conventional in vivo transgenic studies have clearly demonstrated the important concept of “context” for regulatory elements in controlling spatiotemporal patterns of gene expression. For example, in its native genomic context the expression of the endogenous SM22 gene occurs in essentially all smooth muscle cell lineages. When taken out of its normal genomic context, however, the SM22 proximal promoter is active primarily in arterial smooth muscle cells with little or no activity in smooth muscle-rich tissues such as stomach, uterus, and bladder, indicating that elements controlling expression of SM22 in these tissues reside outside of the region analysed.28 In contrast, some in vitro documented promoters/enhancers display little or no activity when cloned in a conventional lacZ plasmid for transgenic mouse studies.29 In another study the authors located an enhancer for the amyloid precursor protein gene (APPb) within the intron of the gene.30 Although the enhancer was active in specific non-neural cells of the notochord when placed with APPb gene promoter proximal elements its function was restricted to, and absolutely required for, specific expression in neurons when juxtaposed with additional far upstream promoter elements of the gene. The authors demonstrated that expression of GFP fluorescence resembling the tissue distribution of APPb mRNA requires both the intron 1 enhancer and ~28 kb of DNA upstream of the gene. The results indicate that tissue-specificity of an isolated enhancer may be quite different from that in the context of its own gene. These examples imply that proper spatiotemporal expression of a gene requires modular cis-regulatory modules (CRMs) that may reside remotely from the core promoter region.
One important innovation of the human genome project was the development of artificial chromosomes that are large capacity cloning vectors harbouring hundreds of kilobases of genomic DNA. Such vectors offer a powerful means of capturing almost all CRMs and their regulatory elements controlling complete spatiotemporal expression of a gene, thus avoiding the incomplete activity profiles observed with most plasmid-based transgenic constructs.31 Distal regulatory elements within CRMs include not only enhancers but also silencers and an array of so-called boundary elements (e.g. insulators and locus control regions) that establish important points of transcriptional control through the establishment of either transcriptional activation or repression complexes.32
Bacterial artificial chromosomes (BACs) are one such specialized plasmids that can be used to clone in large DNA inserts that range in size from 150–300 kb. Large chunks of contiguous genomic sequences have been cloned into BAC vectors and have been used for sequencing of various genomes. Hence BACs are useful tools to modify a genomic region of interest and study a gene and its regulatory elements in its genomic context by introducing a reporter construct into the vector backbone of BACs using homologous recombination in E. coli. When assayed in transgenic animals, classical enhancers present in the genomic insert of any BAC should be able to stimulate the gene promoter and drive transcription of the reporter gene, yielding an in vivo readout of any enhancer activity harboured within a particular BAC. Hence comparing the expression of the reporter to the expression of the endogenous gene allows for the determination of the regulatory elements for the gene that are present in the genomic region cloned in the BAC.33,34 For example the cis-regulatory elements for isl1 gene in zebrafish are non-contiguous and scattered over 100 kb of the genome,35,36 hence to capture all the cis-regulatory elements will require extensive cloning, which is time consuming and in the end may still fail to detect all the functional enhancers. Thus BAC transgenics allows for a less labor-intensive method to detect regulatory elements in comparison to making individual DNA constructs and allows for synergistic activity of enhancers.
BACs have been successfully used to validate cis-regulatory elements for numerous genes in mouse3,37–39 and in certain cases multiple overlapping BACs have been used to locate all the regulatory elements of a gene.40 In this example three distal enhancers controlling Gata2 gene expression in the urogenital system were discovered using a novel “BAC-trapping” approach wherein a series of BACs covering 1 megabase of DNA around the Gata2 locus were systematically analysed in a lacZ-containing BAC vector. BAC modification for transgenics has also been employed for zebrafish genes in the past.5,41–46 The success of this approach lies in the fact that for genes with multiple expression domains, most of these domains can be observed in a single transgenic embryo as opposed to screening for multiple embryos for individual DNA constructs. In zebrafish, the availability of numerous embryos allows researchers to modify overlapping BACs around a gene and simultaneously test them and helps to detect regulatory elements, which at times reside at great distances from the gene.
Approaches in zebrafish BAC modifications
Homologous recombination mediated BAC transgenics
The most efficient way to achieve BAC modification is by the highly efficient homologous recombination technique originally developed by Stewart’ group in 199833,34 and later modified by Copeland’s group in 2001.47 Below is a brief outline of the process that uses the RecE and RecT for the recombination.
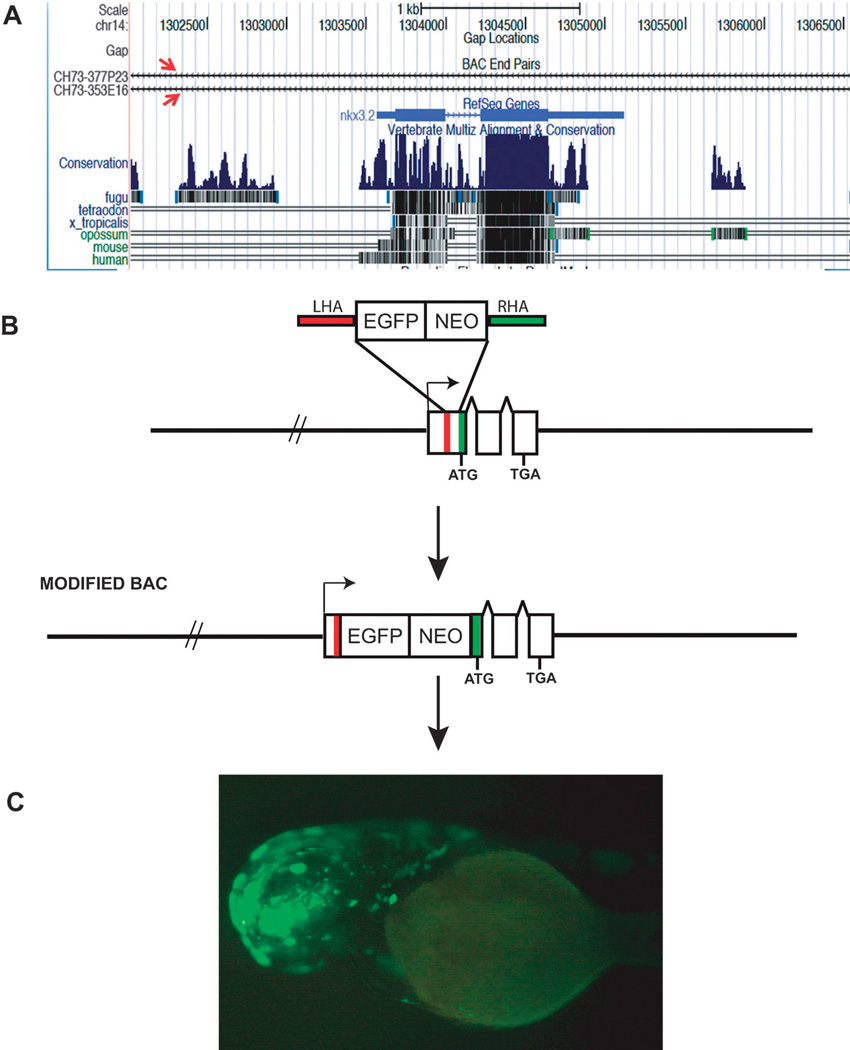
A zebrafish BAC is chosen from the many BAC libraries now available commercially (Table 1). A BAC which contains the largest flanking genomic sequences around the gene is generally selected. This can be easily achieved by visualizing the gene in any of the publicly available browsers. The University of California at Santa Cruz (UCSC) genome browser allows for the visualization of available BAC clones in a specific genomic locus (Fig. 1A), thus allowing for easy selection of a BAC clone for modification. Selecting a proper BAC clone with sufficient flanking regions enables the retention of most of the regulatory elements in their proper genomic context and hence increases the possibility of capturing the maximum number of distal cis-regulatory elements present on either side of the gene. The BAC targeting vector is made by cloning in a drug selection cassette next to a reporter gene (Fig. 1B). Enhanced Green Fluorescent Protein (EGFP) or other fluorescent proteins are the preferred choice, particularly in zebrafish, as they make it easier for direct live visualization under a fluorescent microscope. This region with the reporter gene and the selection cassette can be now PCRed out using 50 bp sequences, which are homologous to the gene. The homology region is generally right on either side of the translational start site. This DNA fragment containing the homology arm can now be used for homologous recombination, thus ensuring that the reporter gene is placed right between the promoter of the endogenous gene and its translational start site. The BAC clones are now sequenced to select a precisely modified BAC and then injected into 1-cell zebrafish embryos48 to detect gene expression (Fig. 1C). Since zebrafish undergoes ex-utero development, it makes it an attractive model system to detect transgene expression at all stages of its development. There are examples of regulatory elements positioned at great distances from the gene they regulate. For example the limb enhancer for Sonic Hedgehog (Shh) in mouse is located 1Mb away from the Shh.49 Similarly the mouse Gdf6 gene has five distant regulatory elements controlling expression of the gene in eleven distinct anatomical structures, spread over hundreds of kilobases. In such cases multiple overlapping BACs are modified and injected to detect all the regulatory elements controlling the gene expression.38 Thus BACs provide us with a quick method to scan large genomic regions to locate multiple regulatory elements.
Table 1.
Different zebrafish BAC libraries available. This table is modified from the Sanger Institute website. www.sanger.ac.uk/Projects/D_rerio/library_details.shtml
| Library | Strain | No. of Clones | Average insert size |
Contact |
|---|---|---|---|---|
| CHORI-211 BAC library | Tubingen | 105 907 | 165 kb | (1) BACPAC Resources, (2) The Max-Planck-Institut fuer Entwicklungsbiologie |
| Daniokey BAC library | Tubingen | 104 064 | 175 kb | (1) Hubrecht laboratory, (2) Keygene N.V. |
| CHORI-73 Doubled haploid BAC library | Tubingen | 297 528 | 110 kb | (1) BACPAC Resources |
| RPCI-71 BAC library | Tubingen | 33 408 | 85 kb | (1) BACPAC Resources, (2) The Max-Planck-Institut fuer Entwicklungsbiologie |
Fig. 1.
(A) The UCSC genome browser showing two BACs (red arrows) spanning the gene nkx3.2 in zebrafish. (B) BAC modification by homologous recombination to insert a reporter gene and a drug selection cassette next to the translation start site of the gene (ATG). (C) A zebrafish carrying the modified BAC for the gene otx1b expressing EGFP in the brain. Abbreviations: EGFP, enhanced green fluorescent protein; NEO, neomycin; LHA, left homology arm; RHA, right homology arm; TGA—stop codon.
Transposon mediated BAC transgenics
Recently Tol2 transposon based BAC modification has also been used in zebrafish.50 One of the advantages of using this method is that only a single copy of the modified BAC is delivered per transgenic embryo. This is important in view of a recent study that showed about half of BAC transgenic mice carried 1 to 5 copies of the injected BAC as concatamers at one genomic locus, and the rest carried more than 5 copies, and up to 48 copies, arranged in various orientations.51 Concatameric transgenes may be associated with silencing, instability, and genetic lesions both inside and around the transgenes52,53 that can sometimes limit experimental applications.
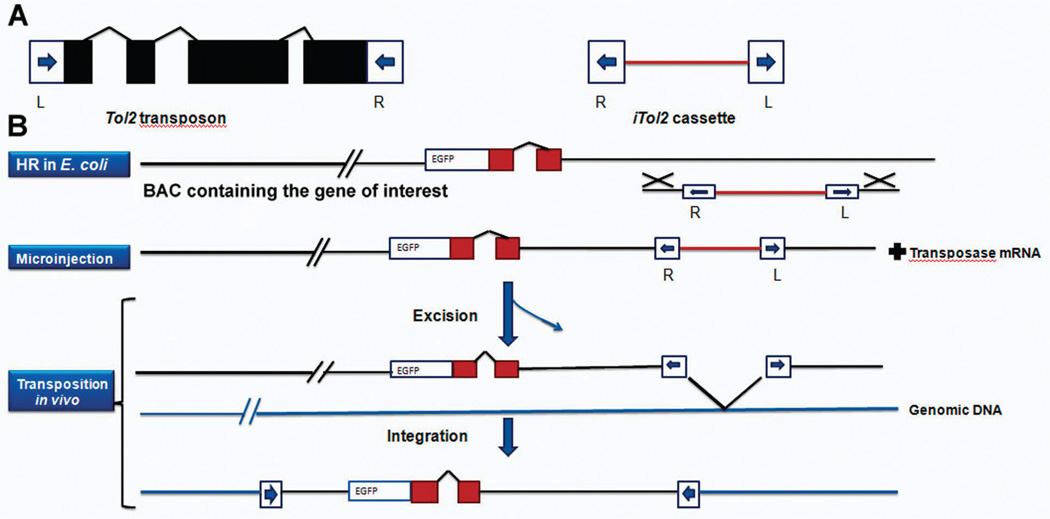
In this method the BAC carrying the gene of interest is first modified by introducing a Gal4FF (a modified version of the yeast transcriptional activator Gal4) in frame with the gene. Thus when this fish is crossed with a UAS:GFP reporter line it only gives GFP expression in the domains of the endogenous gene. To introduce the Tol2 sequence into the BAC plasmid, a cassette containing the minimal cis-sequences of Tol2 in an inverted orientation separated by a ~1 kb spacer is designed. This cassette enables incorporation of the Tol2 cis-sequences essential for transposition into a BAC clone through a single step of homologous recombination. This Tol2:Gal4 BAC can now be injected into 1-cell zebrafish embryos, grown to maturity and crossed with UAS:GFP reporter line to detect expression of the endogenous gene in all or most of its domains. This modification can be in principle done without using a GAL4-UAF system, by direct insertion of an EGFP modified BAC (Fig. 2).
Fig. 2.
(A) Structures of Tol2 and iTol2 cassette. Tol2 encodes a single transposase mRNA. The cis sequences required for transposition are shown as blue arrows (L and R). (B) The iTol2 cassette is integrated into a BAC containing the gene of interest modified with a reporter (EGFP). The modified BAC is then co-injected with the transposase mRNA into one-cell zebrafish embryo, where it integrates into the genomic DNA.
This method clearly has some advantages over the existing ones. First, it can generate single copy integrations that may be analysed and mapped on the genome relatively easily. Second, such integrations may not suffer from problems that have been observed in concatemeric integrations of small linear DNA transgenes; i.e., gene silencing, gross rearrangements at the target loci, unwanted mutant phenotypes, etc. Third, it ensures integration of DNA from end to end without obvious rearrangements inside. All of these features have been observed in transgenesis using transposons with smaller inserts in mice and zebrafish.27,54
BAC dissection to detect cis-regulatory elements
Though injecting whole modified BACs allows for detection of many of the cis-regulatory elements simultaneously, it is still desirable that from the large genomic region of the BAC individual regulatory elements be identified. To this end the large genomic DNA present in the BAC is generally chopped into smaller fragments by digesting with at least two restriction enzymes. This allows for generation of DNA fragments that are overlapping and hence prevents the loss of activity from a functional DNA due to an abrupt break. These fragments can now be individually tested with a reporter construct to detect activity. Though this sounds tedious, the versatility of zebrafish transgenics comes in handy here. In zebrafish unlike most model systems transient transgenics can be efficiently made by co-injecting the putative enhancer fragment with the vector containing the reporter gene.9,22 This allows for a rapid screening of many regulatory elements and gives quick results regarding their control of spatio-temporal expression patterns. Though the method of co-injection is rapid there can be instances where the fragment and the reporter don’t function together as desired giving rise to some false negatives. This dissection can be repeated till the smallest fragments that can drive the reporter gene are found. These fragments can now be used for biochemical analysis to narrow down the binding site. Biochemical assays include the electrophoreticmobility shift assay (EMSA),3,55,56 in which the binding sites (DNA sequences) are incubated with a nuclear protein extract from the specific tissue in which the enhancer was active. If the DNA fragment binds to any of the proteins in the extract, it is retarded in its mobility on a polyacrylamide gel in respect to an unbound DNA. The DNA sequences which form complexes with the proteins in the assay can now be looked at in detail using bioinformatics tools like TRANSFAC (www.gene-regulation.com/index.html),57 to predict potential transcription factors that can bind there. An antibody that recognizes the protein (transcription factor) can be added to this mixture to create an even larger complex with a slower mobility. This method is referred to as a supershift assay, and is used to unambiguously identify a protein present in the protein–nucleic acid complex.
In some cases, instead of digesting the BAC with an enzyme, various regions of the BAC are subcloned into a EGFP reporter vector and tested individually for activity.58 In this study the authors found the minimal enhancer element sufficient for the activity of the eng2a in medial fast-twitch fibres (MFFs) and the slow-twitch muscle pioneers (MPs). They followed it up with a trans-regulation assay to determine that the enhancer element bound Gli2a and activated Smads (pSmads) to carry out its function.
Advancements in BAC modification technologies have also ensured that instead of digesting the BAC with restriction enzymes or subcloning it, direct nested deletions can be carried out on the BAC DNA itself using a loxP transposon. The procedure involves introducing a small (7 kb) loxP transposon plasmid into the BAC or PAC clone by calcium chloride transformation. Transformed colonies are selected for resistance to antibiotic markers carried by both the BAC or PAC and the transposon plasmid. Cloned DNA with transposon insertions are then transduced with P1 phage. Thus, although transposition is equally probable in one of two orientations and only one of these leads to a deletion, the limited packaging capacity of the P1 phage head ensures that deletions are recovered only when the starting BAC or PAC clone is > 110 kb.59 A recent study utilizes this technology to detect multiple enhancers for APPb gene in zebrafish.30
Conclusions
Since the advent of the high-throughput genomic methods to locate cis-regulatory elements in the genome, the need for biological validation of the data has increased manifold. The conventional method for analysing cis-regulatory elements involved an out of genomic context assay wherein a portion of a gene’s promoter or enhancer region is excised from its normal genomic landscape and cloned upstream of a reporter gene in a plasmid-based vector having limited cloning capacity, typically less than 10 kilobases of DNA. This allowed for a limited understanding of gene regulation and severely hindered the elucidation of the large-scale genome wide map of regulation. Modifying large contiguous genomic regions via BACs has helped immensely in both locating and validating distal regulatory elements. These powerful techniques have their limitations however. In zebrafish the rate of germ line transmission of these BACs is very low, compared to the smaller vectors. This increases the time needed to raise stable lines for further studies. Another limitation of these large BAC transgenes, which they share with small vectors, is their susceptibility to genomic silencing and perturbations in genomic landscapes that could confer spurious promoter/CRM activities. This problem has also been somewhat mitigated with the development of transposon based BAC modification, which allows for single copy integration. Despite the above limitations, there have been great strides in defining distal cis-regulatory modules (CRMs) driving tissue-specific expression of a growing number of genes, thus expanding our appreciation of the complexities of gene expression control and providing a foundation for further functional analyses.
Biographies

Sumantra Chatterjee
Sumantra Chatterjee completed his undergraduate and master’s degrees in India, and secured a PhD under a joint program between the National University of Singapore and Genome Institute of Singapore. For his graduate research he focused on locating and validating functional enhancers in the vertebrate genome to better understand transcriptional regulation during development. His long-term interest lies in understanding how genes behave in a concerted manner during embryonic development. When he is not pondering on the mysteries of the genome, he can be found marveling at man-made wonders around the world.

Thomas Lufkin
Thomas Lufkin is a Senior Group Leader in Stem Cell & Developmental Biology, Genome Institute of Singapore; Associate Professor, Dept. Biological Science, National Univ. of Singapore; Associate Professor, School of Biol. Science, Nanyang Technological Univ. He completed postdoctoral training at the LGME, Strasbourg, France, in Molec Embryology (with Pierre Chambon). He received his PhD from Cornell University in Molecular Biology and Virology. He received his A.B. from the University of California, Berkeley in Cell Biology. He received the March of Dimes Basil O’Conner Jr. Faculty Award, was a Lucille B.Markey Scholar in Molecular Biology, received an Alfred P. Sloan Research Fellowship in Neuroscience, an American Cancer Society Postdoctoral Fellowship and a Morton J. Levy Predoctoral Fellowship. He is serving as an editorial board member of 3 reputed journals. He has over 80 publications. Research interests: Embryonic Stem Cell Differentiation, Embryogenesis, Developmental Genomics, Gene regulatory networks, Systems Biology, Regenerative Medicine, Vertebrate Development.
Notes and references
- 1.D’Souza RN, Niederreither K, de Crombrugghe B. J. Bone Miner. Res. 1993;8:1127–1136. doi: 10.1002/jbmr.5650080914. [DOI] [PubMed] [Google Scholar]
- 2.Schlaeger TM, Qin Y, Fujiwara Y, Magram J, Sato TN. Development (Cambridge, UK) 1995;121:1089–1098. doi: 10.1242/dev.121.4.1089. [DOI] [PubMed] [Google Scholar]
- 3.Werner T, Hammer A, Wahlbuhl M, Bosl MR, Wegner M. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 5.Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. Development (Cambridge, UK) 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 6.Shin JT, Priest JR, Ovcharenko I, Ronco A, Moore RK, Burns CG, MacRae CA. Nucleic Acids Res. 2005;33:5437–5445. doi: 10.1093/nar/gki853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanges R, Kalmar E, Claudiani P, D’Amato M, Muller F, Stupka E. Genome Biol. 2006;7:R56. doi: 10.1186/gb-2006-7-7-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AP, Koh EG, Tay A, Brenner S, Venkatesh B. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6994–6999. doi: 10.1073/pnas.0601492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega V, Wong E, Orlov Y, Zhang W, Jiang J, Loh Y, Yeo H, Yeo Z, Narang V, Govindarajan K, Leong B, Shahab A, Ruan Y, Bourque G, Sung W, Clarke N, Wei C, Ng HH. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Petrykowska HM, Vockley C, Elnitski L. Genome Res. 2008;18:1238–1246. doi: 10.1101/gr.073817.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie,M. Kamal X, Lander ES. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11659–11664. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEwen GK, Woolfe A, Goode D, Vavouri T, Callaway H, Elgar G. Genome Res. 2006;16:451–465. doi: 10.1101/gr.4143406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahituv N, Prabhakar S, Poulin F, Rubin E, Couronne O. Hum. Mol. Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- 18.Xie X, Mikkelsen T, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahituv N, Rubin E, Nobrega MA. Hum. Mol. Genet. 2004;13(Spec No 2):R261–R266. doi: 10.1093/hmg/ddh229. [DOI] [PubMed] [Google Scholar]
- 20.Fisher S, Grice E, Vinton R, Bessling S, McCallion AS. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 21.McGaughey DM, Vinton R, Huynh J, Al-Saif A, Beer M, McCallion AS. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller F, Williams D, Kobolak J, Gauvry L, Goldspink G, Orban L, Maclean N. Mol. Reprod. Dev. 1997;47:404–412. doi: 10.1002/(SICI)1098-2795(199708)47:4<404::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Goode DK, Callaway H, Cerda G, Lewis K, Elgar G. Development (Cambridge, UK) 2011;138:879–884. doi: 10.1242/dev.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher S, Grice E, Vinton R, Bessling S, Urasaki A, Kawakami K, McCallion AS. Nat. Protocols. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 25.Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K. Methods Mol. Biol. (Totowa, N. J.) 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 26.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Dev. Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 27.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. Dev. Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Miano J, Mercer B, Olson EN. J. Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miano JM, Kitchen C, Chen J, Maltby K, Kelly L, Weiler H, Krahe R, Ashworth L, Garcia E. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H1793–H1803. doi: 10.1152/ajpheart.00875.2001. [DOI] [PubMed] [Google Scholar]
- 30.Shakes LA, Malcolm T, Allen K, De S, Harewood K, Chatterjee PK. Nucleic Acids Res. 2008;36:6237–6248. doi: 10.1093/nar/gkn628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heaney JD, Bronson SK. Mamm. Genome. 2006;17:791–807. doi: 10.1007/s00335-006-0023-9. [DOI] [PubMed] [Google Scholar]
- 32.Bell AC, West A, Felsenfeld G. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 33.Muyrers JP, Zhang Y, Testa G, Stewart AF. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Buchholz F, Muyrers J, Stewart AF. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 35.Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H. Dev. Biol. 2005;278:587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 36.Higashijima S, Hotta Y, Okamoto H. J. Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong S, Zheng C, Doughty M, Losos K, Didkovsky N, Schambra U, Nowak N, Joyner A, Leblanc G, Hatten M, Heintz N. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 38.Mortlock DP, Guenther C, Kingsley DM. Genome Res. 2003;13:2069–2081. doi: 10.1101/gr.1306003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue YU, Asami J, Inoue T. J. Neurosci. Res. 2009;63:2–9. doi: 10.1016/j.neures.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Khandekar M, Suzuki N, Lewton J, Yamamoto M, Engel JD. Mol. Cell. Biol. 2004;24:10263–10276. doi: 10.1128/MCB.24.23.10263-10276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato Y, Miyasaka N, Yoshihara Y. J. Neurosci. 2007;27:1606–1615. doi: 10.1523/JNEUROSCI.4218-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jessen JR, Meng A, McFarlane R, Paw B, Zon L, Smith G, Lin S. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5121–5126. doi: 10.1073/pnas.95.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Jiang H, Chachainasakul T, Gong S, Yang X, Heintz N, Lin S. Methods. 2006;39:183–188. doi: 10.1016/j.ymeth.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Shin J, Park HC, Topczewska JM, Mawdsley DJ, Appel B. Methods Cell Sci. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- 45.Higashijima S, Masino MA, Mandel G, Fetcho JR. J. Neurosci. 2004;24:5827–5839. doi: 10.1523/JNEUROSCI.5342-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jessen JR, Willett CE, Lin S. Nat. Genet. 1999;23:15–16. doi: 10.1038/12609. [DOI] [PubMed] [Google Scholar]
- 47.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 48.Chatterjee S, Min L, Karuturi RK, Lufkin T. Transgenic Res. 2010;19:299–304. doi: 10.1007/s11248-009-9312-x. [DOI] [PubMed] [Google Scholar]
- 49.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 50.Suster ML, Sumiyama K, Kawakami K. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. Mamm. Genome. 2007;18:693–708. doi: 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrick D, Fiering S, Martin DI, Whitelaw E. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 53.Dorer DR, Henikoff S. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupuy AJ, Clark K, Carlson CM, Fritz S, Davidson AE, Markley KM, Finley K, Fletcher CF, Ekker SC, Hackett PB, Horn S, Largaespada DA. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oberto A, Longone P, Krueger KE. Gene. 1995;167:255–260. doi: 10.1016/0378-1119(95)00686-9. [DOI] [PubMed] [Google Scholar]
- 56.Feik N, Bilic I, Tinhofer J, Unger B, Littman DR, Ellmeier W. J. Immunol. 2005;174:1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- 57.Wingender E, Dietze P, Karas H, Knuppel R. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maurya AK, Tan H, Souren M, Wang X, Wittbrodt J, Ingham PW. Development (Cambridge, UK) 2011;138:755–765. doi: 10.1242/dev.062521. [DOI] [PubMed] [Google Scholar]
- 59.Coren JS, Sternberg N. Gene. 2001;264:11–18. doi: 10.1016/s0378-1119(01)00330-4. [DOI] [PubMed] [Google Scholar]