Abstract
The purpose of this study is to find out the frequency of TP53 mutations in acute myeloid leukemia (AML) patients and correlate sensitivity of drug response with TP53 mutations. In AML more than 90 % of cases comprise of wild type TP53. 94.2 % of TP53 mutations are found within exon 5–8 of which 73 % are point mutations. TP53 mutations were analysed with high resolution melting curve analysis. We analysed 106 AML samples of which we found nine mutations which represents 8.5 % mutation rate and found one rare SNP. The effect of TP53 mutations were studied on the chemosensitivity of two new drugs AZD115 and RHPS4, an Aurora Kinase B inhibitor and Telomerase inhibitor respectively. Four mutations were found out of 17 for RHPS4 stating significant (p = 0.002) increase in sensitivity and no mutation found in AZD1152 database, but need more study to get definite conclusion.
Keywords: TP53, Acute myeloid leukemia, High resolution melting curve analysis, Drug sensitivity
Introduction
Tumour protein 53 (TP53) is a tumour suppressor protein produced by TP53 gene (53 in p53 refers to its mass 53 KDa protein on SDS-PAGE). It is the main regulator of the cell cycle, so called as ‘guardian of the cell’ preventing cancer formation by conserving stability as it prevent genome mutations following DNA damage. The importance of TP53 gene is that it is found to be mutated in almost every cancer in varying frequency and the mutations are heterogenous in nature [1]. Mutations in p53 have been recorded in 50 % of human cancer. The mutations render p53’s DNA binding domain (DBD) non-functional by changing the tertiary structure of the gene product, leading to cancer progression as it destroys the ability of this protein to bind DNA and carry out its function. Even the mutations lead to new proteins with abnormal functions like ability to regulate expression of neoangiogenesis, drug resistance and prevention of tumour initiation and progression. The mutations are most commonly present between exon 5 and 8; 94.2 % of all somatic mutations reside in these codons [2–4]. This is the reason why mutations are selected over deletions and also TP53 was once considered to be oncogene [1].
Acute myeloid leukemia (AML) is a malignant disease of myeloid lineage of hematopoietic cells, with characteristic accumulation of immature myeloblasts which hampers the growth of other myeloid cell lines. It is basically a heterogenous group of disorders with varied morphology, cytogenetics and prognosis basically due to immature hematopoietic progenitor cells. Overall survival at 5 years for AML patients ranges from 3 to 80 % with the cytogenetic profile being the major predictor for prognosis [5].
Specifically, AML with a complex aberrant karyotype has been reported to have a high incidence of TP53 mutations. Tp53 mutations in AML are basically linked with cytogenic aberrations including chromosome 17p monosomy and secondary leukemia which corresponds to resistance to chemotherapy and eventually lower complete remission rates [6].
Study for two novel drugs for the effect of TP53 mutations, which might offer a role in future in AML therapy with drug resistance. Drug AZD1152 is an aurora kinase inhibitor especially of Aurora kinase B. Aurora kinases are chromosomal passenger proteins, located near the spindle and help in various process of mitosis. It has been proved to be novel anticancer drug for acute leukemia [7–11].
RHPS4 is a telomerase inhibitor. Telomerase is required to maintain integrity of telomeres which helps the cell divides without any loss of chromosomal information. In cancer cells telomerase is overexpressed and the cell becomes immortal and divides continuously. G-rich telomeric DNA folds itself into a 4-stranded structure known as G-quadruplex (G4) and if stabilization of this structure occurs by RHPS4 it leads to alteration of telomeres which may lead to cancer cell death. RHPS4 induces telomere damage, disturbing its structure and causing alteration in its function [12–14].
High resolution melting (HRM) curve analysis is a mutation scanning technology which identifies mutations by changes in fluorescence compared to the wild type sequence. This technique has successfully been tested on several other mutations like KIT, BRAF, EGFR, ERBB2 and KRAS genes and has previously been used to detect p53 mutations in various published papers [4].
Methods
Patient Samples
We selected DNA (blood) samples from newly diagnosed patients with AML from City Hospital, Nottingham from Caucasian population. Use of these samples was approved by the Local Research Ethics Committee. DNA extraction had been performed from blood or bone marrow samples using QIAmp® DNA mini/midi kits (QIAGEN, Hilden, Germany) according to manufacturer’s kit.
TP53 Cell Lines
These included OCI-AML3 (wild type) and for gene TP53 with mutations are K-562 (Neubauer,1993,pg. 593-600) and NB4 (Forbes, 2006, pg. 318-322) for exon 5a, u937 for exon 5b, 1992 pg. 2378–2383), BJAB for exon 6 (Farrell, 1991, 1879–2887), KG1A for exon 6 (Sugimoto, 1992, 278–2383), TF1A for exon 7 (Sugimoto, 1992, 278–2383), RAMOS for exon 7 (Gaidano, 1991, 5413–5417), SW480 for exon 8 (Nigro, 1989, pg. 705–708), KARPAS-422 for exon 9 (Forbes, 2006, pg. 318–322) [15–20].
DNA concentration was measured using Nanodrop 2000 spectrophotometer (Thermo Scientific, USA). DNA was diluted to a concentration of 20 ng/μl using nuclease free water.
HRM Primers
To design HRM primers for exons 5 to 8 in TP53 region several factors were taken into consideration.
Due to big size of exon 5 it is divided into two parts to get shorter amplicons (5a and 5b). Several primer pairs were designed by putting exon sequence into primer 3 sequence software (http://frodo.wi.mit.edu/primer3/) and for exon 8 primers were designed by Tm calculator. After designing primers were checked for SNPs in the region of annealing by SNPcheck. (https://ngrl.manchester.ac.uk/SNPCheckV2/snpcheck.htm).
HRM Assays
Using the primers designed for TP53 exon 5A, 5B, 6, 7, 8 and 9. HRM analysis is done on Applied Biosystems® 7500 Fast real time analyser. PCR for HRM analysis was performed in 96-well plates with the help of the specialised fluorescent DNA intercalating dye supplied in HRM Mastermix (Applied Biosystems, Foster City, USA). The 10 μl of Mastermix contains 25 nM of MgCl2, primer concentration of 5 μM of each primer, SYTO® 9 Dye 50 μM, AmpliTaq Gold® 360 DNA Polymerase, genomic DNA 5 ng/μl and deionised water using a MeltDoctor™ HRM Calibration Plate, 96-Well Fast on 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, USA). The software used to analyse the results was high resolution melt (HRM) software v2.0 (Applied Biosystems, Foster City, USA). HRM step is performed after the final PCR cycle.
The thermal cycler protocol as given by instructions manual was stage I: hotstart at 95 °C for 10 min, stage II, step 1: 95 °C for 15 s, step 2: 50 °C for 1 min, step 3: 95 °C for 15 s, step 4: annealing 60 °C for 15 s for a sample volume 20 μl. It is run for 40 cycles and HRM Step from 72 to 95 °C rising 0.1 °C per second to dissociate two stands as instructed in manufacturer’s protocol. Use an AmpliTaq Gold® enzyme; test various primer pairs to determine which one is optimal, including proper controls for PCR and variants, use of triplicates as recommended by manufacturer.
HRM Analysis
High resolution melting curve analysis performed with Applied Biosystems® HRM software v2.0 and analysed by two scientists. Amplification followed by a high resolution melting step using instrumentation capable of a large number fluorescent data points per change in temperature. The temperature is raised by 0.1 °C basically from (60 to 95 °C). Tm is defined as the temperature at which the melt curve shows that the 50 % of DNA is double stranded, and 50 % is single stranded. It is temperature at which fluorescence is also 50 %. The fluorescence was usually seen between 70 and 94 °C. The melt region is selected as pre- and post-melt region for generating normalised and difference plots.
HRM analysis distinguishes DNA sequences based on their length, composition, GC content or strand complementarity. The melting profile enables to differentiate normal from a mutant strand based upon base change. But polymorphisms are to be kept in mind besides some technical error. HRM results were sent for sequencing reaction to confirm the HRM analysis in detection of TP53 mutation in AML patients.
PCR Optimization and DNA Purification
PCR reactions for sequencing were performed in 50 μl reaction mixtures which consist of 1× PCR buffer containing 1.5 mM MgCl2, 04 μl of Taq Pol, 5 ng/μl of each primer pair which was designed for HRM analysis and 2 μl of 1 mM of dNTPs. The samples were run on PTC-100TM Programmable Thermal Controller (MJ Research Inc., Watertown, MA, USA). Amplicons run at 95 °C for 10 min for activation of Taq Pol, followed by 36 cycles of denaturation 95 °C for 1 min, annealing 60 °C for 1 min, extension 72 °C for 1 min and one cycle of final extension of 72 °C for 10 min and then cooling down to 4 °C. PCR products were run on 2 % Agarose gel using 0.5× TBE buffer with 100 bp DNA ladder. The PCR products were than purified using QIAquick PCR Purification kit (QIAGEN, Hilden, Germany) according to manufacturer’s instructions.
DNA Sequencing
All sequences are manually examined, and confirmed results with UCSC GENOME BROWSER (http://genome.ucsc.edu/cgibin/hgBlat?command=start&org=Human&db=hg19&hgsid=203387683). Sequencing reactions were set up with 200 ng of purified PCR product and 3.2 pmol of primer using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). The reactions were electrophoresed using an ABI 3130 Genetic Analyser (Applied Biosystems) and results were analysed using the Sequencing Analysis 5.2 software (Applied Biosystems).
Drug Database
The drug database for AZD1152 got from one our PhD student working on drug sensitivity on AML cancer cells. For AZD11152, phoshphohistone-H3 (pHH3) used as a biomarker instead of usual cell calculation in media. Drug database for RHPS4 was an unpublished data in which it has been calculated on basis of amount of drug concentration require killing cell in colony IC50 RHPS4.
Statistical Analysis
This is performed with the help of SPSS statistical 17.0. The results were represented as scatter plot. Significance for sensitivity data for wild type and mutant were assessed by t-test for unpaired data or Mann–Whitney test depending upon their distribution and p value <0.5 considered to be significant.
Results
Selection of Primer Pairs
Three primer pairs for each exon were analysed with the aim to find a suitable pair for HRM analyses. The same primers were used for analysis of sequencing. 1.5 ng/μl concentration of MgCl2 gave the best results.
HRM Analysis Validation
Literature searches were performed in order to find a cell line with a mutation in each of exons 5–8 to be used a positive control and OCI-AML3 is a p53 wild type cell line used as a wild type control as shown. Exon 5 was big so it is divided into two parts 5a and 5b and then analysed.
The results of the wild type OCI-AML3 with all the appropriate mutant cell line in HRM analysis were good except for exon 5a. From the literature, two different cell lines U937 and NB4 should have been suitable for mutant controls for exon 5 however. In the U937 cell line the mutation is in the 3′ portion of the exon so it was not applicable for exon 5a HRM and the NB4 cell line did not show deviation in the HRM analyses and so was not suitable to set as a mutant control. Subsequent sequencing results for NB4 showed it didn’t have a mutation in exon 5 of p53. Due to this reason exon 5 of 28 patient samples were sequenced without looking at their HRM results and sample number E621 showed a mutation in exon 5 by sequencing which was confirmed by HRM analysis and so it could then be used as a mutant control for exon 5 for further sample analysis. For exon 6 two cell lines BJAB and KG1A were identified from the literature as having a mutation. BJAB gave promising results that match with published data while KG1A showed no mutation in sequencing or HRM analysis. In a similar way RAMOS and TF1A were two potential cell lines for an exon 7 mutation and both show variation in HRM curve which was confirmed by sequencing. Lastly for exon 8 one cell line SW480 gave encouraging results demonstrating a mutant exon 8. The final positive control cell lines for individual exons were as below in Table 1.
Table 1.
sequencing results of mutant cell lines for exon 5–8 of TP53
| Exon | Mutant cell line | Codon | WT codon | Mutation | Event |
|---|---|---|---|---|---|
| 5A | E621 | 175 | CGC | GGC | C->G |
| 5B | U937 | 172 | GTT | DEL46A | Stop at 231 |
| 6 | BJAB | 193 | CAT | CGT | A->G |
| 7 | RAMOS | 254 | ATC | GAC | A->G/T->A |
| TF1A | 251 | ATC | DEL1B | Stop at 344 | |
| 8 | SW480 | 273 | CGT | CAT | G->A |
E621 sample used as mutant control for exon 5A
Confirmation by Sequencing
All the samples which showed variation in curve or shift in curve was sent for sequencing. Also ten samples showing normal curve also sent for sequencing to keep check for false negatives. Nine mutations (exon 5—three mutations, exon 6—1 mutation, exon 7—four mutations, exon 8—one mutation) and one SNP rs1800372 in exon 6 were found. All mutations and SNPs that were found were rechecked in p53 mutation database (http://www-p53.iarc.fr/MutationValidationCriteria.asp).
P53 Mutations and Cytogenetics
Table 2 also shows the cytogenetic profile of the samples that were found to have mutations. The cytogenetics data has been collected from routine cytogenetics analysis performed by the NHS Cytogenetics Department where presentation samples of AML are sent. Out of nine mutations that were found 5 showing confirm complex cytogenetics while one is query complex and one normal. Cytogenetics of two samples was unknown.
Table 2.
Sequencing results of positive samples for mutation/SNPs screened for exon 5–8 of TP53 by HRM
| Sample no | Exon | Amino acid change | Coding description | Genomic description | Frequency of mutation/SNP | Cytogenetics |
|---|---|---|---|---|---|---|
| E355 | 6 | R213R | c.639A>G | g.12708A>G | (SNP- 0.023526) | Complex |
| rs1800372 | ||||||
| E597 | 7 | G244S | c.730G>A | g.13367G>A | 227 | Complex |
| E599 | 7 | G245S | c.733G>A | g.13370G> | 718 | Complex |
| E621 | 5 | R175G | c.523C>G | g.12511C>G | 1,092 | Complex |
| E624 | 7 | R249 K | c.746G>A | g.13383G>A | 573 | Complex |
| E626 | 8 | R273H | c.818G>A | g.13798G>A | 1,425 | ? |
| E627 | 6 | Y205C | c.614A>G | g.12683A>G | 158 | Query complex |
| E629 | 5 | P151A | c.451C>G | g.12439C>G | 210 | Normal |
| E639 | 5 | P151A | c.451C>G | g.12439C>G | 210 | Complex |
| E669 | 7 | R248E | c.743G>A | g.13380G>A | 1,544 | ? |
Frequency of mutation was collected from UMD P53 mutation database
Sensitivity, Specificity and Positive Predictive Value of HRM Technique
The sensitivity and specificity of HRM for all 106 samples for four exons in five reactions (5a, 5b, 6, 7, 8 and 9) calculated were 0.91 [10 true positive/(10 true positive + 3 false negative)], and 0.97 [103 true negative/(103 true negative + 3 false negative)], respectively.
TP53 Mutations and its Sensitivity Affect the of AML Samples to Drugs
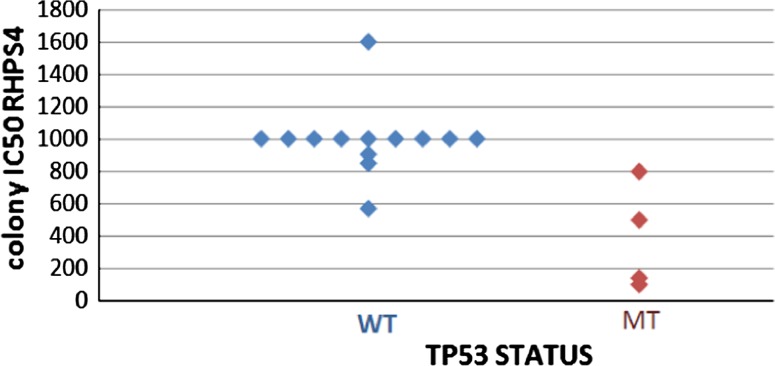
Two drug databases were used to check whether a mutation in p53 in a patient sample affected its response to a particular drug. The first database was contained results pertaining to the sensitivity of 29 AML samples to AZD1152 an aurora kinase B inhibitor. For AZD1152 a biomarker pHH3 (Phosphohistone-H3) was used, indicative of down regulation of pHH3 which correlates with suppression of aurora kinase B. This data was generated by previous study within the department. The second database included chemosensitivity results on 18 patient samples treated with RHPS4 a telomerase inhibitor (unpublished data). For RHPS4 colony counts were used to determine the amount of drug required to suppress the growth of cells. No mutation was found in AZD1152 drug database of 29 samples. For RHPS4 four Samples out of 17 showed mutations and their relation was shown in Fig. 1.
Fig. 1.
Sensitivity of RHPS4 and TP53 status
Statistical Analysis
Mann–Whitney test for two independent samples has performed as the drug sensitivity data for WT sample did not show normal distribution as suggested by Shapiro–Wilk test (p = 0.000). The results suggested that there is a statistically significant difference between the underlying distributions of the drug sensitivity scores of WT and MT samples for TP53 (z = −3.065, p = 0.002). This states that the samples with a mutant p53 are more sensitive to RHPS4.
Discussion
Development of newer technique HRM is a new step towards detection of single base changes. There are many methodologies to detect base changes with sequencing method considered to be the best among all. However, even sequencing cannot detect rare allele with frequency less than 10 % [21]. HRM a post-PCR technique proved to be very simple, fast and cost-effective technique for screening samples with unknown mutations or SNPs on a large scale with rapid turn-around time requiring just PCR, DNA dye and melting instrument. It can reduce unnecessary load of sequencing of samples [22]. It is a closed technique so very less chances of contamination and also it is non-destructive so the same sample can be used to analyse further. The development of new fluorescent dye makes this method more attractive and target oriented for mutation scanning [23–25].
In this study, ten mutations were found out of 106 samples within exon 5–8 of which one turned out to be SNP indicating 8.5 % mutation frequency of TP53 in AML which is nearly same as the literature says [6]. Some of the mutations had extremely high frequency. All mutations were validated with TP53 mutation database which adds an additional confirmation to HRM results by providing information on codon number, frequency and exact location of mutation/SNP on gene. We found one A>G SNP rs1800372 with a frequency of 0.023, with allele A as an ancestral allele. HRM can detect rare mutations or alleles without prior knowledge of its exact location on gene. Furthermore, out of nine mutations five showed complex cytogenetics and other three unknown and one fell into normal category. This indicates that most of the p53 mutations were associated with complex cytogenetics.
Sensitivity testing is necessary to set up HRM assays for detection of mutations. Sensitivity and specificity of 0.91 and 0.97 were achieved for this assay respectively. Smaller amplicons have better sensitivity as they give better differentiation. The error rate depends upon length of amplicon and type of base change but not on the position of base change [26, 27]. DNA contamination was thought to be the reason for showing false positive results in study.
During the course of work, I come across few weaknesses of HRM. The sensitivity testing results showed variation of replicates of wild type melt curve profile which ultimately affects ability to distinguish mutant samples from wild type ones affecting sensitivity of whole technique. Also samples with pure mutant DNA pose problem to analyse by HRM. Have to rely on another method for confirmation. Additional limitation is less flexibility while selecting primer pair. Data from other studies mentioned that deletion/insertion of single base do not change melt profile compared to heterozygous sample [4].
Results of the drug sensitivity testing indicated significant difference (p = 0.002) of drug response for RHPS4 between wild type and mutant showed by lower concentration of drug required for cell death in colony IC50 RHPS4 in mutant cells. No mutations were found in drug database for AZD1152. This points that the mutation of TP53 increases drug sensitivity supporting role of drug RHPS4 in AML and cannot conclude anything for AZD1152.
In conclusion, HRM validates its use for screening TP53 mutations but need to confirm screening results by sequencing. TP53 mutations in AML were same as in other literature proving reliability of HRM technique and those mutations found linked with complex cytogenetics which is associated with poor survival of AML patients [28]. Detection of TP53 mutations help in better selection of drug and newer drugs like RHPS4 provide promising results in those cases. However, more database for drugs AZD1152 and RHPS4 require for concluding relation between mutation and drug sensitivity.
Acknowledgments
I thank my supervisor Dr Claire Seedhouse (Haematology department, city hospital) for support and help during the entire project and Martin Grundy for providing drug database. I am obliged to all my colleagues and my family who supported me to accomplish my project successfully.
Footnotes
This article has been retracted at the request of the second author of the paper, Claire Seedhouse, whose name was included in the paper without her permission. Further, the data in the paper has been published without any approvals from the department or the institution, where the corresponding author, Ankur Shah, was working at the time of submitting this paper.
An erratum to this article is available at http://dx.doi.org/10.1007/s12291-014-0422-8.
References
- 1.Iwakuma T, Adhikari AS. Mutant p53 gain of oncogenic function: in vivo evidence, mechanism of action and its clinical implications. Fukuoka Acta Med. 2009;100(6):217–228. [PubMed] [Google Scholar]
- 2.Stojnev Slavica, Golubović Mlađan, Babović Petar. TP53 gene mutations—from guardian of the genome to oncogene. Acta Medica Medianae. 2010;49(1):59–63. [Google Scholar]
- 3.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60(24):6788–6793. [PubMed] [Google Scholar]
- 4.Krypuy M, et al. High resolution melting for mutation scanning of TP53 exons 5–8. BMC cancer. 2007;7:168. doi: 10.1186/1471-2407-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimwade D, Hills RK. Independent prognostic factors for AML outcome. Hematology. 2009;2009(1):385–395. doi: 10.1182/asheducation-2009.1.385. [DOI] [PubMed] [Google Scholar]
- 6.Ånensen N. p53 protein biosignatures in acute myeloid leukemia. Bergen: The University of Bergen; 2006. [Google Scholar]
- 7.Tao Y, et al. The aurora B kinase inhibitor AZD1152 sensitizes cancer cells to fractionated irradiation and induces mitotic catastrophe. Cell Cycle. 2009;8(19):3172–3181. doi: 10.4161/cc.8.19.9729. [DOI] [PubMed] [Google Scholar]
- 8.Ikezoe T, et al. p53 is critical for the aurora B kinase inhibitor-mediated apoptosis in acute myelogenous leukemia cells. Int J Hematol. 2010;91(1):69–77. doi: 10.1007/s12185-009-0462-7. [DOI] [PubMed] [Google Scholar]
- 9.Grundy M, et al. The FLT3 internal tandem duplication mutation is a secondary target of the aurora B Kinase inhibitor AZD1152-HQPA in acute myelogenous leukemia cells. Mol Cancer Ther. 2010;9(3):661–672. doi: 10.1158/1535-7163.MCT-09-1144. [DOI] [PubMed] [Google Scholar]
- 10.Curry J, et al. Aurora B kinase inhibition in mitosis: strategies for optimizing the use of aurora kinase inhibitors such as AT9283. Cell Cycle. 2009;8(12):1921–1929. doi: 10.4161/cc.8.12.8741. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, et al. AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood. 2007;110(6):2034–2040. doi: 10.1182/blood-2007-02-073700. [DOI] [PubMed] [Google Scholar]
- 12.Rizzo Angela, et al. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucl Acid Res. 2009;37:5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvati E, et al. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Investig. 2007;117(11):3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biroccio A, et al. TRF2 inhibition triggers apoptosis and reduces tumorigenicity of human melanoma cells. Eur J Cancer. 2006;42:1881–1888. doi: 10.1016/j.ejca.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Forbes S, et al. Cosmic 2005. Br J Cancer. 2006;94(2):318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto K, et al. Frequent mutations in the P53 gene in human myeloid-leukemia cell-lines. Blood. 1992;79(9):2378–2383. [PubMed] [Google Scholar]
- 17.Farrell PJ, et al. p53 is frequently mutated in Burkitt’s lymphoma cell lines. EMBO J. 1991;10(10):2879–2887. doi: 10.1002/j.1460-2075.1991.tb07837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaidano G, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Nat Acad Sci USA. 1991;88(12):5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nigro JM, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342(6250):705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 20.Neubauer A, He M, Schmidt CA, et al. Genetic alterations in the p53 gene in the blast crisis of chronic myelogenous leukemia: analysis by polymerase chain reaction based techniques. Leukemia. 1993;7:593–600. [PubMed] [Google Scholar]
- 21.Ogino S, et al. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7(3):413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liew M, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50(7):1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 23.Garritano S, et al. Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 2009;10(1):5. doi: 10.1186/1471-2156-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Bosquet J, et al. Detection of somatic mutations by high-resolution DNA Melting (HRM) analysis in multiple cancers. PLoS ONE. 2011;6(1):e14522. doi: 10.1371/journal.pone.0014522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krypuy M, et al. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC cancer. 2006;6(1):295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liew M, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50(7):1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 27.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 28.Haferlach C, et al. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22(8):1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]