Abstract
In our previous studies, ZKSCAN3 was demonstrated to be over-expressed in invasive colonic tumor cells and their liver metastases, but minimally expressed in adjacent non-transformed tissues. Further preliminary data showed that ZKSCAN3 was expressed in a majority of prostate cancer patient samples, but not in normal prostate tissues. Moreover, the ZKSCAN3 protein is highly expressed in the PC3 prostate cancer cell line, which has high metastatic potential, but little expression was observed in non-metastatic prostate cancer cell lines. Thus, we hypothesized that ZKSCAN3 could participate in tumor metastasis by regulating tumor cell migration. To test this hypothesis, ZKSCAN3 mRNA was knocked down by ZKSCAN3 specific shRNA in PC3 cells and a significant decrease in cell motility was observed. In contrast, when ZKSCAN3 cDNA was overexpressed in PC3 cells, cell detachment was observed and suspension culture induced apoptosis was greatly decreased, suggesting that ZKSCAN3 is able to enhance PC3 cell survival under anoikis stress. Additional wound healing and invasion assays showed that cell migration was enhanced by ZKSCAN3 expression. Interestingly, the ZKSCAN3 gene was amplified in 26% (5/19) of metastatic prostate cancers and 20% (1/5) of lymph node metastases, but there was no amplification found in primary prostate cancers, further supporting the role of ZKSCAN3 in tumor cell migration. In vivo studies using orthotopic tumor models indicated that overexpression of ZKSCAN3 significantly enhanced tumorigenicity. Taken together, we provide evidence that ZKSCAN3, a zinc finger transcription factor, plays a critical role in promoting prostate cancer cell migration.
Keywords: ZKSCAN3, prostate cancer, cell metastasis, cell migration, tumorigenicity
1. Introduction
Prostate cancer is a major health problem in the United States, with over 200,000 new cases diagnosed each year and approximately 30,000 deaths per year (Howlader et al., 2011). Despite an increased awareness of the disease, which has led to earlier detection and treatment of the primary tumor, many patients still die of metastases that are resistant to conventional therapies, including hormonal ablation (Tu and Lin, 2004, Taylor et al., 2006). Improving the therapy for metastatic prostate cancer now depends on improving our understanding of the biology of this disease. Metastasis is a multistep process that includes detachment of cancer cells from the primary tumor, active migration, adhesion and invasion of tumor cells into the blood vessels, extravasation and subsequent growth in a distant tissue (Chambers, 1999). In order to metastasize, cancer cells have to acquire various properties; these include releasing their normal cell-cell adhesion, producing proteolytic enzymes and acquiring migratory properties. Several earlier reports demonstrated a correlation between motility of tumor cells and their metastatic potential (Grimstad, 1987, Stracke et al., 1991). It was recently shown that the cell surface glycoprotein endoglin is a regulator of cell adhesion, motility and invasion in human prostate cells (Liu et al., 2002).
In prostate cancer, three genes (KAI1, CD44 and MAPK kinase 4) have been identified that act as metastasis suppressor genes without affecting primary tumor growth (Kauffman., 2003). MAPK kinase 4 appears to suppress metastasis by inhibiting cancer cell growth at the secondary site (Kauffman., 2003). Loss of expression has been demonstrated for all of these genes during the clinical progression of prostate cancer to metastasis (Hendrix, 2003). Although progress has been made toward understanding genetic alterations that lead to prostate cancer cell metastasis, much remains to be learned about how cancer cells acquire the property of migration, a prerequisite for invasion and metastasis. Identification of the rate limiting steps of metastasis and their underlying molecular mechanisms may lead to new therapeutic targets and also allow more accurate risk stratification for clinical metastases.
Previously, we demonstrated that ZKSCAN3, a novel zinc finger transcription factor, is overexpressed in colon tumors and multiple myeloma (MM), and activates a gene expression program driving colon cancer and MM progression (Yang et al., 2008a , Yang et al., 2011). More importantly, ZKSCAN3 is highly expressed in deep invasive tumor cells compared to superficial non-invasive tumor cells, suggesting that it plays a very important role in driving tumor cell migration and invasion (Yang et al., 2008a). Interestingly, ZKSCAN3 resides on chromosome 6p22.1. The 6p21-23 region is commonly amplified in multiple solid tumors (Santos et al., 2007, Kallioniemi et al., 1992). Recently, chromosome 6p22 was linked to a high risk pedigree with aggressive prostate cancer (Schaid et al., 2006, Janer et al., 2003). Thus it is of particular interest to investigate if ZKSCAN3 plays a role in modulating prostate cancer cell metastasis.
In this study, we report that ZKSCAN3 is highly expressed in prostate tumor tissues but not normal adjacent tissues. ZKSCAN3 not only enhanced prostate cancer cell detachment, motility and migration, but caused a robust induction of tumorigenicity in vivo as well, supporting a role of ZKSCAN3 in regulating prostate cancer cell migration.
2. MATERIALS AND METHODS
2.1. Array Comparative Genomic Hybridization (aCGH) Analysis
DNA from prostatectomy cases was isolated and prepared as described previously (Paris et al., 2003). aCGH was performed using BAC arrays containing 2,460 BAC clones printed at UCSF. The BAC aCGH was performed as described in Paris et al (Paris et al., 2003, Paris et al., 2007) with a male reference DNA (Promega). Regions of copy number gain and loss for the BAC aCGH data were identified by creating sample specific thresholds (Paris et al., 2004).
2.2. Cell culture and Transfections
PC3 and CWR22r cells were kindly provided by Dr Isaiah J Fidler (The University of Texas M.D. Anderson Cancer Center, Houston, TX, USA), and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were transfected with Lipofectamine 2000 (as per Invitrogen instructions). Flag-tagged ZKSCAN3 was sub-cloned into the pIRES2-EGFP bicistronic vector and PC-3 cells were transfected with this Flag-tagged ZKSCAN3 expression construct. The DNA lipofectamine complex was added to ~ 107 cells and after 48 hours, 2 mg/ml G418 was added to select stably-transfected cells. Knockdown of ZKSCAN3 with ZKSCAN3 specific shRNA was performed according to previously described procedures (Yang et al., 2011). For transient transfections to identify optimal shRNA sequences, we used the procedure described by Invitrogen. (http://www.invitrogen.com-/downloads/HCT116_-stealthrnai_tsf_protocol.pdf).
2.3. Western blotting
ZKSCAN3 protein expression in prostate cancer cell lines was measured by western blotting (Yang et al., 2008a, Yang et al., 2008b). Cells were harvested and rinsed twice with ice-cold PBS and homogenized in buffer containing 10 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 5mol/L edetic acid, 1 mmol/L PMSF, 0.28 U/L aprotinin, 50 mg/L leupeptin, 1 mmol/L benzamidine, 7 mg/L pepstain A. Protein concentration was determined using the BCA kit (Thermo Scientific Pierce, Rockford, IL). Thirty micrograms of protein from each sample was subjected to electrophoresis on a 10-12% SDS-PAGE gel using a constant current. Proteins were transferred to nitrocellulose membranes and incubated with Tris-buffered saline containing 0.2% Tween-20 (TBST) and 3% nonfat dry milk for 3 hours in the presence of one of the following antibodies: 0.1 μg/ml of the affinity-purified anti-ZKSCAN3 antibody which was as described previously (Yang et al., 2008a) or a 1:2000 dilution of the anti-Flag M2 antibody from Sigma-Aldrich (Saint Louis, MO, USA). Membranes were washed and incubated with horseradish peroxidase (HPR)-conjugated anti-mouse IgG (1:10,000) in TBST containing 3 % nonfat dry milk for 1 hour. Immunoreactivity was detected with the Amersham enhanced chemiluminescent (ECL) kit (Amersham, Buckinghamshire, UK) according to the manufacturer’s instructions.
2.4. Immunohistochemistry
For immunohistochemistry analysis, prostate cancer tissue array slides were purchased from Biomax US (Catalog number: PR802), in which each tissue microarray contained 78 cases of prostate carcinoma, 2 cases normal tissue with single core per case. After de-waxing and antigen retrieval, endogenous peroxidase was inactivated with H2O2, and slides were blocked with 5% normal horse serum, 1% normal goat serum. Sections were incubated with affinity-purified anti-ZKSCAN3 and then with a horseradish peroxidase-conjugated secondary antibody. Immunoreactivity was detected with the DAB chromogen (Research Genetics). For negative controls, the anti-ZKSCAN3 antibody was substituted with an equivalent amount of pre-immune IgG.
Immunoreactivity was quantitated as follows: five independent fields (100 × magnification; no counterstain) were digitized and immunoreactivity pixel area determined using ImageJ (v1.34s) software (http://rsb.info.nih.gov/ij/).
2.5. Flow cytometric analysis
For flow cytometric analysis, PC3 cells transfected with shNon-targeting or shZKSCAN3 for 48 hours were trypsinized, washed in PBS, and resuspended in ice-cold 80% ethanol. Cells were permeabilized with 0.25% Triton X-100, and were then blocked with 10% normal goat serum (NGS) for 15 min. Cells were incubated with the indicated antibodies: 0.1 μg/ml affinity-purified anti-ZKSCAN3 antibody (Yang et al., 2008a) or the isotype matched control mineral oil plasmacytoma-10 from Sigma-Aldrich (Saint Louis, MO, USA), followed by incubation with goat anti-mouse IgG1-FITC (Southern Bio-technology, Birmingham, AL). All of the staining was done on ice for 45 min followed by three washes in HBSS + 5% FBS. The stained cells were filtered through the cell strainer caps of Falcon polystyrene round-bottomed tubes. Fluorescence was analyzed on a Becton Dickinson FACScan (Becton Dickinson, San Jose, CA). Data were collected from at least 10,000 cells. Using the Coulter software, the percentage of positive cells and their Mean Fluorescence Intensity (MFI) were determined by comparison with the isotype matched control stained cells.
2.6. Cell motility and invasion assays
For the cell motility assay, the upper and lower surfaces of the membrane in transwell inserts (Costar) were coated with collagen I at 4°C overnight. To prepare for the invasion assay, Matrigel (Collaborative Research; 0.5 μg) was diluted with cold water and dried onto each transwell insert overnight at room temperature. On the following day, transwell membranes were blocked with DMEM for 1 hour at 37°C. Cells were trypsinized and resuspended in serum-free DMEM. A total of 105 cells were added to the upper chamber of each well. Lysophosphatidic acid (LPA; 100 ng/mL) was added to the lower chambers as a chemoattractant. Inserts were incubated for 2 to 3 hours, and non-migrating cells were mechanically removed with cotton swabs. The cells attached to the bottom side of the membrane were stained with crystal violet and counted. Assays were done in triplicate and repeated five times. Cell motility and invasion were quantified by counting the cells that migrated to the lower surface of the membrane per square milliliter using bright-field optics.
2.7. PC3 cell detachment
PC3-empty-vector and PC3-ZKSCAN3 stably transfected cells were seeded into 24 well plates at 5×105/well. After 48 hours, the cells were treated with 2mM EDTA for 5 to 25 minutes. After incubation with EDTA, the cells were washed with PBS and then incubated with (3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide (MTT) reagent diluted in RPMI medium (5 mg/ml) at 37°C. After 2 hours the formazan crystals produced by metabolism of MTT in live cells were reconstituted in DMSO. The optical density at 570 nm was determined using an ELISA reader. The percentage of detached cells was calculated using this formula: Percentage of detached cells = 100 - [(OD570 of EDTA treated cells) / (OD570 of untreated cells) × 100%].
2.8. Cell Migration Assay
Cells were sub-cultured in 6-well plates and incubated in RPMI with 10% FBS. PC3 cells were transfected with shNon-targeting or shZKSCAN3 for 48 hours and CWR22r cells were transfected with empty-vector or ZKSCAN3 for 48 hours. A 1 mm thick wound was created using a 10 μL micropipette tip. Cells were washed and incubated in serum free medium. Cell migration was monitored and photographed after a 24 hour incubation.
2.9. Intraprostatic Injection of PC3 Cells
Two independent PC3 cell clones stably overexpressing ZKSCAN3 (PC3-ZKSCAN3#1 and PC3-ZKSCAN3#2) and PC3 cells stably transfected with the empty vector were harvested from subconfluent cultures and resuspended in Ca2+- and Mg2+-free Hanks’ balanced salt solution (HBSS). Cell viability was determined by Trypan Blue dye exclusion and only single-cell suspensions of >95% viability were used to inject mice. Three groups of nude mice (5 mice/group) were anesthetized with Nembutal (0.5 mg/g body weight) (Abbott Laboratories, North Chicago, IL), and intraprostatic injection of 25 μL/mouse of cell suspension (1×105 cells) from either empty vector or Flag-ZKSCAN3 cDNA was performed as described previously (Yang et al., 2008b). The animals tolerated the surgical procedure well, and no anesthesia-related deaths occurred. After 4 weeks mice were sacrificed, and tumors were circumscribed, surgically removed, and weighed by electronic balance. The abovementioned experiments were repeated three times. All experiments were approved by the Institutional Animal Care and Use Committee.
2.10. Cell cycle assays
For flow cytometric analysis, PC3 cells stably transfected with empty vector or ZKSCAN3 for 48 hours were trypsinized, washed in PBS, and resuspended in ice-cold 80% ethanol. Briefly, 2.5×105 fixed cells were incubated in 250 μL propidium iodide solution (500 mg/mL propidium iodide in 3.8 mol/L sodium citrate at pH 7.0) and 250 μL RNase A (10 mg/mL prepared in 10 mmol/L Tris-HCl at pH 7.5) for 30 minutes at 37°C in the dark. The stained cells were filtered through the cell strainer caps of Falcon polystyrene round-bottomed tubes. DNA content was analyzed on a Becton Dickinson FACScan (Becton Dickinson, San Jose, CA). Percent cell cycle phase was determined using Cell Fit software (Becton Dickinson). Data were collected from at least 20,000 cells.
3. RESULTS
3.1. ZKSCAN3 is overexpressed in prostate tumors
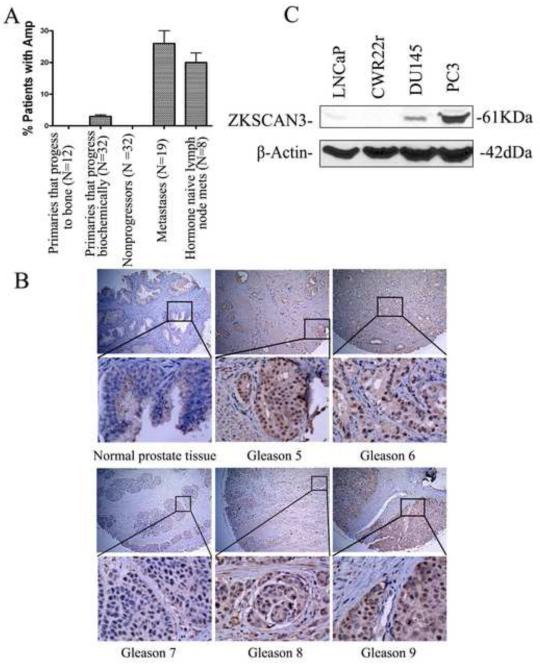
Because the ZKSCAN3 gene has been shown to be amplified in colon cancer and multiple myeloma cells, we investigated whether the ZKSCAN3 gene was amplified in prostate cancer as well. ZKSCAN3 was mapped to the BAC clone RP11-2P4 on the UCSF array. aCGH was performed using BAC arrays containing 2,460 BAC clones printed at UCSF, and regions of copy number gain and loss for the BAC clone RP11-2P4 were identified by creating sample specific thresholds (Paris et al., 2004). As shown in Fig. 1A, the region for the ZKSCAN3 gene was amplified in 26% of bone metastatic prostate cancers (5/19) and 20% of lymph node metastases (1/5), and there was no amplification of the ZKSCAN3 gene observed in primary tumors, suggesting that ZKSCAN3 may play a role in prostate cancer metastasis.
Fig. 1.
Expression of ZKSCAN3 in primary prostate cancer, metastatic prostate cancer, and prostate cancer cell lines. A: ZKSCAN3 loci are amplified in metastatic prostate cancer. BAC arrays were used to detect ZKSCAN3 gene amplification using DNA isolated from primary or metastatic prostate cancers. The prostate cancers were divided into 5 groups: primaries that progress to bone (N=12), primaries that progress biochemically (N=32), nonprogressors (N=32), bone metastases (N=19) and hormone naïve lymph node metastases (N=8). B: Expression and localization of the ZKSCAN3 protein to the tumor cells in resected prostate cancer. Normal prostate tissue and 5 different gleasons cancer tissue were arrayed by immunohistochemistry using the affinity-purified anti-ZKSCAN3 antibody. C: The expression of ZKSCAN3 in representative prostate cancer cell lines with differential metastatic potential. ZKSCAN3 expression was analyzed in nuclear extracts from LNCaP, CWR22r, DU145 and PC3 cells by Western blotting with the anti-ZKSCAN3 antibody.
To demonstrate that the ZKSCAN3 protein is expressed in prostate cancer, we performed immunohistochemistry (IHC) on a prostate cancer tissue array with an anti-ZKSCAN3 antibody generated during our previous studies (Yang et al., 2008a). While little staining was seen in normal prostate tissue, 48.7% of tumor tissues (38/78) showed strong reactivity predominantly localized to the nucleus (Fig. 1B), a pattern similar to many zinc finger-containing proteins, supporting the hypothesis that ZKSCAN3 may play an important role in driving prostate cancer progression.
Since our previous studies suggested that ZKSCAN3 could participate in tumor cell invasion (Yang et al., 2008a), endogenous ZKSCAN3 protein expression was compared in the prostate cancer cell lines LNCaP, CWR22r, DU145, and PC3, which represent different metastatic potentials. Western blot analysis of nuclear extracts from these cell lines demonstrated that the level of ZKSCAN3 expression correlated with the metastatic potential of the cell lines, with the highest expression level found in PC3 cells and no detectable expression in non-metastatic CWR22r cells (Fig. 1C). The DU145 and LNCaP cell lines expressed ZKSCAN3 at a lower level than PC3 cells, which correlated with their lower metastatic potential.
3.2. Exogenous expression of ZKSCAN3 increases prostate cancer cell detachment and motility
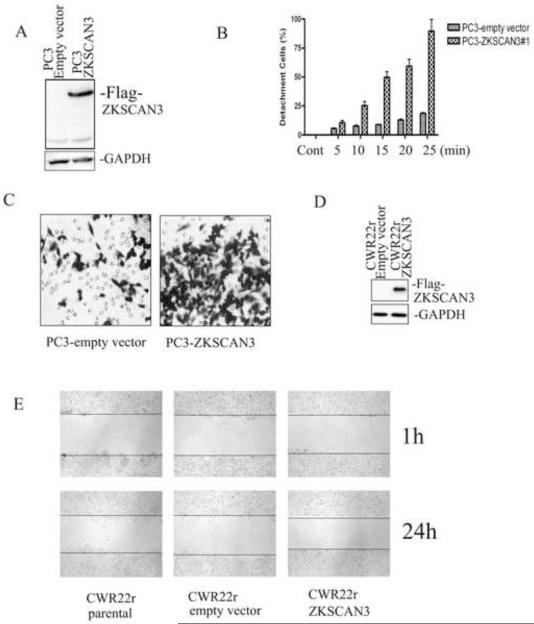
Differential expression of ZKSCAN3 in invasive and non-invasive tumor cells suggests a role for ZKSCAN3 in promoting tumor metastasis (Yang et al., 2008a). It has been suggested that both the ability of tumor cells to detach as well as an increase in motility are associated with the development of metastatic potential in prostate cancer. Thus, the next experiments were designed to determine if ZKSCAN3 participates in the process of prostate cancer cell detachment and motility. PC3 cells were transfected with a Flag-tagged ZKSCAN3 expression construct, and transfected cells were selected with 0.5 mg/ml G418. After 2 weeks, a G418-resistant GFP-positive pool was harvested and analyzed for ZKSCAN3 expression by Western blotting (Fig. 2A) using the anti-Flag M2 antibody. The Flag-tagged ZKSCAN3 protein was expressed in the PC3-ZKSCAN3 cell lysate, but not in the empty-vector transfected PC3 cell lysate.
Fig. 2.
Exogenous expression of ZKSCAN3 increases prostate cancer cell detachment and motility. A: Generation of stable PC3-ZKSCAN3 cell lines. PC3 cells were transfected with the Flag-tagged ZKSCAN3 expression construct. Cells were selected with 0.5 mg/ml G418 and after 2 weeks, a G418-resistant pool was harvested and analyzed for ZKSCAN3 expression by Western blotting using the anti-Flag M2 antibody. B: ZKSCAN3 enhanced PC3 cell detachment. PC3-vector and PC3-ZKSCAN3 stable cells were seeded in 24 well plates. After 48 hours, the cells were treated with 2mM EDTA for 5 to 25 minutes. The cells were washed with PBS and then incubated with MTT reagent diluted in RPMI medium (5 mg/ml) at 37°C for 2 hours. The formazan crystals produced by metabolism of MTT were reconstituted in DMSO, and the optical density at 570 nm was determined using an ELISA reader. The percentage of detached cells was calculated using the formula: 100 - [(OD570 of EDTA treated cells) / (OD570 of untreated cells) × 100%]. C: ZKSCAN3 enhanced PC3 cell motility in a transwell migration assay. PC3-vector and PC3-ZKSCAN3 stable cells were added to the upper chamber of each well and allowed to migrate through the collagen coated membrane for 2 to 3 hours. Cell motility was quantified by counting the cells that migrated to the lower surface of the membrane per square milliliter using bright-field optics. D: Overexpression of Flag-tagged ZKSCAN3 was confirmed in CWR22r cells. CWR22r cells were transfected with the Flag-tagged ZKSCAN3 expression construct, and ZKSCAN3 expression was analyzed by Western blotting using the anti-Flag M2 antibody. E: Overexpression of ZKSCAN3 enhanced CW22Rr cell migration in a wound healing assay. CWR22r cells were transfected with empty-vector or ZKSCAN3 for 48 hours. A 1 mm thick wound was created using a 10 μL micropipette tip, and cell migration was photographed after 24 hours.
We next examined the effect of overexpression ZKSCAN3 on PC3 cell proliferation and found that the proliferation rate of the PC3-ZKSCAN3 cells was similar to that of the PC3-empty vector cells, suggesting that ZKSCAN3 has little effect on PC3 cell proliferation in monolayer culture conditions (data not shown). However, we observed that the PC3-ZKSCAN3 cells were sensitive to incubation with detaching buffer (i.e. 2mM EDTA, trypsin), suggesting that ZKSCAN3 might have an impact on cell detachment. When 2mM EDTA was added to PC3 cells that had been adhered for 48 hours to 24 well plates, the PC3-ZKSCAN3 cells were significantly more sensitive to EDTA-induced detachment than the PC3-empty vector cells (Fig. 2B).
Since ZKSCAN3 enhanced cell detachment, we next investigated whether ZKSCAN3 regulated cell motility using a transwell migration assay. As shown in Fig.2C, PC3-ZKSCAN3 cells showed greater motility compared to PC3-empty vector cells. There was a three fold increase in the number of cells migrating through the membrane in the PC3-ZKSCAN3 cultures compared to the PC3-empty vector controls.
Because ZKSCAN3 greatly enhanced cell detachment and motility in PC3 cells, we next studied whether ZKSCAN3 regulated cell migration. CWR22r cells were transfected with the Flag-tagged ZKSCAN3 expression construct, and expression of Flag-tagged ZKSCAN3 was confirmed by Western blotting (Fig.2D). Wound healing assays were performed by creating a 1 mm wide wound in a monolayer of CWR22r cells transfected with either Flag-tagged ZKSCAN3 or empty vector. After 24 hours, CWR22r cells transfected with ZKSCAN3 had migrated farther than cells transfected with the empty vector (Fig. 2E), indicating that overexpression of ZKSCAN3 enhanced CWR22r cell migration, even though the CWR22r cells were originated from a primary xenograft without metastatic potential.
3.3. ZKSCAN3 knockdown decreases PC3 cell migration
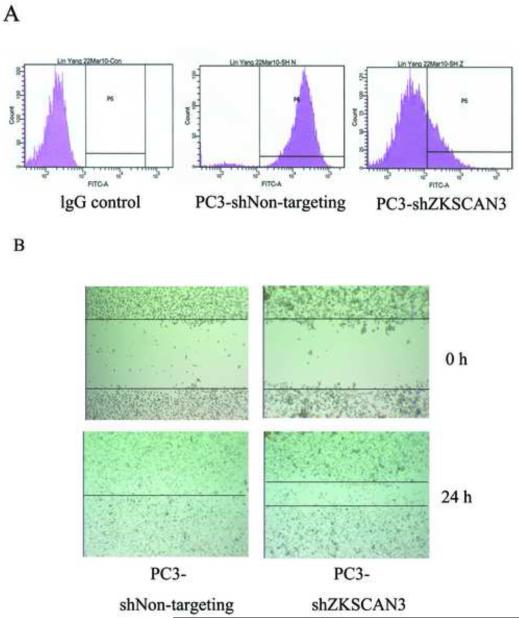
Since expression of ZKSCAN3 enhanced cell migration, we next studied whether knockdown of ZKSCAN3 could inhibit cell migration. PC3 cells were transfected with shZKSCAN3, an shRNA construct targeting ZKSCAN3. Flow cytometry was used to detect the expression of ZKSCAN3 in PC3 cells after treatment with shZKSCAN3 for 48 hours. ZKSCAN3 protein expression was greatly inhibited by shZKSCAN3 compared to the non-targeting shRNA control (Fig 3A).
Fig. 3.
Knockdown of ZKSCAN3 decreased PC3 cell migration A: The expression of ZKSCAN3 in PC3 cells was greatly reduced by shZKSCAN3. Flow cytometry analysis was used to detect ZKSCAN3 expression in PC3 cells treated with shNon-targeting and shZKSCAN3. Permeabilized cells were stained with either an anti-ZKSCAN3 antibody or an isotype matched control antibody, followed by anti-mouse IgG1 conjugated to FITC. The expression of ZKSCAN3 was greatly inhibited by shZKSCAN3 compared to the shNon-targeting control. B: Knockdown of ZKSCAN3 enhanced PC3 cell migration in a wound healing assay. PC3 cells were transfected with shNon-targeting control or shZKSCAN3 for 48 hours. A 1 mm thick wound was created using a 10 μL micropipette tip, and cell migration was photographed after 24 hours.
Wound healing assays were performed by creating a 1 mm thick wound in a monolayer of PC3 cells transfected with either shZKSCAN3 or shNon-targeting vectors. As shown in Fig. 3B, PC3 cells transfected with shZKSCAN3 demonstrated a significantly decreased migration 24 hours after the wound was created compared to PC3-shNon-targeting cells, indicating that knockdown of ZKSCAN3 decreased PC3 cell migration.
3.4. Exogenous expression of ZKSCAN3 increases cell cycle progression in suspension-cultured prostate cancer cells
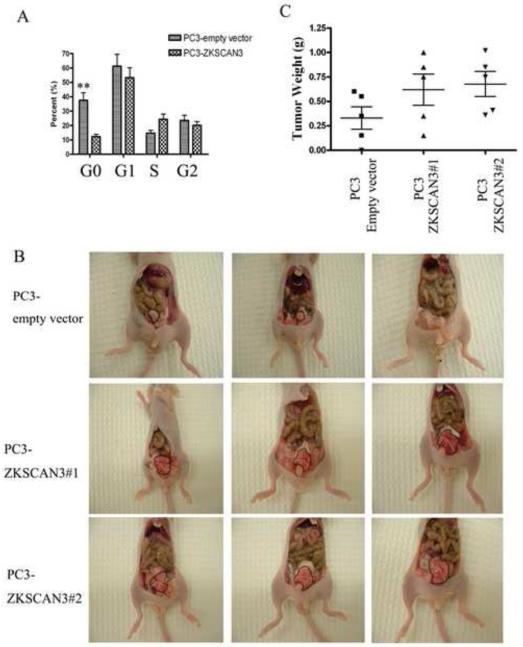
Our initial observation that overexpression of ZKSCAN3 in PC3 cells cultured in monolayer did not alter the proliferation rate was unexpected. However, if ZKSCAN3 indeed represents a critical factor in regulating cell migration, it would be expected to enhance cell viability and proliferation under conditions of anoikis stress, a situation that can be approximated in vitro by culturing prostate cancer cells in suspension. To determine if an enhanced cell proliferation rate contributed to the increased migration ability of the ZKSCAN3 transfected cells, cell cycle analysis was performed on PC3 cells transfected with ZKSCAN3 and the empty vector control under cell suspension culture conditions that mimic anoikis in vitro. When PC3-empty vector cells were grown in suspension culture, about 37% of the cells apoptotic were found in the sub-G1 portion of the cell cycle profile (Fig 4A). However, overexpression of ZKSCAN3 in PC3 cells decreased the apoptotic cells to about 12%, and increased the S phase cell population, suggesting that ZKSCAN3 is able to enhance PC3 cell survival and proliferation under anoikis stress, further supporting the role of ZKSCAN3 in regulating prostate cancer migration.
Fig. 4.
Effect of ZKSCAN3 on cell cycle and orthotopic tumor growth. A: Cell cycle profiles were obtained in PC3 cells transfected with either ZKSCAN3 or the control empty vector by staining with propidium iodide followed by flow cytometry analysis. A total of 20,000 events were collected from each of three replicate experiments, and cell cycle distributions were analyzed using Cell Fit software (Becton Dickinson). Statistical comparisons of the differences in the cell cycle distributions were carried out by ANOVA followed by Dunnett’s t-test. **p< 0.01 PC3-ZKSCAN3 group vs. PC3-empty vector group. B: Effect of ZKSCAN3 overexpression on orthotopic tumor growth. PC3 empty vector or PC3 ZKSCAN3 transfected cells (1×105 cells/25μl) were injected into the prostate of each nude mouse. After 4 weeks mice were sacrificed, and tumors were photographed. The tumors are circumscribed in the photographs. C: Orthotopic tumors were surgically removed and weighed.
3.5. Effect of ZKSCAN3 on PC3 cell tumor formation in vivo
As migration and motility of tumor cells are prerequisites for the development of prostate cancer metastasis, and the tumor micro-environment plays a critical role in affecting tumorigenesis, it is essential to evaluate the role of ZKSCAN3 in vivo with an orthotopic model. Two independent PC3 cell clones stably overexpressing ZKSCAN3 (PC3-ZKSCAN3#1 and PC3-ZKSCAN3#2) and PC3 cells stably transfected with the empty vector were orthotopically injected into nude mice. As expected, ZKSCAN3 caused a robust induction of tumorigenicity (Fig. 4B) as evidenced by a 2-fold increase in tumor weight four weeks after injection (Fig. 4C). The weight of the tumors increased from 0.3 ± 0.1g with PC3-empty vector cells to 0.7 ± 0.3g with PC3-ZKSCAN3(PC3-ZKSCAN3#1 and PC3-ZKSCAN3#2) cells, and the difference was statistically significant (p<0.05; n = 5). Statistical comparisons were carried out by ANOVA followed by Dunnett t-test. The experiment was repeated three times with similar results.Taken together, these data demonstrate that overexpression of ZKSCAN3 contributes to the tumorigenic phenotype.
4. DISCUSSION
Cancer remains one of the leading causes of death in developed countries. However, much remains to be learned about the mechanisms controlling cancer cell growth and metastasis, especially the molecular events in gene regulation in tumor cells that contribute to cancer metastasis. In our previous studies, we identified the novel zinc finger transcription factor ZKSCAN3 as a new driver for colorectal cancer and multiple myeloma progression (Yang et al., 2008a , Yang et al., 2008b, Yang et al., 2011). In particular, previous studies demonstrated that ZKSCAN3 is selectively expressed in invasive tumor cells and colon cancer liver metastases compared to non-invasive tumor cells (Yang et al., 2008a). To examine the possibility that ZKSCAN3 is involved in regulating tumor metastasis, we first investigated the impact of ZKSCAN3 on tumor cell migration, a prerequisite for tumor metastasis. In this work, we demonstrate that the expression level of ZKSCAN3 correlates with the metastatic potential of the prostate cancer cell lines, with the highest expression level in PC3 cells, which have high metastatic potential, but no expression in the primary xenograft derived CWR22r cells. Functional assays showed that knock down of ZKSCAN3 significantly decreased prostate cancer cell migration, while overexpression of ZKSCAN3 enhanced prostate cancer cell detachment in monolayer cultures treated with EDTA and enhanced survival under suspension culture conditions.
However, the mechanism by which ZKSCAN3 contributes to prostate cancer migration has not been elucidated. Previous studies from our group determined that ZKSCAN3 coordinated a gene expression program including vascular endothelial growth factor (VEGF), integrin β4, and cyclin D2; that it was an important “driver” for colon cancer and multiple myeloma progression; and that its small interfering (si) RNA-mediated down-regulation slowed tumor growth in vivo (Yang et al., 2011). Gene expression profiling data showed that EGFR (epidermal growth factor receptor), cyclin D1, NF-κB, MEK2 (mitogen-activated protein kinase kinase 2), and IGF-2 (insulin-like growth factor-2) may represent ZKSCAN3 downstream targets as well. Since most of these genes have been demonstrated to promote tumorigenesis, including cancer metastasis (Ueno et al., 2011, Li et al., 2006, Su et al., 2010, Suvasini et al., 2011), it would be scientifically significant to further analyze the gene expression program regulated by ZKSCAN3 during the process of enhancing prostate cancer cell migration.
Gene amplification could be one of the factors that contribute to increased ZKSCAN3 expression as prostate cancer progresses to a metastatic phenotype because amplification of the ZKSCAN3 gene was observed in metastatic prostate cancer samples and not in primary samples. Analysis of prostate cancer patient samples by aCGH array found that the region closest to ZKSCAN3 was amplified in 26% of metastatic prostate cancer samples (Fig. 1A). Indeed, our previous studies also observed that the ZKSCAN3 gene was amplified in colorectal cancer and multiple myeloma (Yang et al., 2008a , Yang et al., 2011), which contributed to ZKSCAN3 expression in these malignant cells.
In conclusion, we have described ZKSCAN3 as a driver of prostate tumor progression by modulating cell detachment, migration and motility. Moreover, ZKSCAN3 caused a robust induction of tumorigenicity in in vivo models. Further investigation of the upstream signal regulation of ZKSCAN3 will provide new insights into the mechanisms accommodating or contributing to prostate cancer cell metastasis, thereby unveiling new strategies for tumor therapy.
Acknowledgments
Grant support: This work was supported, in whole or in part, by the Priority Academic Program Development of Jiangsu Higher Education Institutions, NIH SPORE (P50 CA142509-DRP), Science and Technology Support Program Project of Jiangsu Province (BE2010649, BE2011682), Technological Plan of Society Development of Yunnan Province (2009CC026), and Postdoctoral Science Foundation of China (2011M500949).
We thank Dr. Pamela L. Paris (UCSF Comprehensive Cancer Center, CA, USA) for the high-resolution aCGH analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Chambers A. The metastatic process: basic research and clinical implications. Oncol Res. 1999;11:161–168. [PubMed] [Google Scholar]
- Grimstad I. Direct evidence that cancer cell locomotion contributes importantly to invasion. Exp Cell Res. 1987;173:515–523. doi: 10.1016/0014-4827(87)90291-6. [DOI] [PubMed] [Google Scholar]
- Hendrix M. Editorial: potential clinical use of exploiting metastasis suppressor genes to regulate prostatic cancer. J Urol. 2003;169:1134. doi: 10.1097/01.ju.0000047314.77231.f3. [DOI] [PubMed] [Google Scholar]
- Howlader N, Noone A, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER Cancer Statistics Review, 1975-2008. National Cancer Institute; Bethesda: 2011. [Google Scholar]
- Janer M, Friedrichsen D, Stanford J, Badzioch M, Kolb S, Deutsch K, et al. Genomic scan of 254 hereditary prostate cancer families. Prostate. 2003;57:309–319. doi: 10.1002/pros.10305. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi O, Sudar D, Rutovitz D, Gray J, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kauffman E, Robinson V, Stadler W, Sokoloff M, Rinker-Schaeffer C. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 2003;169:1122–33. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiao X, Wang C, Ju X, Yinan L, Yuan L, et al. Cyclin D1 induction of cellular migration requires p27KIP1. Cancer Res. 2006;66:9986. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jovanovic B, Pins M, Lee C, Bergan R. Over expression of endoglin in human prostate cancer suppresses cell detachment, migration and invasion. Oncogene. 2002;21:8272–8281. doi: 10.1038/sj.onc.1206117. [DOI] [PubMed] [Google Scholar]
- Paris P, Albertson D, Alers J, Andaya A, Carroll P, Fridlyand J, et al. High-resolution analysis of paraffin-embedded and formalin-fixed prostate tumors using comparative genomic hybridization to genomic microarrays. Am J Pathol. 2003;162:763–770. doi: 10.1016/S0002-9440(10)63873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris P, Sridharan S, Scheffer A, Tsalenko A, Bruhn L, Collins C. High resolution oligonucleotide CGH using DNA from archived prostate tissue. Prostate. 2007;67:1447–1455. doi: 10.1002/pros.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris P, Andaya A, Fridlyand J, Jain AN, Weinberg V, Kowbel D, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum. Mol. Genet. 2004;13:1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- Santos GC, Zielenska M, Prasad M, Squire JA. Chromosome 6p amplification and cancer progression. J. Clin. Pathol. 2007;60:1–7. doi: 10.1136/jcp.2005.034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D, McDonnell S, Zarfas K, Cunningham J, Hebbring S, Thibodeau S, et al. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120:471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- Stracke M, Aznavoorian S, Beckner M, Liotta L, Schiffmann E. Cell motility, a principal requirement for metastasis. EXS. 1991;59:147–162. doi: 10.1007/978-3-0348-7494-6_10. [DOI] [PubMed] [Google Scholar]
- Su B, Bu Y, Engelberg D, Gelman H. I. SSeCKS/Gravin/AKAP12 Inhibits Cancer Cell Invasiveness and Chemotaxis by Suppressing a Protein Kinase C- Raf/MEK/ERK Pathway. J. Biol. Chem. 2010;285:4578–4586. doi: 10.1074/jbc.M109.073494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvasini R, Shruti B, Thota B, Shinde S, Friedmann-Morvinski D, Nawaz Z, et al. Insulin Growth Factor-2 Binding Protein 3 (IGF2BP3) Is a Glioblastoma-specific Marker That Activates Phosphatidylinositol 3-Kinase/Mitogen-activated Protein Kinase (PI3K/MAPK) Pathways by Modulating IGF-2. J. Biol. Chem. 2011;286:25882–25890. doi: 10.1074/jbc.M110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B, Varambally S, Chinnaiyan A. Differential proteomic alterations between localised and metastatic prostate cancer. Br J Cancer. 2006;95:425–430. doi: 10.1038/sj.bjc.6603274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Lin S. Clinical aspects of bone metastases in prostate cancer. Cancer Treat Res. 2004;118:23–46. doi: 10.1007/978-1-4419-9129-4_2. [DOI] [PubMed] [Google Scholar]
- Ueno S, Mojic M, Ohashi Y, Higashi N, Hayakawa Y, Irimura T. Asialoglycoprotein Receptor Promotes Cancer Metastasis by Activating the EGFR-ERK Pathway. Cancer Res. 71:6419. doi: 10.1158/0008-5472.CAN-11-1773. [DOI] [PubMed] [Google Scholar]
- Wan X, Kim S, Guenther L, Mendoza A, Briggs J, Yeung C, et al. Beta4 integrin promotes osteosarcoma metastasis and interacts with ezrin. Oncogene. 2009;28:3401–3411. doi: 10.1038/onc.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Hamilton SR, Sood A, Kuwai T, Ellis L, Sanguino A, et al. The Previously Undescribed ZKSCAN3 (ZNF306) Is a Novel “Driver” of Colorectal Cancer Progression. Cancer Res. 2008a;68:4321–4330. doi: 10.1158/0008-5472.CAN-08-0407. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang H, Kornblau S, Graber D, Zhang N, Matthews J, et al. Evidence of a role for the novel zinc-finger transcription factor ZKSCAN3 in modulating Cyclin D2 expression in multiple myeloma. Oncogene. 2011;30:1329–1340. doi: 10.1038/onc.2010.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang L, Wu Q, Boyd DD. Unbiased Screening for Transcriptional Targets of ZKSCAN3 Identifies Integrin {beta}4 and Vascular Endothelial Growth Factor as Downstream Targets. J. Biol. Chem. 2008b;283:35295–35304. doi: 10.1074/jbc.M806965200. [DOI] [PMC free article] [PubMed] [Google Scholar]