Abstract
Tyrosine phosphorylation is an essential element of signal transduction in multicellular animals. Although tyrosine kinases were originally regarded as specific to the metazoan lineage, it is now clear that they evolved prior to the split between unicellular and multicellular eukaryotes (≈ 600 million years ago). Genome analyses of choanoflagellates and other protists show an abundance of tyrosine kinases that rivals the most complex animals. Some of these kinases are orthologs of metazoan enzymes (e.g., Src), but others display unique domain compositions not seen in any metazoan. Biochemical experiments have highlighted similarities and differences between the unicellular and multicellular tyrosine kinases. In particular, it appears that the complex systems of kinase autoregulation may have evolved later in the metazoan lineage.
Keywords: tyrosine kinase, phosphorylation, evolution, choanoflagellate, SH2 domain, SH3 domain, autoinhibition
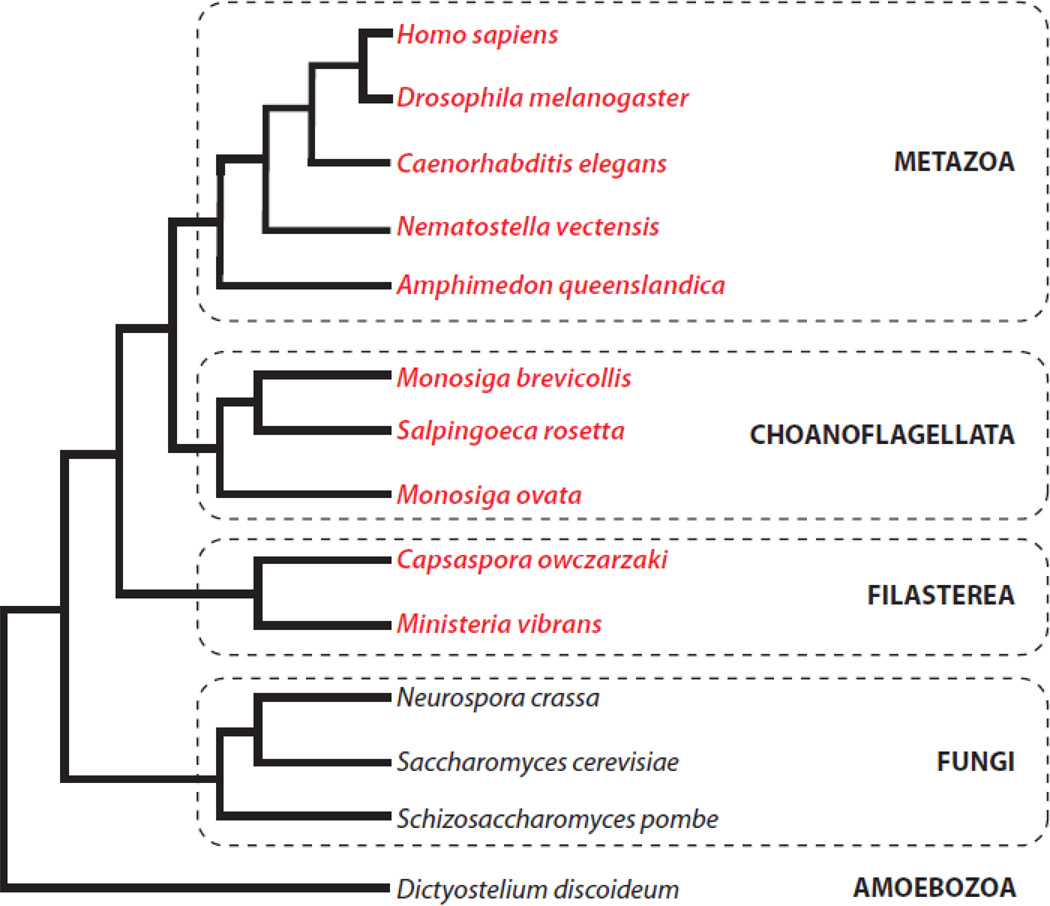
Protein phosphorylation is a sine qua non of signal transduction in both prokaryotes and eukaryotes. While prokaryotic phosphorylation is dominated by phospho-histidine signaling, phosphorylation of serine, threonine, and tyrosine predominates in eukaryotes [1, 2]. Eukaryotic tyrosine phosphorylation is catalyzed by a specific group of protein kinases (designated TK-group kinases in this review). The emergence of these enzymes appears to be a relatively recent innovation in the history of life. TK-group kinases are absent in prokaryotes, although some prokaryotes have unique tyrosine kinases (designated BY kinases) that are related to the nucleotide triphosphatase superfamily [3]. Eukaryotic-like tyrosine kinases are also absent in plants, in yeast and other fungi, and in the slime mold Dictyostelium discoideum (Fig. 1) [1, 4]. Thus, the view emerged that TK-group kinases were metazoan-specific signaling enzymes. Tyrosine kinases play essential roles in the regulation of growth and differentiation, and in cell-cell communication, in multicellular organisms [2].
Fig. 1. Distribution of tyrosine kinases.
The eukaryotic tree of life is shown schematically. Species containing eukaryotic-like tyrosine kinases are shown in red.
A gap of over 1.5 billion years separated the appearance of single-celled eukaryotes and the evolution of the first multicellular eukaryotes [5–7]. While all organisms require signal transduction systems to respond to external cues, one critical requirement for multicellularity was the development of signaling mechanisms that enabled intercellular communication and coordination. Hunter and Cooper, in the first comprehensive review of tyrosine kinases (which appeared in 1985, only six years after the discovery of tyrosine phosphorylation), stated that “a clear prerequisite for a multicellular organism is a means of cell-cell signaling and the protein tyrosine kinase nature of many of the growth factor receptors may be pertinent” [8]. The requirement for intercellular signaling mechanisms raises the possibility that tyrosine kinases might be found in an organism close to the branch point with metazoans. This was confirmed in 2001 by King and Carroll in their studies of the unicellular choanoflagellate Monosiga brevicollis. These workers discovered the first TK-group kinase (a receptor tyrosine kinase designated MBRTK1) outside of the Metazoa [9]. A subsequent paper confirmed the existence of multiple active tyrosine kinases in this organism [10], leading to the suggestion that unicellular tyrosine kinases (and other key signaling molecules such as adhesion proteins) may have facilitated the evolution of multicellular animals [5].
Choanoflagellates are a group of protists that are believed to be the closest single-celled relatives to metazoans (Fig. 1) [11, 12]. Many choanoflagellates can form colonies, suggesting that they may represent a transitional form between unicellular and multicellular organisms. Strikingly, genomic analyses have revealed that choanoflagellates contain numbers of tyrosine kinase genes that are comparable to (or even exceed) the numbers in the most complex multicellular animals [13]. In Monosiga brevicollis, the number of tyrosine kinases has been estimated to be as high as 128, although estimates differ depending on the methodology used for identification [14, 15]. Tyrosine kinases have also been identified in the choanoflagellates Monosiga ovata [16] and Salpingoeca rosetta [17]. Recent genome analyses indicate that tyrosine kinases are present in even more ancient opisthokonts (the eukaryotic supergroup that includes Fungi and Metazoa [7, 18, 19]). The filasterean Capsaspora owczarzaki, a member of a sister group to choanoflagellates and metazoans [20], has a rich and diverse tyrosine kinome (103 tyrosine kinase genes), and tyrosine kinases are present in Ministeria vibrans, a related filasterean (H. Suga and I. Ruiz-Trillo, personal communication). Thus, the familiar tyrosine phosphorylation-based signaling system probably evolved before the divergence of C. owczarzaki from the branch including choanoflagellates and metazoans (Fig. 1). This innovation, which occurred ≈ 600 million years ago, may have been a critical event enabling the development of multicellular animals. Analysis of the genome of the sponge, Amphimedon queenslandica, one of the simplest and earliest branching metazoans, shows a dramatic expansion in the number of tyrosine kinases [21]. Remarkably, there are over 150 likely receptor tyrosine kinases in the sponge, including members of animal families such as the epidermal growth factor and Met receptors.
In present-day animal cells, the complete ‘toolkit’ of pTyr-based signaling consists of three components: (1) tyrosine kinases, which catalyze the modification; (2) protein tyrosine phosphatases (PTPs), which catalyze the opposing reaction (dephosphorylation of tyrosine residues); and (3) modular pTyr-binding domains (SH2, PTB). These three components have been designated “writers,” “erasers,” and “readers,” respectively, by Wendell Lim and colleagues [14, 22]. Regulatory circuits containing these three components play critical roles in the growth and differentiation of most metazoan cells. Based on the sequence of appearance of the three components, a model can be constructed for the evolution of pTyr signaling [22]. Genome analyses indicate that tyrosine phosphatases were the first to emerge in evolution. PTPs have been identified in several evolutionarily divergent protozoans that lack TK-group kinases, including Leishmania and Trypanosoma [23]. While the budding yeast Saccharomyces cerevisiae contains no tyrosine kinases and one primitive SH2 domain (which lacks the ability to bind p-Tyr), it has a few simple PTPs that are enzymatically active. These PTPs may have evolved to control phosphorylated tyrosine residues modified by yeast dual specificity kinases. (A caveat in considering S. cerevisiae as an example of a simple signaling system is that this organism lost a large number of genes since the radiation from the common ancestor with S. pombe) [24]. A more sophisticated p-Tyr signaling system is observed in organisms that contain “reader” domains in addition to PTPs. For example, the slime mold Dictyostelium discoideum has a repertoire of 13 proteins containing SH2 domains but lacks metazoan-like tyrosine kinases [15, 22]. The expansion of putative dual-specificity kinases in this organism, coupled with the lack of TK-group kinases, again suggests that SH2 domains initially evolved to control pathways involving dual-specificity catalytic domains.
The number and identity of signaling domains with which tyrosine kinases, PTPs, and SH2 domains are combined also serve as clues to the history of p-Tyr signaling [14, 25]. In animal cells, p-Tyr signaling proteins tend to be large molecules composed of several modular signaling domains. This architecture is believed to reflect the importance of domain shuffling and recombination in the evolution of phosphotyrosine-based signaling. In contrast, the PTPs found in simple eukaryotes such as Tetrahymena thermophile, Saccharomyces cerevisiae, and Dictyostelium discoideum possess “stripped-down” architectures consisting of single-domain proteins or fusions with rhodanese domains [14, 15, 22]. Similarly, the 13 “reader” SH2 domains found in Dictyostelium discoideum cluster into 5 basic architectures. The domain combinations in Dictyostelium are fairly limited, and do not display the kind of diversity observed in metazoan SH2 domain-containing proteins.
In contrast, with the emergence of the “writer” domain (i.e., the tyrosine kinase catalytic domain) closer to the advent of multicellular organisms, there was a flowering of domain combinations involving kinase, PTP, and SH2 domains. Choanoflagellates and metazoans have 30–50 PTP domains and up to 100–150 SH2 domains, about a 10-fold expansion as compared to Saccharomyces or Dictyostelium [14, 15, 22, 25]. The number and variety of pairwise domain combinations are also greatly expanded in lineages containing tyrosine kinase domains. This reflects the increasing sophistication of pTyr-dependent regulatory circuits, in which the multidomain proteins can serve to integrate multiple signals, to bifurcate pathways, or to modulate the strength or duration of signaling [26, 27]. Novel signaling systems based on tyrosine kinases, such as those observed in the nervous, vascular, and immune systems, arose at a later stage [28]. This explains the lack of kinases dedicated to these functions (e.g., immune cell tyrosine kinases such as Syk and ZAP-70) in choanoflagellates.
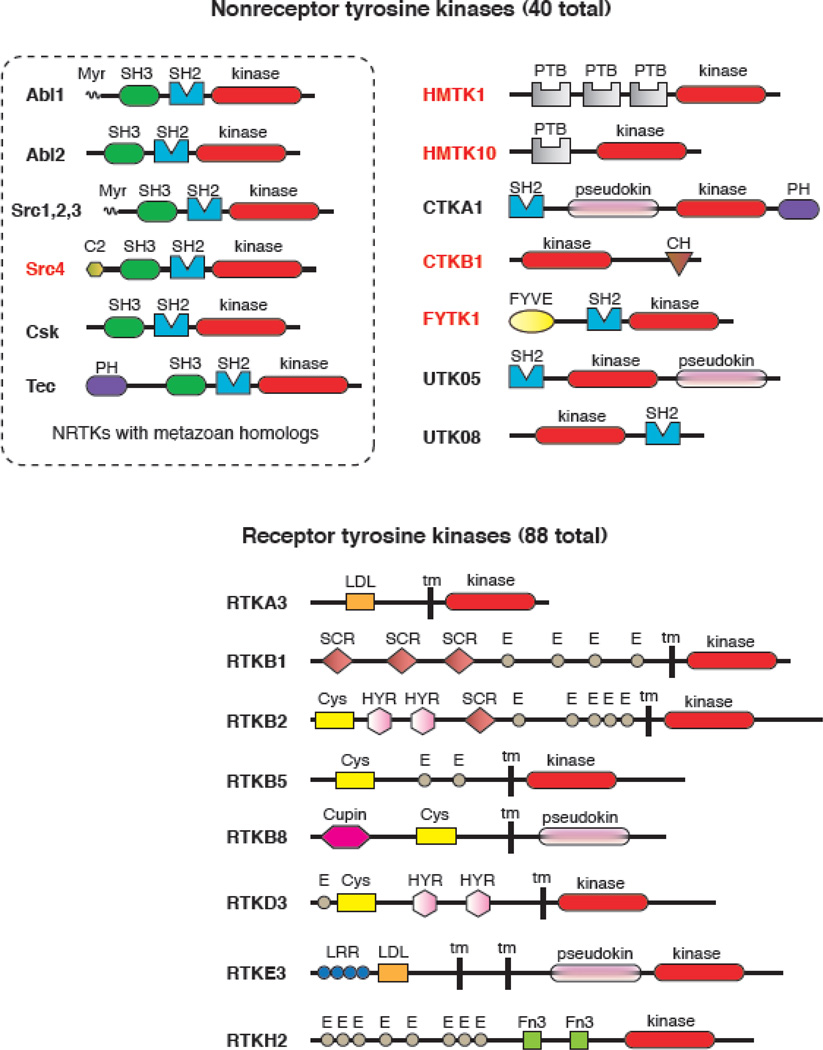
The domain architectures of the 128 tyrosine kinases in Monosiga brevicollis are more varied than those seen in complex metazoans (Fig. 2) [15]. This reinforces the idea that tyrosine kinases evolved before the split between choanoflagellates and metazoans. The study of the Monosiga kinome affords a glimpse into an early stage in the development of tyrosine kinase-mediated signaling. Based on predicted signal peptides and transmembrane sequences, 88 receptor tyrosine kinases are present in Monosiga. Most of these proteins show the typical type I transmembrane layout of mammalian RTKs, and contain extracellular domains composed of modular units, similar to mammalian RTKs (e.g., Cys-rich regions, EGF-like repeats). The majority of the Monosiga RTKs have one or more tyrosines in the activation loop of the catalytic domain, suggesting that they are activated by phosphorylation at this position. Despite their similarity in overall domain architecture to metazoan RTKs, the Monosiga RTKs are only weakly related by sequence to their metazoan counterparts. The kinome of the sponge Amphimedon queenslandica presents a hybrid case, in which there are RTKs from six known animal families, but others with unique extracellular domains and no obvious animal counterparts [21].
Fig. 2. Tyrosine kinases in Monosiga brevicollis.
The domain arrangements of representative nonreceptor tyrosine kinases (NRTKs, top) and receptor tyrosine kinases (RTKs, bottom) are shown. Domain predictions are based on the analysis in ref. [15], and a complete list of kinases is given in the supporting information to that paper. Tyrosine kinase catalytic domains are shown in red. At the top left, all Monosiga brevicollis NRTKs with clear metazoan homologs (based on kinase domain sequence similarity) are boxed. Names given in red are NRTKs with domain combinations that have not been observed in metazoans. List of domain abbreviations: Myr, myristoylation sequence; SH3, Src homology 3; SH2, Src homology 2; C2, phospholipid binding domain; PH, pleckstrin homology; PTB, phosphotyrosine binding domain; pseudokin, predicted inactive kinase domain; CH, calponin homology; FYVE, Fab1/YOTB/Vac1/EEA1 zinc-finger domain; LDL, low density lipoprotein receptor motif; tm, transmembrane sequence; SCR, short consensus repeat/complement control protein domain; E, epidermal growth factor repeat; Cys, cysteine-rich region; HYR, repeats similar to hyaline domains; Cupin, barrel-like fold seen in cupin family proteins; LRR, leucine-rich repeats; Fn3, fibronectin type 3 domain.
The diversity of tyrosine kinases in Monosiga brevicollis is most evident in the 40 non-receptor tyrosine kinases (NRTKs). The Monosiga genome contains clear orthologs of several metazoan NRTKs (Src, Abl, Tec, and Csk) [15], but most of the cytoplasmic tyrosine kinases have no obvious orthologs in metazoans. Many of these kinases contain combinations of signaling domains that are not observed in multicellular animals (Fig. 2). For example, the two FYTK kinases have inositol lipid-binding FYVE domains in addition to SH2 domains, and 10 of the 15 HMTK kinases contain phosphotyrosine-binding (PTB) domains. A Src variant (MbSrc4) has a lipid-binding C2 domain at its amino terminus. None of these domains is found to be associated with a tyrosine kinase domain in metazoans. Many of the domain combinations in Monosiga involving PTP and SH2 domains are also not found in any metazoan genome [13, 14]. The differences in domain combinations between choanoflagellates and metazoans suggests that tyrosine kinases arose shortly before the split between these lineages, and that the divergent expansion occurred subsequently.
The earliest demonstration of an active TK-group kinase outside Metazoa was in lysates from Monosiga brevicollis cells. Western blotting with anti-phosphotyrosine antibodies detected 15–18 proteins under conditions of nutrient depletion [10]. When the Monosiga culture was propagated in seawater infused with cereal grass and live Enterobacter aerogenes (a food source), increases and decreases in protein tyrosine phosphorylation were observed. Furthermore, treatment of Monosiga cultures with genistein (a general tyrosine kinase inhibitor) or PP2 (a Src family kinase inhibitor) led to decreases in cell proliferation, suggesting that pTyr-based signaling plays a functional role in choanoflagellates. Because of the limited homology of Monosiga RTK extracellular domains with known metazoan counterparts, and because the ligands for Monosiga RTKs are currently unknown, it is not possible to identify which RTK (or combination of RTKs) was activated in these whole-cell experiments.
A number of nonreceptor tyrosine kinases from unicellular choanoflagellates have been cloned, expressed, and demonstrated to be active. Three Src-family kinases and one Csk kinase were identified in Monosiga ovata (a choanoflagellate derived from a distinct clade from M. brevicollis). When expressed in mammalian cells and analyzed by Western blotting with anti-phosphotyrosine antibody, these kinases were demonstrated to have varying levels of activity [16]. Similarly, a Src-family kinase (MbSrc1) and a Csk ortholog from Monosiga brevicollis were active in mammalian cells. The Src-family kinase could functionally substitute for mammalian Src in a reporter gene assay that depends on Src phosphorylation of STAT proteins [13, 29]. Furthermore, purified MbSrc1 displayed a turnover number that was comparable to the value for a mammalian Src-family kinase [29]. Two Monosiga brevicollis NRTKs with unusual domain combinations have also been shown to be active: the C2 domain-containing MbSrc4 [30], and HMTK1, which contains three tandem PTB domains N-terminal to the tyrosine kinase catalytic domain [31]. Each of these M. brevicollis NRTKs contains a single tyrosine in the predicted activation loop, a flexible segment between the two lobes of the kinase domain that is a site for regulatory phosphorylation in many kinases. Biochemical experiments confirmed that these kinases were regulated by autophosphorylation [29–31], consistent with the idea that the unicellular kinase domains share the important structural features found in metazoan NRTKs.
In cases where they have been examined, the associated signaling domains of choanoflagellate NRTKs have similar functions to their counterparts in animal cells. Most Monosiga Src-family kinases possess the same architecture as observed in multicellular organisms: from N-terminus to C-terminus, they contain a membrane-targeting myristoylation sequence, a unique region, SH3, SH2, and tyrosine kinase domains, and a C-terminal regulatory sequence. Ligand binding to the isolated SH3 and SH2 domains of Monosiga brevicollis MbSrc1 was studied in vitro by isothermal titration calorimetry. These experiments demonstrated that the SH2 domain bound to a pTyr-containing peptide, and the SH3 domain bound to a proline-rich peptide derived from Sam68; in each case, the binding affinity was comparable to the value observed in mammalian Src family kinases [29]. In vitro experiments on the purified C2 domain from M. brevicollis MbSrc4 demonstrated that it bound to large unilamellar vesicles, confirming the prediction that it acts as a membrane targeting module [30]. A PTB domain derived from M. brevicollis HMTK1 bound to a series of tyrosine-phosphorylated and unphosphorylated peptide motifs in vitro [31]. These examples suggest that there are a common set of constraints in the design of pTyr-based signaling pathways in choanoflagellates and metazoans.
The common themes in the design of pTyr signaling proteins suggest a number of examples of convergent evolution between choanoflagellates and metazoans. Both groups contain numerous transmembrane RTKs with similar overall domain architectures, yet the kinase domain sequences do not display specific homologies within families [15]. M. brevicollis lacks orthologs of the SH2-containing Ras guanine nucleotide exchange factor (RasGEF) and Rho GTPase activating protein (RhoGAP) found in animal cells. Unrelated variants with different combinations of these domains are present, however, suggesting that they may have arisen independently in the two lineages [15, 25]. The domain architectures of several Monosiga NRTKs also show evidence of functional convergence. While three out of the four Src-family kinases retain the N-terminal myristoylation sequence for membrane anchoring, the sequence is replaced in MbSrc4 by a C2 domain. The C2 domain targets MbSrc4 to membranes in vitro and in cells, suggesting that this alternative lipid-binding mechanism arose to fulfill a similar function [30]. In mammalian Src-family kinases, one function of the SH3 and SH2 domains is to target the enzymes to specific substrates containing the appropriate ligands [32–34]. The combination of PTB and tyrosine kinase domains in the M. brevicollis HMTK family enzymes, which is not seen in metazoans, appears to exist to facilitate substrate targeting [31].
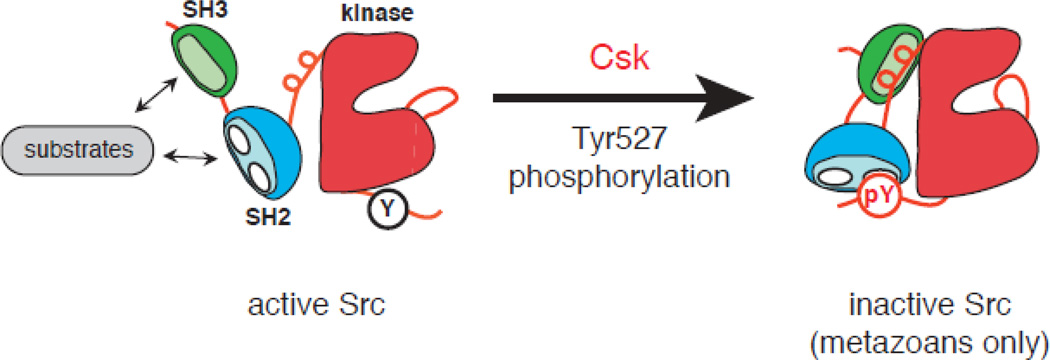
A hallmark of mammalian tyrosine kinases is that their activity is tightly controlled; aberrant activation can lead to unregulated cell growth [35]. In unstimulated cells, basal RTK and NRTK activity is suppressed by elaborate autoinhibitory interactions that stabilize inactive conformations of the enzymes. Src-family NRTKs are regulated by two important intramolecular interactions: (1) Csk catalyzes the phosphorylation of a C-terminal negative regulatory tyrosine (Y527) in Src, which produces an interaction with Src’s SH2 domain; (2) the SH3 domain binds to a polyproline type II helix present in the linker between SH2 and kinase domains. Disruption of these interactions stimulates phosphorylation in the activation loop (Y416) and increases Src kinase activity [36–38].
Experiments in two choanoflagellate systems suggest that Csk-imposed negative regulation of Src is relatively weak in these unicellular organisms. Monosiga ovata Src kinases were active and induced cell transformation when expressed in mammalian fibroblasts, even in the presence of M. ovata Csk [16]. Mass spectrometry experiments on purified Monosiga brevicollis Src confirmed the ability of M. brevicollis Csk to phosphorylate Src at the C-terminal negative regulatory site. This phosphorylation did not inhibit Src, despite the fact that the SH2 domain was functional and could bind to a synthetic phosphopeptide mimicking the C-terminal tail [29]. Co-expression of M. brevicollis Csk did not inhibit M. brevicollis Src tyrosine kinase activity in mammalian cells, or alter the ability of Src to induce gene expression. These experiments suggested that the autoinhibitory interactions necessary for maintaining the down-regulated state of Src kinases are absent in M. ovata and M. brevicollis. Thus, these organisms should possess high levels of basal Src activity; Western blotting experiments in M. ovata were consistent with this prediction [16]. The situation is even more pronounced in the amoeba Capsaspora owczarzaki, where the Csk ortholog has no measurable tyrosine kinase activity toward C. owczarzaki Src or any other substrate (K. Schultheiss and W.T.M, unpublished observations). Substrate targeting via the SH3 domain appears to be present in M. brevicollis Src, as measured in vitro [29]. This leads to a model in which the substrate recruitment function of the SH3 and SH2 domains (and, more generally, of noncatalytic domains of tyrosine kinases) arose before their roles in enzyme autoinhibition [31, 39]. Further refinements to the Src-Csk systems were required to establish the autoregulatory interactions observed in metazoans. Intramolecular regulation of the Crk adaptor protein is another example of pTyr-dependent signaling that was added relatively recently in metazoan evolution [28].
The biological purpose of extremely high numbers of tyrosine kinases, PTPs, and SH2-containing proteins in unicellular organisms such as M. brevicollis and C. owczarzaki remains enigmatic. The number and diversity of RTKs suggests that pTyr-based signaling is present to allow the cells to respond to their extracellular environments; this suggestion is consistent with the observed changes in M. brevicollis tyrosine phosphorylation in response to nutrient availability [10]. Experiments with inhibitors suggest that tyrosine kinase activity is required for M. brevicollis proliferation [10], yet RTKs and NRTKs may be involved in the responses to other extracellular cues, such as the presence of various nutrients, ions, or chemical messengers, or in the mating process. Detailed functional analyses of individual tyrosine kinases in choanoflagellates or C. owczarzaki will be required to shed light on this puzzle. A current obstacle is the lack of methodology to manipulate gene function in these organisms. Nonetheless, genomic comparisons with metazoans have given important glimpses into the early evolution of pTyr-based signaling, and helped to identify genes that may have facilitated the transition to multicellularity.
Fig. 3. Csk-mediated inhibition of Src.
In the active conformation of Src (left), Tyr527 in the C-terminal tail is unphosphorylated, and the SH2 and SH3 domains are disengaged from their intramolecular ligands. The SH3 and SH2 domains bind to ligands on potential substrates, targeting them for phosphorylation by the kinase domain. Phosphorylation of Tyr527 (red circle) produces an intramolecular interaction with the SH2 domain of Src. This interaction, together with an interaction between the SH3 domain and a polyproline type II helix in the linker region, stabilizes the autoinhibited conformation of Src. In the choanoflagellates Monosiga ovata and Monosiga brevicollis, the Csk homologs phosphorylate the residue equivalent to Tyr527, but this does not repress Src activity [16, 29]. The domain structure of Src family kinases and other NRTKs may have arisen initially to fulfill the substrate targeting function [31, 39].
Acknowledgements
I thank Kira Schultheiss, Markus Seeliger, Gerard Manning, and Wendell Lim for helpful comments on the manuscript. Work on kinase evolution in the author’s laboratory is supported by NIH grant R01 CA58530.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends in biochemical sciences. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Tyrosine phosphorylation: thirty years and counting. Current opinion in cell biology. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee DC, Jia Z. Emerging structural insights into bacterial tyrosine kinases. Trends in biochemical sciences. 2009;34:351–357. doi: 10.1016/j.tibs.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 5.King N. The unicellular ancestry of animal development. Developmental cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Rokas A. The molecular origins of multicellular transitions. Current opinion in genetics & development. 2008;18:472–478. doi: 10.1016/j.gde.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Hunter T, Cooper JA. Protein-tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 9.King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 11.Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 12.Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Molecular biology and evolution. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- 13.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci U S A. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segawa Y, Suga H, Iwabe N, Oneyama C, Akagi T, Miyata T, Okada M. Functional development of Src tyrosine kinases during evolution from a unicellular ancestor to multicellular animals. Proc Natl Acad Sci U S A. 2006;103:12021–12026. doi: 10.1073/pnas.0600021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Molecular biology and evolution. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 19.Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PloS one. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz-Trillo I, Inagaki Y, Davis LA, Sperstad S, Landfald B, Roger AJ. Capsaspora owczarzaki is an independent opisthokont lineage. Curr Biol. 2004;14:R946–R947. doi: 10.1016/j.cub.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreeva AV, Kutuzov MA. Protozoan protein tyrosine phosphatases. International journal for parasitology. 2008;38:1279–1295. doi: 10.1016/j.ijpara.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer BJ. Clues to the evolution of complex signaling machinery. Proc Natl Acad Sci U S A. 2008;105:9453–9454. doi: 10.1073/pnas.0804669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they're apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 28.Liu BA, Shah E, Jablonowski K, Stergachis A, Engelmann B, Nash PD. The SH2 Domain-Containing Proteins in 21 Species Establish the Provenance and Scope of Phosphotyrosine Signaling in Eukaryotes. Science signaling. 2011;4:ra83. doi: 10.1126/scisignal.2002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Young SL, King N, Miller WT. Signaling Properties of a Non-metazoan Src Kinase and the Evolutionary History of Src Negative Regulation. J Biol Chem. 2008;283:15491–15501. doi: 10.1074/jbc.M800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Scarlata S, Miller WT. Evidence for convergent evolution in the signaling properties of a choanoflagellate tyrosine kinase. Biochemistry. 2009;48:5180–5186. doi: 10.1021/bi9000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prieto-Echague V, Chan PM, Craddock BP, Manser E, Miller WT. PTB domain-directed substrate targeting in a tyrosine kinase from the unicellular choanoflagellate Monosiga brevicollis. PloS one. 2011;6:e19296. doi: 10.1371/journal.pone.0019296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller WT. Determinants of substrate recognition in nonreceptor tyrosine kinases. Acc Chem Res. 2003;36:393–400. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 34.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Current opinion in cell biology. 2009;21:147–153. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 36.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [see comments] [DOI] [PubMed] [Google Scholar]
- 37.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Current opinion in structural biology. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 38.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 39.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]