Abstract
Resting state functional connectivity MRI (rsfc-MRI) reveals a wealth of information about the functional organization of the brain, but poses unique challenges for quantitative image analysis, mostly related to the large number of voxels with low signal-to-noise ratios. In this study, we tested the idea of using a prior spatial parcellation of the entire brain into various structural units, to perform an analysis on a structure-by-structure, rather than voxel-by-voxel, basis. This analysis, based upon atlas parcels, potentially offers enhanced SNR and reproducibility, and can be used as a common anatomical framework for cross-modality and cross-subject quantitative analysis. We used Large Deformation Diffeomorphic Metric Mapping (LDDMM) and a deformable brain atlas to parcel each brain into 185 regions. To investigate the precision of the cross-subject analysis, we computed inter-parcel correlations in 20 participants, each of whom were scanned twice, as well as the consistency of the connectivity patterns inter- and intra-subject, and the intersession reproducibility. We report significant inter-parcel correlations consistent with previous findings, and high test-retest reliability, an important consideration when the goal is to compare clinical populations. As an example of the cross-modality analysis, correlation with anatomical connectivity is also examined.
Keywords: resting state, fMRI, rsfc-fMRI, atlas, connectivity
Introduction
Spontaneous low-frequency fluctuations in blood oxygenation level-dependent (BOLD) signal display spatial distributions concordant with known functional areas and networks (Biswal et al., 1995; De Luca et al., 2006; Fox et al., 2005; Greicius et al., 2003; Hampson et al., 2002; Hampson et al., 2004; Lowe, Mock, Sorenson 1998), suggesting that they may reflect fluctuations in ongoing neuronal activity, rather than mere physiological noise. Seed-based connectivity (SBC) (Biswal et al., 1995) and independent component analysis (ICA) (Calhoun et al., 2001a; Calhoun et al., 2001b) have been used to characterize the intrinsic connectivity networks (ICNs) observable while participants are “resting” (Damoiseaux et al., 2006; De Luca et al., 2006; van de Ven et al., 2004). Compared to task-activation fMRI studies, the “resting state” reduces demands on participant compliance. Importantly, the ICN maps appear to match task activation networks (Smith et al., 2009) and the spatial distribution of abnormalities seen in some pathological conditions (Greicius et al., 2004; Luo et al., 2011; Seeley et al., 2009; Sheline et al., 2010; Song et al., 2011).
While the resting state functional connectivity MRI (rsfc-MRI) reveals information about the functional organization, it also poses unique challenges for image analysis, mostly related to low signal-to-noise ratio (SNR) and the large number of voxels. Even with 3mm isotropic voxel resolution, there are more than 60,000 voxels in a typical brain and, consequently, almost 1,800,000,000 potential inter-voxel correlations. While this provides a large amount of spatial information, the signal from each voxel is noisy, and statistical analysis is challenging. A common approach to ameliorate this problem is to perform spatial smoothing, which, however, increases partial volume effects. In the original fMRI data, the complex cortical anatomy was not quite resolved. After smoothing, information from the CSF, white matter, and adjacent (spatially close but anatomically distant) cortex becomes further mixed. Recently, an alternative approach has been reported in which brains are parceled into structural units prior to the connectivity analysis (Achard et al., 2006; Eickhoff et al., 2005; Hagmann et al., 2008; He et al., 2009; Tzourio-Mazoyer et al., 2002; Wang et al., 2009). Particularly, an approach to parcel the entire brain using a deformable atlas is called atlas-based analysis (ABA)(Lancaster et al., 2000; Maldjian et al., 2003; Tzourio-Mazoyer et al., 2002). In this ABA, the automated parcellation works as an anatomical filter, reducing the dimensionality of the data from hundreds of thousands of voxels to hundreds of regions.
There are two main motivations to test the ABA as a promising tool. First, if one wants to use rs-fMRI as a diagnostic tool or biomarker, a simple quantitative approach with a high level test-retest reproducibility and small cross-subject variability is essential. The huge reduction of the dimensionality from dozens of thousands of voxels to mere few hundred structures would make the seed-based correlation (SBC) analysis much more straightforward. Second, it allows us to perform multi-modal analysis within the same anatomical framework, such as tissue atrophy (morphology), relaxation parameters (e.g. T1, T2, MTR), diffusion properties (FA, MD, axial and parallel diffusivity), and anatomical connectivity by tractography.
In this study, we parceled the brain into 185 structures, which can reduce the multi-modal measurements into 185-element vectors for each scalar measurement (e.g. volumes, T2, FA, MD, etc) and 185×184 matrices for anatomical and functional connectivity. Based on this parcellation, we first investigated the reproducibility of the parcel-to-parcel functional correlation using data from 20 normal participants each of whom were scanned twice. We report significant inter-parcel correlations consistent with previous findings, and a high test-retest reliability, an important consideration for application in clinical populations. The idea of multi-modal analysis was then demonstrated by integrating the population-based anatomical and functional connectivity analyses.
Methods
Data acquisition and co-registration
Twenty healthy volunteers with no history of neurological conditions (10 M/10 F, 22–61 years old, mean: 31 years old) participated in this study. Local Institutional Review Board approval and written, informed consent were obtained prior to examination. This is the same data used by Landman et al., 2011(Landman et al., 2011), where details of the protocol can be found. Subjects were scanned twice using a 3T MR scanner (Achieva, Philips Healthcare, Best, The Netherlands). Anatomical images were acquired using a 3D MPRAGE sequence with a 1.0×1.0×1.2 mm3 resolution. For resting state functional MRI, we used a sequence identical to that used for BOLD functional MRI studies of task activation (a 2D EPI sequence with fat suppression and SENSE partially parallel image acceleration to obtain a 3×3 mm (80 by 80 voxels) in-plane resolution in thirty-seven 3 mm transverse slices with a 1 mm slice gap. The rs-fMRIs were slice-time-corrected to adjust for differences in the acquisition time between slices, realigned to the first image using rigid body registration to adjust for motion, and then co-registered to the MPRAGE using SPM5 rigid body transformation (www.fil.ion.ucl.ac.uk/spm).
LDDMM-based Parcellation
We used a single-subject template segmented into 176 3D parcels (Mori et al., 2008; Oishi et al., 2008). As shown in Fig. 1, each participant's MPRAGE, co-registered with their rsfc-MRI data, was skull-stripped and normalized to the template, using a 12-parameter linear algorithm (AIR, (Woods et al., 1998)) and then LDDMM (Miller et al., 2005). Using the deformation fields from LDDMM and the inverse linear matrix, we then warped the parcellation map from the template to each subject's space, using the software DiffeoMap (www.mristudio.org). The details of the LDDMM operation are described elsewhere (Ceritoglu et al., 2009; Oishi et al., 2008). For each participant, peripheral white matter and cortical grey matter were separated using Statistical Parametric Mapping (SPM) segmentation. As a result, we obtained a matrix of 185 regions by 210 time points for each participant, in each session.
Figure 1.
Schematic representation of the normalization procedure. An atlas containing hundreds of structures manually defined is mapped to each subject brain using a linear algorithm (AIR) and LDDMM. The parcellation and MPRAGE-based tissue segmentation can be overlaid on the fMRIs. Averaging voxel values within each parcel results in a matrix of 210 time points × 185 parcels, for each run in each subject, that can be used to study inter-parcel, inter-run, and inter-subject correlations.
Nuisance Removal and Filtering
One of the primary sources of artifacts in functional connectivity is due to correlation of signals from non-neuronal physiological activity, such as respiration and cardiac pulsation. We used a recently described approach, COMPCOR (Behzadi et al., 2007), to minimize these effects. COMPCOR uses signal from the deep white matter and ventricles to estimate these artifacts and then remove them from grey matter time courses, and has been shown to match the performance of approaches requiring respiratory and cardiac monitoring data (Glover, Li, Ress 2000). The principal components that explained 80% of the variance derived from the time courses of 11 parcels defined as ventricles, 34 parcels defined as “deep white matter,” and of a ROI defining the whole brain along with realignment parameters, were used as the “nuisance parameters” to regress and remove the nuisance from the other parcel time courses. Finally, a time-domain band-pass filter (0.01 – 0.1 Hz) was applied.
Parcellation-based analysis
From the 40 matrices (20 participants × 2 sessions) containing the parcel time courses for each individual (185 parcels × 210 time points), we calculated the matrices of inter-parcel correlation coefficients (r). The inter-parcel r-matrices were used to study parcel-to-parcel correlations and the inter- and intra-subject variability (or reproducibility), as shown in the Results section. We also used hierarchical clustering to cluster parcels according the similarities of their inter-parcel correlation pattern.
Results
Examples of time-domain data and parcel-to-parcel correlation
Fig. 2 shows time domain data and correlation plots for selected pairs of parcels. In Fig. 2A and 2B, examples of correlations between the left and right motor cortices, and the white matter immediately beneath them, are shown. The motor cortex parcels show high correlation (r = 0.70) between the two hemispheres, while the white matter beneath the motor cortex does not (r = 0.17). Some pairs of parcels showed negative correlations, as shown in Fig. 2C.
Figure 2.
Time courses and correlation patterns between parcels. High correlation coefficients were observed between homotopic cortical parcels (A), while the white matter beneath these parcels had only very weak correlation (B). Some negative correlations were observed (C).
Connectivity patterns and correlation strength
In Fig. 3, several examples of spatial correlation patterns are shown (average of all 20 subjects × 2 sessions). For a given parcel of interest (POI), parcels with high correlations (r<−0.3 or r>0.3) were color-coded according to their correlation coefficient. In general, a given POI had a high correlation with the contralateral (or homotopic) parcel, and with adjacent parcels. There were cases, however, where high correlations were found with distant ipsilateral parcels (e.g., left angular and left middle frontal gyrus, r=0.62). Most white matter parcels, as well as the ventricles, remained uncorrelated. Figure 4 shows the degree of correlation among all 46 cortical parcels hierarchically clustered. Again, strong correlations with homotopic and adjacent parcels are evident, while some high correlations with distant parcels in the ipsilateral hemisphere can also be found. Reproducible negative correlation patterns can also be seen.
Figure 3.
Average of inter-parcels correlation among the 20 participants scanned twice, color-coded by the correlation coefficient, r; parcels with r<−0.3 or r>0.3 are colored. Each row of images shows parcels correlated with the labeled parcel. Generalizing, adjacent and contralateral parcels are generally correlated, although r can be high between distant parcels (such as between the medium frontal and angular gyrus at left). Negative correlations occurred mostly with distant parcels.
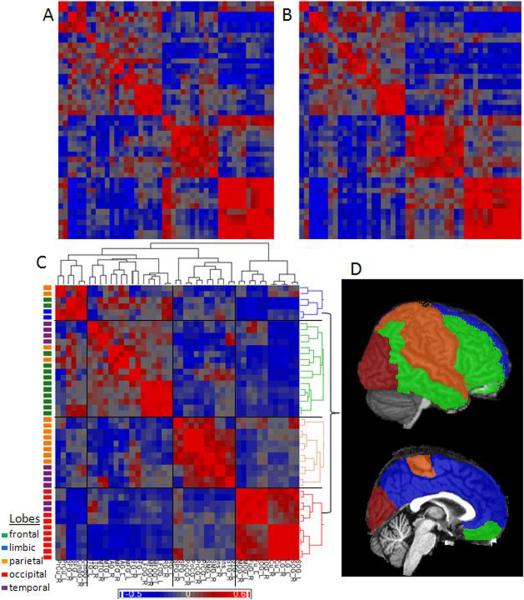
Figure 4.
Inter-cortical parcel correlations in one participant (first session, A, and second session, B), and in the average of 20 subjects (C), grouped by hierarchical clustering. Note that the pattern obtained in the group average is reproducible in the individual. At left, the parcels are color-coded according the lobe they belong to, indicating that the clustering is partially spatially related. At right, the dendrogram is color-coded according to the grouping found at four-level clustering, and these clusters are anatomically overlaid at the template brain used (D). Again, it evidences a tendency to “small world architecture” and “anti-correlated” patterns. This is a preliminary illustration of how this approach can be developed to investigate possible parcel-based networks. Abbreviations: R: right; L: left; PrCu: Pre-cuneus; Cu: cuneus; SFG, MFG, and IFG: superior, middle and inferior frontal gyrus, respectively; Cing: cingulum, STG, MTG, and ITG: superior, middle and inferior temporal gyrus, respectively; AG: angular gyrus; MFOG, LFOG: middle and lateral fronto-orbital gyrus, respectively; RG: retus gyrus; SPG: superior parietal gyrus; PoCG, PrCG: post and pre central gyrus, respectively; SMG: supra marginal gyrus; Ins: Insula; SOG, MOG, and IOG: superior, middle and inferior occipital gyrus, respectively; Fu: fusiform gyrus; LG: lingual gyrus.
Reproducibility measures
The parcel-to-parcel correlation patterns in a single subject (two trials; Fig. 4A and 4B), as well as the group average results shown in Fig. 4C, give a qualitative view of the test-retest and cross-subject consistency of our parcellation-based analysis. The strength of the intra- and inter-subject correlation varies depending on the areas. For example, the homotopic correlations tend to be strong; for r>0.3 (Fig. 3), the average r of the symmetric cortical areas (n=23) was 0.67 and the r standard deviation was 0.11. The average of intra-subject r difference between the two repetitions was also 0.11. These numbers for the correlations between neighbors and distant parcels were 0.45/0.14/0.16 and 0.35/0.14/0.16. The negative correlations tended to be weaker: −0.31/0.06/0.17. The detailed information about the specific correlation coefficients can be found in the supplemental material.
While this approach can provide information for inter-parcel correlation, another interesting question is the consistency of the global connectivity pattern shown in Fig. 4. To calculate the consistency of these correlation patterns across subjects, we converted the matrices of 46×45 r-values, as shown in Fig. 4, into 2,070-element vectors, and investigated the Pearson correlations between such vectors inter- and intra-subjects. The intra-subject consistency of the cortical correlation patterns was, on average, r=0.71+/−0.11, while the corresponding inter-subject consistency was r=0.57+/−0.085. The mean intersession reliability, calculated by the intraclass correlation coefficient (ICC), was 0.35+/−0.18, with the ICC values normal distributed. Two % of the connection had excellent reliability (ICC: 0.75–1); 8% had good (ICC: 0.6–0.75); 30% had fair (ICC: 0.4–0.6); 29% had low (ICC: 0.2–0.4); and 31% had poor reliability (ICC: 0–0.2). Positive and negative parcel-to-parcel correlations were significantly but weakly correlated with their reliability ICC indices (r=0.01, p=0.03; and r=0.13, p=0.002, respectively).
Multi-modal analysis: Combination with anatomical connectivity studies
In our past publication, we applied the same brain parcellation scheme to diffusion tensor images (DTI) data and performed a population-based tractography study, in which tracts were exhaustively searched between the 46 cortical parcels (Zhang et al., 2010). It is straightforward to identify a pair of parcels with high functional connectivity (rsfc-MRI) and high probability of anatomical connection (DTI-tractography with 100% population probability, meaning identifiable in all tested subjects). An example of such an analysis is shown in Fig. 5, in which the functional and anatomical connectivity information is integrated for the left superior parietal gyrus (SPG). In this figure, the trajectories of the tracts with 100% population-probability are shown between two parcels with high functional correlation. If parcel-pairs are categorized into two classes based on tractography, one with 100% identifiable tracts and the other with 0% probability, the average rsfc-MRI correlations (r) of these two classes are 0.4±0.18 and 0.1±0.08, respectively.
Figure 5.
An example of the multi-modal analysis in which DTI-based anatomical connectivity information is combined with the rs-fMRI data. The left superior parietal gyrus (SPG) is the “seed” and regions correlated with this area by r>0.3 (absolute value) are color-coded. Since DTI and rs-fMRI are co-registered, we can use rs-fMRI information to investigate white matter connections among regions functionally correlated. Both the short cortico-cortical fibers and the long inter-hemispheric association fibers (between left and right SPG) were automatically traced (following the protocol described by Zhang et al. (Zhang et al., 2010) according the same parcellation map used for the rs-fMRI analysis.
Discussion
In this study, we reduced data dimensionality of rs-fMRI data from voxels to structure-based parcels and performed a parcel-by-parcel functional connectivity analysis. While this approach could potentially be applied to task-activation fMRI studies, rsfc-MRI is a natural application for the parcel-by-parcel approach, because connectivity analysis intrinsically involves the square of the number of the voxels, which usually requires spatial dimensionality reduction. A widely used strategy to tackle the problem of dimensionality in functional connectivity is to employ spatially independent component analysis (ICA) (Calhoun et al., 2001a; Calhoun et al., 2001b; McKeown et al., 1998; van de Ven et al., 2004), which yields a set of networks each comprising a collection of voxels with similar time courses. While this approach has been widely used for revealing functional brain networks without requiring a priori temporal or spatial assumptions, the interpretation and quantification of ICA results can be challenging. In clinical research, where the differences among groups are important, the question may arise as to whether all groups' spatial distribution of the networks is sufficiently similar as to not violate the assumptions behind the analysis.
We employed SBC based on whole brain parcellation as an alternative approach to reduce spatial dimensionality. Our current brain atlas parcels the brain into 185 structures; this can be considered structure-specific spatial filtering, as voxels that belong to the same structures are grouped into parcels. In fMRI studies, one of the central questions is to identify voxels that are associated with a specific brain functions. On the other hand, in the parcel-based approach, brain structures are defined and voxels are clustered a priori. This relegates one of the most important roles of fMRI study, but in return it potentially offers enhanced SNR and reproducibility. This could be an important tradeoff if one wants to use fMRI as a tool to characterize clinical populations. After the brain is parceled, the data dimensionality is reduced to a 185×184 matrix. This allows rapid and quantitative calculation of inter-parcel temporal correlation, such as that shown in Figures 2, 3, and 4. The inter-parcel grey matter spatial correlation pattern shown in Fig. 4 can be treated as a connectivity signature of each individual or group of subjects. This connectivity signature can be readily submitted to statistical analyses if one has data from two populations. It would, thus, be of great interest to see how this pattern changes in pathological conditions.
To use this approach to investigate a pathological condition, the reproducibility of the measurements should first be determined. Based on the data from 20 normal subjects, we could investigate both test-retest reproducibility and cross-individual variability. As mentioned in the Results and showed in the Supplemental Material, the intra-subject correlations were, as expected, higher than the inter-subject correlations, and both global and regional (parcel-by-parcel) correlations revealed high intra- and inter-individual consistency. Functional networks have been found to be robust using seed-based correlations (Shehzad et al., 2009), ICA (Zuo et al., 2010) and graph theoretical approaches (Wang et al., 2011) within (Anderson et al., 2011) and across subjects (Damoiseaux et al., 2006; Shehzad et al., 2009). Our ICCs were slightly higher than those found in previous studies and, recently, by Wang et al. (Wang et al., 2011), when comparing the test-retest reliability of functional connectivity for 3 atlases: Anatomical Automatic Labeling atlas (AAL), Harvard-Oxford atlas (HOA) and Dosenbach's 160 spheres. We found poor to low ICCs in 60% of the connections (against 70–80% reported by Wang et al.) and fair to excellent ICCs in 40% of the connections (against 20–30% reported by Wang et al). In agreement with this previous study, we found weak but significant correlations between the strength of positive and negative connections and their reliability, while Wang et al. did not find significant correlations between ICCs and negative connections using AAL atlas. This indicates that the reliability of functional connectivity was partly determined by their strength, but the functional connectivity strength has limited predictive ability to their reliability. The power analysis can be performed based on this information, but the power is dependent on specific combinations of the parcels. For example, on average, a 10% reduction in the correlation among the 23 pairs of homotopic parcels, which are in general strongly correlated, can be statistically detected with 46 samples.
Our reliability across scans was higher than that described by Honey et al., 2009 (Honey et al., 2009), probably because of the increased SNR provided by our method. However, we found the same phenomenon they observed regarding the strength and variability of the correlations: pairs of parcels spatially close or with evidence of structural connection (“neighbors” or homotopic parcels) exhibit less variability and higher correlations than pairs of parcels spatially distant or with no evidence of structural connections. As shown in Fig. 4, our ABA pointed to the same “small world architecture” previously described by voxel-based and simulation studies, in which neighboring regions show stronger connections than regions that lie further apart (Bullmore and Sporns 2009). Nevertheless, as in previous studies, the inter-parcel analysis demonstrated correlations that could not be purely anatomical or distance-determined, because some spatially remote regions, such as the frontal and angular gyrus, showed strong correlation (Fig. 3 and 4).
We also found negative-correlated parcels, consistent with earlier reports. Although the meaning and mechanisms of anti-correlated networks are still under investigation, reports of anti-correlation of “task-positive” areas and the default mode network are widespread (Anderson et al., 2010; Fransson 2006; Lin et al., 2011). As shown in Fig. 4D, the groups of parcels we found at four-level clustering are spatially similar to those initially described by Fox et al. (Fox et al., 2005) in humans and by Honey et al. (Honey et al., 2007) in macaques and computational simulations: the red (occipital: visual region) and orange clusters (parietal and temporal superior: motor and sensorial regions) involve areas consistently activated in cognitive tasks, while the green and blue parcels (frontal medial, temporal inferior, cingulate and pre-cuneus: cognitive, memory, emotional, and behavioral regions), with an opposite pattern of correlation (Fig. 4C), are similar to the “task-negative” networks.
A variety of atlases have been used to automatically parcel the cortex, such as AAL, HOA, WFU_PickAtlas, Talairach, MNI-structural, Jülich histological, Allen Institute atlas, etc. Compared with those, advantages of our atlas include that it parcels the entire 3D brain volume, which allows, for example, to confirm the regional specificity of cortico-cortical correlation by examining the connectivity of the white matter beneath the cortex of interest, as shown in Fig. 2; to use the parcels to correct physiological nuisances; to scrutiny possible connectivity patterns in the deep gray matter; and to make exploratory analysis in the whole brain. Indeed, one of the most exciting extensions of our method is to provide a common platform to integrate multimodal data and to perform both anatomical and functional connectivity analysis (Fig. 5). Multiple imaging modalities, including T1-based morphometry, DTI, rs-fMRI, MR Spectroscopy, PET, etc, can be analyzed and correlated among the modalities, across subjects, or to clinical data, once the ABA allows the incorporation of all image-based information into the same anatomical space (Fig 6). For individuals with dementia or neurodevelopmental conditions, for example, one can investigate atrophy and MR parameter abnormalities (e.g. T2, FA, MD, etc) of about two hundred structures (Faria et al., 2010; Faria et al., 2011; Oishi et al., 2009) and directly correlate the functional profiles with fMRI data.
Figure 6.
Schematic example illustrating how the atlas-based analysis can unify the multi-modal imaging quantification. T1-WI, DTI, rs-fMRI, MR Spectroscopy (MRS), and other imaging modalities not represented here (such as T2-WI, Susceptibility-WI, Magnetization transfer images, PET, etc..) can be parceled into hundreds of structures biological meaningful and the various image parameters can be quantified. Therefore, it is possible to characterize each subject by a matrix of regions by anatomic features that carries complementary information in different image domains. This multi-modal quantification at the structural basis enables comparisons at group and/or individual level, facilitates longitudinal analysis, and makes easier and more object the search of anatomo-functional correlations. The tables contain a subsample of quantitative data from subjects included in this study (normal adults). SFL = superior frontal lobe, CingL = Cingulum left, PrCuL= Pre Cuneus left. Volume is in mm3; MD is in mm/s2(e10−4); metabolites (Cr, Cho, NAA) are concentrations in mM; the rsfc-MRI correlation vector represents the vector of correlations of a given region of interest with all the other regions defined in the parcellation map; GE (global efficiency), LE (local efficiency), PL (path length) and CC (cluster coefficient) are derived from graph analysis of rsfc-MRI.
Although our parcellation-based approach has yielded a promising method for dimensionality reduction, it has a few drawbacks. One of the most important questions is how the brain is parcelled. It has been shown (Achard et al., 2006; He et al., 2009; Wang et al., 2009) that the topological organization of brain networks estimated from rsfc-MRI data is affected by the choice of parcellation strategy. If one parcel contains two areas within it with different time-dependent characteristics, the averaging of these areas would lead to information degeneration. For instance, the temporoparietal junction can be divided into three distinct subregions based on their structural and functional connectivity (Mars et al., 2011): a dorsal area that overlaps with the middle part of the inferior parietal lobule and is concerned with directing attention based on current goals and preexisting top-down information (Fox et al., 2006); and two ventral areas which can be distinguished on the basis of their participating in different cortical networks involving ventral prefrontal cortex / anterior insula, concerned with reorienting of attention in response to behaviorally relevant events from the environment (Corbetta and Shulman 2002), and anterior medial prefrontal cortex. Likewise, axonal tracings in macaque monkeys revealed distinct patterns of anatomical connectivity for three subdivisions of the precuneus (Buckwalter et al., 2008; Colby et al., 1988; Leichnetz 2001; Morecraft et al., 2004; Pandya and Seltzer 1982). Zhang et all. (Zhang and Li 2011) showed that these subdivisions have distinct patterns of rsfc-MRI, can be grouped under different networks, and are involved on different functions. These examples, in addition to showing a certain level of disagreement between “broad” anatomical definitions and “fine” functional connectivity patterns, reinforce that the combination of multiple imaging modalities, such as rsfc-MRI and DTI/tractography, can contribute to the refinement of the mapping of brain connections.
Our current atlas is based purely on anatomical information, which could differ from functional information. In addition, some of the parcels in our atlas are large (more than 60,000 mm3) where no consistent anatomical clues allow further division. Craddock et al. (Craddock et al., 2011) evaluated the suitability of the parcellation atlas against four ROI atlases (Talairach and Tournoux, Harvard-Oxford, Eickoff-Zilles, and Automatic Anatomical Labeling) and a random parcellation approach. They observed that ROI size and hence the number of ROIs in a parcellation had the greatest impact on their suitability for FC analysis. With 200 or fewer ROIs anatomically homologous, parcellations offer increased interpretability while those containing higher numbers of ROIs most accurately represent connectivety patterns present at the voxel scale. They proposed a data-driven method for generating an ROI atlas by parcellating whole brain rsfc-MRI data into spatially coherent regions of homogeneous connectivity. Hence, refinement of the atlas (by dividing large parcels into subparcels) may be important for its application to rsfc-MRI; reciprocally, subparcellation of the atlas based on resting-state functional connectivity may enhance more broadly the power of ABA of multimodal MRI data.
Supplementary Material
Appendix Table 1: Mean of correlations (r) between cortical parcels (inferior left) and standard deviations of r (superior right), threshold for r>0.3 or r<−0.3. ns = non-significant correlations according one-sample t-test (p-value < 0.05, corrected for multiple comparisons with FDR). For the abbreviations, see legend of figure 4.
Appendix Table 2: Mean correlation between participants, based on cortical parcels. For the labels, the first number represents the participant ID; the second number is the session (01 or 02).
Acknowledgments
This publication was made possible by NIH grants UL1 RR025005 from NCRR and NIH Roadmap for Medical Research (AVF); RO1AG20012, and RO1NS058299 (SM), R21AG033774, and P50AG005146 (KO); and P41 EB015909 from NCRR/NIBIB. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of any of these Institutes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2006;26(1):63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D. Reproducibility of single-subject functional connectivity measurements AJNR. American Journal of Neuroradiology. 2011;32(3):548–555. doi: 10.3174/ajnr.A2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Druzgal TJ, Lopez-Larson M, Jeong EK, Desai K, Yurgelun-Todd D. Network anticorrelations, global regression, and phase-shifted soft tissue correction. Human Brain Mapping. 2010 doi: 10.1002/hbm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37(1):90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar. MRI Magnetic Resonance in Medicine : Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Buckwalter JA, Parvizi J, Morecraft RJ, van Hoesen GW. Thalamic projections to the posteromedial cortex in the macaque. The Journal of Comparative Neurology. 2008;507(5):1709–1733. doi: 10.1002/cne.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems Nature Reviews. Neuroscience. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapping. 2001a;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Human Brain Mapping. 2001b;13(1):43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceritoglu C, Oishi K, Li X, Chou MC, Younes L, Albert M, Lyketsos C, van Zijl PC, Miller MI, Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. NeuroImage. 2009;47(2):618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby CL, Gattass R, Olson CR, Gross CG. Topographical organization of cortical afferents to extrastriate visual area PO in the macaque: A dual tracer study. The Journal of Comparative Neurology. 1988;269(3):392–413. doi: 10.1002/cne.902690307. [DOI] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain Nature Reviews. Neuroscience. 2002;3(3):201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Human Brain Mapping. 2011 doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. NeuroImage. 2006;29(4):1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Eickhoff SB, Stephan KE, Mohlberg H, Grefkes C, Fink GR, Amunts K, Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage. 2005;25(4):1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Faria AV, Hoon A, Stashinko E, Li X, Jiang H, Mashayekh A, Akhter K, Hsu J, Oishi K, Zhang J, et al. Quantitative analysis of brain pathology based on MRI and brain atlases--applications for cerebral palsy. NeuroImage. 2011;54(3):1854–1861. doi: 10.1016/j.neuroimage.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria AV, Zhang J, Oishi K, Li X, Jiang H, Akhter K, Hermoye L, Lee SK, Hoon A, Stachinko E, et al. Atlas-based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. NeuroImage. 2010 doi: 10.1016/j.neuroimage.2010.04.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(26):10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P. How default is the default mode of brain function? further evidence from intrinsic BOLD signal fluctuations. Neuropsychologia. 2006;44(14):2836–2845. doi: 10.1016/j.neuropsychologia.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magnetic Resonance in Medicine : Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes alzheimer's disease from healthy aging: Evidence from functional MRI. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(1):253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biology. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Olson IR, Leung HC, Skudlarski P, Gore JC. Changes in functional connectivity of human MT/V5 with visual motion input. Neuroreport. 2004;15(8):1315–1319. doi: 10.1097/01.wnr.0000129997.95055.15. [DOI] [PubMed] [Google Scholar]
- Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Human Brain Mapping. 2002;15(4):247–262. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y, et al. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PloS One. 2009;4(4):e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Kotter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(24):10240–10245. doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(6):2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated talairach atlas labels for functional brain mapping. Human Brain Mapping. 2000;10(3):120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman BA, Huang AJ, Gifford A, Vikram DS, Lim IA, Farrell JA, Bogovic JA, Hua J, Chen M, Jarso S, et al. Multi-parametric neuroimaging reproducibility: A 3-T resource study. NeuroImage. 2011;54(4):2854–2866. doi: 10.1016/j.neuroimage.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichnetz GR. Connections of the medial posterior parietal cortex (area 7m) in the monkey. The Anatomical Record. 2001;263(2):215–236. doi: 10.1002/ar.1082. [DOI] [PubMed] [Google Scholar]
- Lin P, Hasson U, Jovicich J, Robinson S. A neuronal basis for task-negative responses in the human brain. Cerebral Cortex (New York, N.Y.: 1991) 2011;21(4):821–830. doi: 10.1093/cercor/bhq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe MJ, Mock BJ, Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. NeuroImage. 1998;7(2):119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Luo C, Li Q, Lai Y, Xia Y, Qin Y, Liao W, Li S, Zhou D, Yao D, Gong Q. Altered functional connectivity in default mode network in absence epilepsy: A resting-state fMRI study. Human Brain Mapping. 2011;32(3):438–449. doi: 10.1002/hbm.21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage. 2003;19(3):1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Mars RB, Sallet J, Schuffelgen U, Jbabdi S, Toni I, Rushworth MF. Connectivity-based subdivisions of the human right “temporoparietal junction area”: Evidence for different areas participating in different cortical networks. Cerebral Cortex (New York, N.Y.: 1991) 2011 doi: 10.1093/cercor/bhr268. [DOI] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Human Brain Mapping. 1998;6(3):160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Beg MF, Ceritoglu C, Stark C. Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9685–9690. doi: 10.1073/pnas.0503892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morecraft RJ, Cipolloni PB, Stilwell-Morecraft KS, Gedney MT, Pandya DN. Cytoarchitecture and cortical connections of the posterior cingulate and adjacent somatosensory fields in the rhesus monkey. The Journal of Comparative Neurology. 2004;469(1):37–69. doi: 10.1002/cne.10980. [DOI] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Huang H, Yoshioka T, Ying SH, Zee DS, Zilles K, Amunts K, Woods R, Toga AW, Pike GB, et al. Superficially located white matter structures commonly seen in the human and the macaque brain with diffusion tensor imaging. brain connectivity. Brain Connectivity. 2011;1(1):37–47. doi: 10.1089/brain.2011.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, Hsu JT, Miller MI, van Zijl PC, Albert M, et al. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: Application to normal elderly and alzheimer's disease participantstlas. NeuroImage. 2009;46(2):486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Zilles K, Amunts K, Faria A, Jiang H, Li X, Akhter K, Hua K, Woods R, Toga AW, et al. Human brain white matter atlas: Identification and assignment of common anatomical structures in superficial white matter. NeuroImage. 2008;43(3):447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya DN, Seltzer B. Intrinsic connections and architectonics of posterior parietal cortex in the rhesus monkey. The Journal of Comparative Neurology. 1982;204(2):196–210. doi: 10.1002/cne.902040208. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62(1):42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, et al. The resting brain: Unconstrained yet reliable. Cerebral Cortex (New York, N.Y.: 1991) 2009;19(10):2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Price JL, Yan Z, Mintun MA. Resting-state functional MRI in depression unmasks increased connectivity between networks via the dorsal nexus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(24):11020–11025. doi: 10.1073/pnas.1000446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, et al. Correspondence of the brain's functional architecture during activation and rest. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Du H, Wu N, Hou B, Wu G, Wang J, Feng H, Jiang T. Impaired resting-state functional integrations within default mode network of generalized tonic-clonic seizures epilepsy. PloS One. 2011;6(2):e17294. doi: 10.1371/journal.pone.0017294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van de Ven VG, Formisano E, Prvulovic D, Roeder CH, Linden DE. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Human Brain Mapping. 2004;22(3):165–178. doi: 10.1002/hbm.20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang L, Zang Y, Yang H, Tang H, Gong Q, Chen Z, Zhu C, He Y. Parcellation-dependent small-world brain functional networks: A resting-state fMRI study. Human Brain Mapping. 2009;30(5):1511–1523. doi: 10.1002/hbm.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Zuo XN, Gohel S, Milham MP, Biswal BB, He Y. Graph theoretical analysis of functional brain networks: Test-retest evaluation on short- and long-term resting-state functional. MRI data PloS One. 2011;6(7):e21976. doi: 10.1371/journal.pone.0021976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. general methods and intrasubject, intramodality validation. Journal of Computer Assisted Tomography. 1998;22(1):139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li CS. Functional connectivity mapping of the human precuneus by resting state fMRI. NeuroImage. 2011 doi: 10.1016/j.neuroimage.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, Akhter K, Rosa-Neto P, Pike B, Evans A, et al. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage. 2010;52(4):1289–1301. doi: 10.1016/j.neuroimage.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. Reliable intrinsic connectivity networks: Test-retest evaluation using ICA and dual regression approach. NeuroImage. 2010;49(3):2163–2177. doi: 10.1016/j.neuroimage.2009.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix Table 1: Mean of correlations (r) between cortical parcels (inferior left) and standard deviations of r (superior right), threshold for r>0.3 or r<−0.3. ns = non-significant correlations according one-sample t-test (p-value < 0.05, corrected for multiple comparisons with FDR). For the abbreviations, see legend of figure 4.
Appendix Table 2: Mean correlation between participants, based on cortical parcels. For the labels, the first number represents the participant ID; the second number is the session (01 or 02).