Abstract
OleA is a thiolase superfamily enzyme which has been shown to catalyze the condensation of two long-chain fatty-acyl-Coenzyme A (CoA) substrates. The enzyme is part of a larger gene cluster responsible for generating long-chain olefin products – a potential biofuel precursor. In thiolase superfamily enzymes, catalysis is achieved via a ping-pong mechanism. The first substrate forms a covalent intermediate with an active site cysteine which is followed by reaction with the second substrate. For OleA, this conjugation proceeds by a non-decarboxylative Claisen condensation. The OleA from Xanthomonas campestris has been crystallized and its structure solved, along with inhibitor bound and xenon derivatized structures, to better understand substrate positioning in the context of enzyme turnover. OleA is the first characterized thiolase superfamily member that has two long-chain alkyl substrates that need to be bound simultaneously, and therefore uniquely requires an additional alkyl binding channel. The location of the fatty acid biosynthesis inhibitor, cerulenin, that possesses an alkyl chain length in the range of known OleA substrates, in conjunction with a single xenon binding site, leads to the putative assignment of this novel alkyl binding channel. Structural overlays between the OleA homologs, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase and the fatty acid biosynthesis enzyme FabH, allow assignment of the remaining two channels; one for the thioester-containing pantetheinate arm and the second for the alkyl group of one substrate. A short β-hairpin region is ordered in only one of the crystal forms and that may suggest open and closed states relevant for substrate binding. Cys143 is the conserved catalytic cysteine within the superfamily, and the site of alkylation by cerulenin. The alkylated structure suggests that a glutamic acid residue (Glu117β) likely promotes Claisen condensation by acting as the catalytic base. Unexpectedly Glu117β comes from the other monomer of the physiological dimer.
The impetus to discover microbial pathways capable of producing new, high-energy molecules has increased in response to a dwindling fossil fuel supply. Consequently, microbes have been isolated that are capable of generating such compounds as aliphatic isoprenoid compounds and alkanes from fatty acid deformylation (1, 2). The biocatalyzed generation of energy rich long-chain olefins by microbes across multiple phyla produces hydrocarbons ranging in size from C27-C31 (3). Future large scale production of such compounds would provide a higher-energy biofuel in comparison to current technologies such as the ethanol fermentation process (4).
The ability of microbes to generate long-chain hydrocarbons has been well documented (5–9). Consistent amongst the microbial olefinic hydrocarbon products is a double bond located at the median carbon (6). This transformation is initiated by a “head-to-head” condensation; so called because the carboxyl carbon on one fatty acyl group reacts with the α-carbon of another fatty acyl group to generate a new carbon-carbon bond (10). Recent work has demonstrated that a dedicated gene ensemble (oleABCD for olefin biosynthesis)1 is required for the production of olefins formed through head-to-head condensation (11, 12). Sequence homology has assigned the oleBCD gene products as an α/β-hydrolase, AMP-dependent ligase, and short-chain dehydrogenase respectively (3). The oleA gene product was predicted to belong to the thiolase superfamily which contains members known to catalyze carbon-carbon bond condensation reactions.
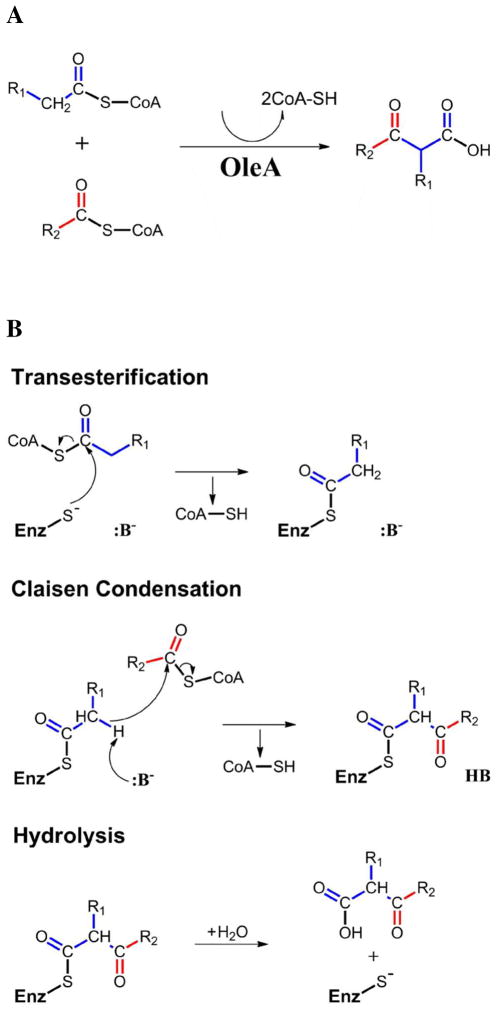
Heterologous oleA gene expression and in vitro characterization of purified OleA has confirmed the enzyme is capable of initiating olefin biosynthesis (3, 13). Specifically, the enzyme can condense two coenzyme A (CoA) charged fatty acids to produce a long-chain β-ketoacid via a non-decarboxylative Claisen condensation reaction (Figure 1A) (13). The catalytic cycle begins with transesterification of the first fatty acyl-CoA group to an active site cysteine (Figure 1B). The second fatty acyl-CoA then binds, and proton abstraction from the cysteine-tethered acyl group is thought to generate a β-carbanion capable of nucleophilic attack on the CoA thioester. The final step is hydrolysis of the cysteine acyl to free the β-ketoacid product. In vitro studies have shown that the β-ketoacid product can then be converted to an olefin by incubation with OleC and OleD (13).
Figure 1.
OleA catalyzed condensation with CoA-charged substrates. (A) The overall reaction. (B) The three steps of the catalytic cycle.
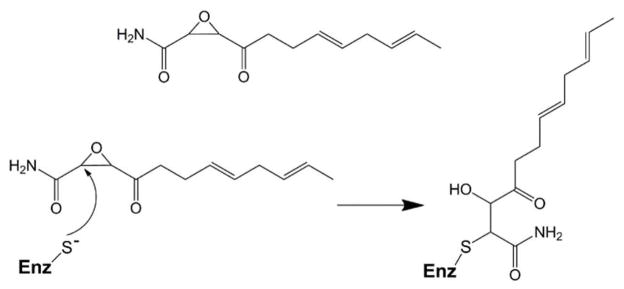
A decarboxylative Claisen condensation mechanism is utilized by other enzymes in the thiolase superfamily, such as the fatty acid biosynthetic (Fab) enzymes. The Fab enzymes condense fatty acid acyl-CoA substrates with malonic acid, which is typically charged with an Acyl Carrier Protein (ACP) rather than CoA (14). Crystal structures have been determined for members of the Fab enzyme family (FabB, FabP, and FabH), and the homologous 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase that employs a similar catalytic mechanism to OleA (15–21). These enzymes are functional homodimers, with an active site in each monomer containing the proposed reactive cysteine. Structures of FabH (also known as β-ketoacyl-acyl carrier protein synthase III) and HMG-CoA have been determined in complex with substrates, substrate analogues, and inhibitors that define substrate binding in these enzymes (18–21). A frequently used inhibitor is the antibiotic cerulenin, a natural product that irreversibly inhibits fatty acid biosynthesis by forming a covalent adduct with the active site cysteine (Figure 2) (22, 23). HMG-CoA synthase condenses two acetyl-CoA molecules to yield acetoacyl-CoA, in a ping-pong mechanism that requires a single substrate binding channel to bring each CoA activated substrate sequentially to the active site (20, 21). Fab enzymes, in particular FabH from Mycobacterium tuberculosis that elongates long-chain fatty acyl substrates (C16-C22) by two carbons, require an additional binding channel for the alkyl chain prior to Claisen condensation (18, 19). However, OleA is unusual in that both its substrates possess long alkyl chains (C10-C16). In that context, an additional alkyl binding channel is required. In the present study, we have solved the structure of OleA alone, in complex with cerulenin, and in complex with cerulenin and xenon, to better understand how this atypical thiolase enzyme may accomodate both long-chain fatty acyl-CoA substrates during turnover.
Figure 2.
Inhibition of OleA through covalent modification of the active site cysteine by cerulenin.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
The expression and purification of Xanthomonas campestris OleA were carried out as described previously (13). Protein stocks were concentrated between 220–300 μM in 500 mM NaCl, 20 mM Hepes pH 7.4. To covalently bind cerulenin to OleA, the protein stock was reacted with 6 mM cerulenin for one hour, resulting in complete inhibition of the enzyme (Figure 2). Initial crystallization trials of ~1600 conditions for both OleA and cerulenin-treated OleA were conducted at the Hauptman-Woodward Medical Research Institute (24). Multiple conditions gave crystals for both enzyme forms, with the final condition selected for ease of growth, reproducibility and diffraction quality. All crystals were grown via hanging drop vapor diffusion at 20°C. OleA was crystallized in drops containing 1 μl protein plus 1 μl of mother liquor (16–21% w/v PEG 8000, 70–100 mM potassium phosphate dibasic, and 100 mM sodium citrate pH 4.2), while OleA-cerulenin co-crystals were grown in drops containing 1 μl protein plus 1 μl of mother liquor (13–18% w/v PEG 4000, 80–100 mM manganese chloride, and 100 mM MES 6.0). OleA crystals were cryoprotected in a mother liquor solution containing 20% PEG 400, whilst the OleA-cerulenin co-crystals were cryoprotected in a mother liquor solution containing 25% glycerol, prior to flash freezing in liquid nitrogen.
Xenon Derivitization of OleA-Cerulenin Co-Crystals
OleA-cerulenin crystals were harvested and washed in their respective cryoprotectant solutions. Crystals were then transferred into a pressure cell for derivatization (4DX Systems). Crystals were pressurized with xenon (Airgas Inc.) at ~130 PSI for fifteen minutes. Crystals were then immediately flash frozen in liquid nitrogen.
X-ray Data Collection, Processing, and Refinement
The datasets using 1.03 Å X-rays were collected at GM/CA-CAT beamlines 23-ID-D and 23-ID-B of the Advanced Photon Source (APS), Argonne National Laboratory, Argonne IL at 100 K using a MARmosaic 300 CCD. The beam size was adjusted to match the crystal size and orientation, and attenuated by 100–250-fold; 200° of diffraction data were collected on a single crystal, during which time no significant degradation of diffraction quality was observed. The dataset at a wavelength of 2.29 Å was collected at the University of Minnesota using a MicroMax-007 chromium rotating anode X-ray generator with VariMaxCr optics and a R-AXIS IV++ detector with a helium cone (Rigaku) at 100 K. Data collection statistics are given in Table 1.
Table 1.
Data collection and refinement statisticsa.
| OleA unbound | OleA-cerulenin co-crystal | OleA-cerulenin co-crystal Xe Pressurized |
OleA-cerulenin co-crystal Xe Pressurized |
|
|---|---|---|---|---|
| Data Collection | ||||
| Wavelength (Å) | 1.03 | 1.03 | 1.03 | 2.29 |
| Space Group | P212121 | P3221 | P3221 | P3221 |
| Unit Cell (Å) | 82.2 × 85.4 × 102.7 | 90.5 × 90.5 × 69.8 | 90.0 × 90.0 × 69.5 | 90.0 × 90.0 × 69.5 |
| Resolution (Å) | 50.0-1.85 (1.88-1.85) | 50.0-1.70 (1.73-1.70) | 50.0-1.95 (1.98-1.95) | 50.0-2.46 (2.50-2.46) |
| Measured Reflections | 509308 | 270278 | 267054 | 113152 |
| Unique Reflections | 61685 | 36319 | 24115 | 11836 |
| Completeness (%) | 98.8 (97.9) | 99.0 (100) | 99.9 (99.8) | 97.5 (90.5) |
| Rmerge (%)b | 5.5 (38.5) | 4.4 (42.3) | 5.8 (40.8) | 6.7 (30.6) |
| I/σI | 40.2 (5.3) | 44.5 (5.0) | 55 (7.1) | 48.7 (9.8) |
| Multiplicity | 8.2 (8.1) | 7.4 (7.4) | 11.1 (11.1) | 8.8 (9.6) |
| Wilson B-factor (Å2) | 21.4 | 23.3 | 27.4 | |
| Refinement | ||||
| Resolution (Å) | 44.0-1.85 (1.89-1.85) | 27.6 – 1.70 (1.75-1.70) | 29.5 – 1.95 (2.03-1.95) | |
| No.reflections; work/test | 57593(2364)/3059(123) | 34442(2646)/1852(130) | 22513(2344)/1224(139) | |
| Rwork (%)c | 16.0 (19.4) | 17.8 (22.4) | 18.0 (19.9) | |
| Rfree (%)d | 19.1 (28.6) | 20.9 (25.2) | 21.82 (25.2) | |
| Protein atoms | 5244 | 2590 | 2614 | |
| Other atoms | 594 | 249 | 146 | |
| Ramachandran Statistics | ||||
| Allowed (%) | 100 | 100 | 100 | |
| Outliers (%) | 0 | 0 | 0 | |
| RMS deviation | ||||
| Bond lengths (Å) | 0.007 | 0.007 | 0.008 | |
| Bond angles (°) | 1.038 | 1.013 | 1.027 | |
| Average B-factor (Å2) | 21.2 | 23.5 | 27.1 | |
| Coordinate Error (ML based)f | 0.18 | 0.18 | 0.22 | |
| Protein Data Bank ID | 3ROW | 3S21 | 3S23 |
Data in parentheses are for the highest resolution shell.
Rmerge = Σi |Ihkl,i − (Ihkl)| /Σhkl Σi Ihkl, i, where I is the observed intensity, and (Ihkl) is the average intensity of multiple measurements.
Rwork = Σ|Fo|-|Fc| / Σ|Fo|, where |Fo| is the observed structure factor amplitude, and |Fc| is the calculated structure factor amplitude.
Rfree is the R factor based on 5% of the data excluded from refinement keeping the same 5% between isomorphous datasets.
Based on values obtained from MolProbity (33).
Estimated coordinate error based on maximum likelihood estimate in Phenix 1.7-650 (26).
OleA and OleA-cerulenin co-crystals crystallized in distinct space groups (P212121 and P3221 respectively). The orthorhombic OleA structure was solved by molecular replacement. The crystal structure of the annotated FabH from Xanthomonas oryzae without solvent (Protein Data Bank (PDB) ID 3FK5) was used as a search model (sequence identity 91% to X. campestris OleA). The X. oryzae enzyme has not been biochemically characterized, and the high sequence identity with X. campestris OleA, coupled with the gene being found within an ole gene cluster (3), suggests that this gene is misannotated and actually codes for an OleA. Molecular replacement with PHASER placed a dimer within the asymmetric unit (AU). For all structures cycles of model building with COOT (25) and restrained refinement using PHENIX 1.7-650 (26) with TLS were performed until all interpretable regions of the 2Fo-Fc and Fo-Fc electron density maps were explained (PDB ID 3ROW).
OleA-cerulenin co-crystals were in a different space group (P3221), and so the structure was solved by molecular replacement using PHASER and the OleA model without solvent as the search model (27). The correct enantiomorph, P3221, was confirmed by a strong translation function Z-score (TFZ) peak of 30.6 (TFZ, 8.5 for P3121) with a single monomer in the AU. The physiological dimer was generated through the two-fold crystallographic axis. The final structure was consistent with cerulenin bound at 100% occupancy (PDB ID 3S21). X-ray diffraction data from a xenon pressured OleA-cerulenin co-crystal were collected at wavelengths of 1.03 Å and 2.29 Å; the latter leading to a strong anomalous signal due to the xenon (λ=2.29 Å; Xe f″=11.9 electrons). Difference Fourier was used to solve the 1.03 Å wavelength data, whilst the PHENIX maps program suite was used to generate anomalous peak maps using the 2.29 Å wavelength data (26). Both used the OleA-cerulenin co-crystal structure (without solvent) for phasing. Two xenon sites were identified (occupancy 45% and 35%) (PDB ID 3S23). Final refinement statistics for all structures can be seen in Table 1.
RESULTS
Overall Structures of the Unbound and Inhibited OleA Dimer
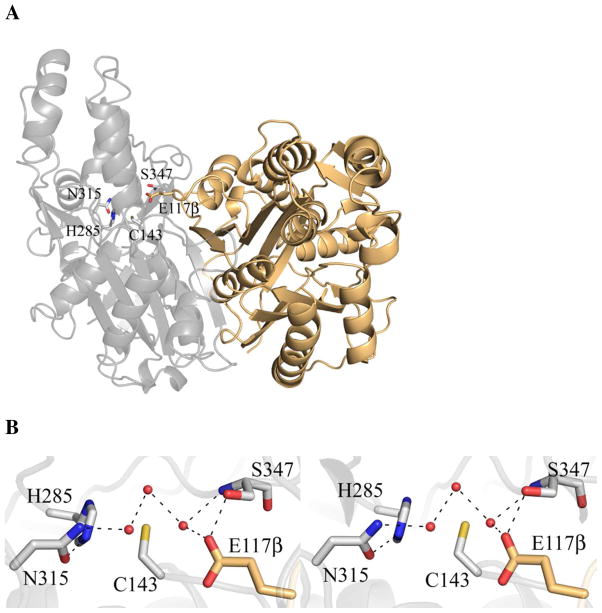
The crystal structures of unbound OleA and OleA-cerulenin were determined to 1.85 and 1.70 Å resolution, respectively (Table 1). The unbound structure of OleA is a dimer consistent with the oligomeric state of most characterized thiolase superfamily members including FabH and HMG-CoA synthase (Figure 3A) (28). The monomer consists of two domains, with the dimer interface (2135 Å2) being formed between a single equivalent domain from each monomer (29). The OleA-cerulenin structure contains a single monomer in the AU with a crystallographic two-fold axis running directly through the dimer. Nevertheless, the gross structures of the three OleA structures are similar, with the largest overall rmsd between any pair being ≤ 0.7 Å across all matching atoms (Figure S1).
Figure 3.
OleA dimer and active site. (A) The physiological dimer of OleA. The two monomers are drawn in gray and tan cartoon. Each monomer contains one active site. The gray cartoon active site residues are drawn in stick. Note that E117β derives from the neighboring monomer. (B) Stereo view of the OleA active site. The 2Fo-Fc electron density map is contoured to 1.0 σ. Ordered solvent molecules are represented by red spheres. This figure was produced using PyMOL (http://www.pymol.org/).
An eleven residue β-hairpin (239-249) is ordered in only a single monomer of the P212121 crystal form and is disordered in the P3221 crystal form. The β-hairpin contains hydrophobic residues (Leu243, Met246, and Val247) that pack against OleA when ordered.
Active Site of OleA
The active site of each monomer contains a single catalytic cysteine residue (Cys143) resting at the base of a solvent accessible channel (Figure 3B). A novel feature of OleA is a glutamate (Glu117β) in the active site that originates from a loop in the neighboring monomer. Immediately above Cys143 are two pockets positioned to stabilize tetrahedral, oxyanionic intermediates generated during turnover. The first is formed by the side-chains of His285 and Asn315 (oxyanion hole 1), and the second by the main chain amides of Ser347 and the catalytic Cys143 (oxyanion hole 2). Ordered waters interlink these residues, and a hydrogen bond between the carboxylate side chain of Glu117β with the hydroxyl of Ser347 (Figure 3B) may be important for positioning Glu117β within the active site.
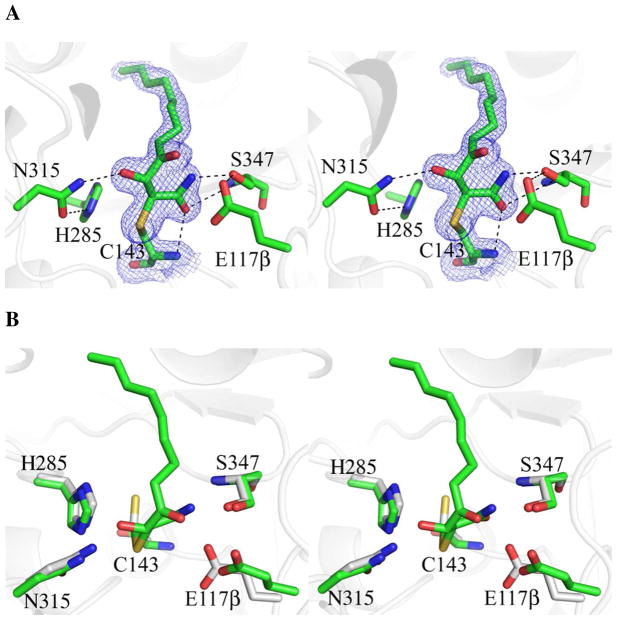
Active Site Changes Induced by Reaction with Cerulenin
Cerulenin treatment leads to the formation of a covalent adduct with Cys143 of OleA (Figures 2 and 4A). The entire cerulenin molecule is well resolved with clear positioning of the amide and hydroxyl moieties, and is at 100% occupancy. The amide of the bound cerulenin forms hydrogen bonds with the backbone amide nitrogen of Cys143 and Ser347, and the hydroxyl of Ser347. The adduct displaces the water network and causes movement and increased mobility in the side chain of Glu117β, which only retains a single hydrogen bond to the hydroxyl of Ser347 (Figure 4A). The cerulenin hydroxyl is hydrogen bonded to Asn315 of oxyanion hole 1. To accommodate the cerulenin, Cys143 rotates approximately 120° from its position in the unbound form of the enzyme (Figure 4B). The imidazole ring of His285 in the unbound and cerulenin-complexed structures is rotated by 180° in χ2 (flipped) compared to the common conformer observed in other thiolase superfamily members, and hydrogen bonds directly to Asn315 of oxyanion hole 1.
Figure 4.
Stereo view of the OleA active site with cerulenin bound to Cys143. Active site residues and inhibitor are drawn in stick. Solvent molecules are represented by red spheres. Hydrogen bond contacts are indicated by dashed lines. Blue mesh illustrates the 2Fo-Fc electron density maps contoured at 1.0 σ. (A) Active site of OleA co-crystallized with cerulenin (carbon green). (B) Overlay of OleA active sites in the unbound (carbon gray), and cerulenin bound states. The overlay was generated in PyMOL using the program SUPER. This figure was produced using PyMOL (http://www.pymol.org/).
A New Long-Chain Alkyl Binding Pocket Defined in OleA
The cerulenin complex experimentally identifies an undescribed alkyl channel accessible in OleA. When compared to the substrates of X. campestris OleA (C10, C12, C14, C16, and C16:1) cerulenin is equivalent to a C11:2 fatty acid (the two double bonds are trans, Figure 2). To seek further evidence an OleA-cerulenin crystal was pressurized with xenon gas to identify hydrophobic sites that might further delineate this new channel, and suggest compatibility with longer X. campestris OleA substrates. 2Fo-Fc maps indicated the presence of an electronically dense atom at the base of the channel originating from the alkyl chain of Cys143 tethered cerulenin (Figures 5A). The xenon site did not refine to full occupancy (45%) but anomalous maps collected on a Cr anode X-ray source allowed for unambiguous assignment. Taken together, the position of the cerulenin C11:2 alkyl chain (whose length is in the range of X. campestris OleA substrates) and this xenon site suggest a continuous channel beginning at the enzyme active site and terminating at a group of bulky hydrophobic residues (Figures 5A). Residues 252-272 and 289-301 are a pair of α-helices that form the ceiling of the channel, while a β-sheet consisting of residues 280-284, 339-346, and 349-355 forms the base. The interior of the channel contains hydrophobic side chains creating a favorable environment for binding of a long alkyl chain. The channel (designated channel A) extends beyond the length of the cerulenin alkyl chain to the xenon site, and is compatible with the C10-C16 fatty acyl substrates of OleA (13).
Figure 5.
Proposed OleA channel A. (A) The position of the cerulenin alkyl chain and xenon (Xe) atom are consistent with a continuous alkyl binding channel. The cerulenin bound to Cys143 and the xenon are drawn in space-filling colored by atom (carbon, green; Xe, pale yellow). Electron density for the Xe is included (2Fo-Fc contoured at 1.0 σ, blue mesh; anomalous contoured at 9.0 σ, pink mesh). The OleA secondary structural elements that form the channel and contact the cerulenin and Xe are represented in cartoon and colored orange. The bulky hydrophobic side chains that line the most distal part of the channel from Cys143 are also colored orange and drawn in stick. (B) Two helices that line channel A of OleA (gray cartoon) are more closely packed in FabF (PDB ID 1B3N, carbon cyan), FabB (PDB ID 1FJ8, carbon brown), and FabH (PDB ID 2QX1, carbon pink). This figure was produced using PyMOL (http://www.pymol.org/).
DISCUSSION
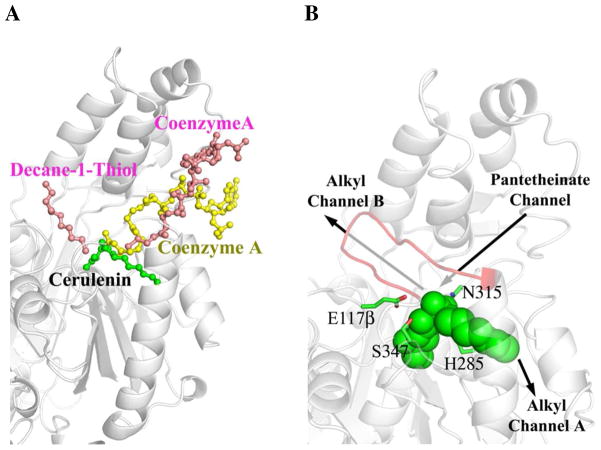
OleA initiates long-chain olefin biosynthesis by condensing two fatty acyl-CoA substrates at a cysteine-centered active site (Figures 1 and 3). The pantetheinate channel that accommodates CoA- or ACP-charged substrates have been well characterized and structurally visualized in the thiolase superfamily enzymes FabF, FabB, FabH and HMG-CoA synthase, and the channel is conserved in OleA (Figure 6A; Table 2) (16, 17, 19, 21). However, OleA is distinct in requiring two additional channels to simultaneously accomodate two long alkyl chains. HMG-CoA synthase substrates do not have long alkyl chains, while members of the Fab enzyme family need to accommodate a single long alkyl chain (18, 19). The hydrophobic channel A defined by cerulenin and xenon in the OleA structure has not been previously described (Figure 5A). In vitro data on X. campestris OleA demonstrate the enzyme can use C10, C12, C14, C16, and C16:1 as substrates (13). Modeling an extended alkyl chain from the cerulenin is consistent with channel A being able to accomodate C16 alkyl chains, although the exact length tolerance remains unclear.
Figure 6.
Illustration of OleA binding channels. (A) OleA bound with cerulenin overlaid with FabH (bound with decane-1-thiol and coenzyme A, PDB ID 2QX1) and HMG-CoA synthase (bound with coenzyme A, PDB ID 1TXT). OleA is drawn in gray cartoon and cerulenin as a green ball-and-stick model. For clarity, the monomers of FabH and HMG-CoA synthase are omitted and their bound ligands are drawn in ball-and-stick (pink and yellow respectively). Decane-1-thiol occupies the alkyl channel B in FabH. Coenzyme A occupies the pentetheinate channel in both FabH and HMG-CoA synthase. (B) Putative assignment of OleA binding channels. Active site residues are drawn in stick (carbon green) and bound cerulenin drawn in green space-filling. Labeled arrows indicate the position of the alkyl and pantetheinate channels in the OleA monomer (gray cartoon). An eleven residue (239-249) β-hairpin that could only be modeled in one monomer of the P212121 crystal form is drawn as red cartoon. This figure was produced using PyMOL (http://www.pymol.org/).
Table 2.
Structures used in overlays with OleA.
| Organism | Enzyme | PDB ID | RMSDa from OleA (Å) | Ligands | Channels Bound |
|---|---|---|---|---|---|
| Mycobacterium tuberculosis | FabH | 2QX1 (18) | 2.01 | Decane-1-Thiol, CoA-SH | Alkyl Channel B, Pantetheinate Channel |
| Staphylococcus aureus | HMG-CoA Synthase | 1TXT (20) | 2.62 | CoA-SH | Pantetheinate Channel |
| Escherichia coli | FabB | 1FJ8 (17) | 3.93 | Cerulenin | Alkyl Channel B |
| Escherichia coli | FabF | 1B3N (16) | 4.04 | Lauric Acid | Alkyl Channel B |
Root-mean-square-deviation calculated over all matched atoms between structures by SUPER in pymol (http://www.pymol.org/).
While the alkyl chain of cerulenin lies in OleA channel A, the crystal structure of the Escherichia coli FabB-cerulenin complex shows the cerulenin alkyl chain bound in the opposing direction (Figure S2; Table 2) (17). This channel (known as B) has been shown to bind alkyl chains in other Fab enzymes and thus appears to be the preferential binding site for the long-chain alkyl substrate utilized in fatty-acid biosynthesis (Figure 6A; Table 2). The two helices of OleA channel A are spread apart when compared to the closely packed helices observed in the Fab enzymes (Figure 5B), consistent with the idea that alkyl chains can more easily thread between the β-sheet and two helices that define channel A.
Channel B in OleA appears very similar to channel B in the Fab enymes. A β-hairpin (residues 239-249) that defines the ceiling of channel B is ordered in only one of the monomers within the P212121 crystal form, and disordered in the OleA-cerulenin structure. In M. tuberculosis FabH, this region is similarly disordered and ordered in different crystal structures, and has been proposed to represent an open state allowing easy access for the long alkyl chain of a CoA-charged substrate, and a closed state with an ordered β-hairpin optimized for productive catalytic turnover (19). As cerulenin binds in the newly defined channel A, this is likely the site where the OleA first substrate binds. The OleA second long-chain fatty acyl-CoA substrate would then bind in channel B. This would favor ordering of the β-hairpin to attain the closed state, as suggested for FabH (18), promoting the Claisen condensation and hydrolysis (Figure 1B). This model is also consistent with the observed release of fatty acids as aborted reaction products in OleA turnover (13). Acylated Cys143 in the open conformation is likely more vulnerable to premature hydrolysis within the time-frame required to bind the second fatty acyl-CoA substrate.
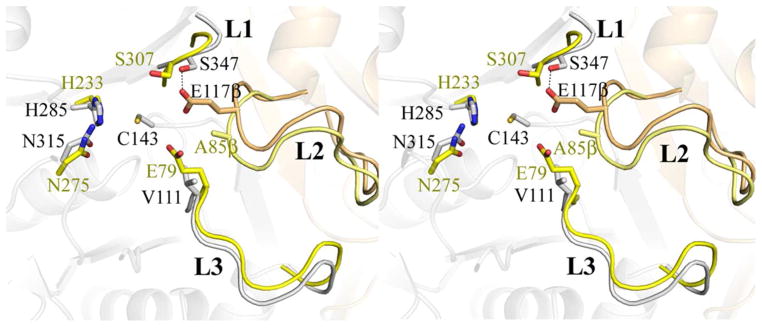
The OleA active site reveals a spatial arrangement of chemical groups conserved within the thiolase superfamily (Figure 3). Oxyanion holes are formed on either side of the centralized Cys143 and the active site is fully occupied by solvent (Figure 3B). In OleA, cerulenin-binding reveals hydrogen bonding partners in each pocket that are likely important during turnover (Figure 4A). In HMG-CoA synthase, biochemical and structural studies have identified the histidine as the general base that deprotonates the cysteine to initiate the nucleophilic attack on the first substrate that leads to acetylation (21, 30). By analogy, His285 likely acts as the general base in this part of the OleA reaction.
The subsequent activation of the acetylated Cys to initiate condensation with the second substrate requires an additional base that in biosynthetic thiolases is another Cys, and in HMG-CoA synthase is a Glu (31, 32). OleA does not have a second Cys in the active site, but does contain a glutamate (Glu117β). The residue is in a loop element (L2) from the neighboring monomer, and is the first superfamily member in which both monomers contribute to the active site. The glutamic acid base of HMG-CoA synthase (Glu79) originates from loop element L3 within the same monomer that contributes the other active site residues (Figure 7). In OleA, the sequential and structural equivalent of the HMG-CoA synthase Glu79 is Val111, whereas the equivalent of OleA Glu117β in HMG-CoA synthase is an alanine (Ala85β, Figure 7). In the cerulenin complex, Glu117β is relatively disordered and is not in contact with the cerulenin, being ~ 4 Å away at closest approach. Having Glu117β in a loop that is not structurally part of the core protein fold containing the active site may provide a more flexible environment in which to spatially coordinate the chemistry between the two long-chain fatty acid acyl-CoA substrates during OleA turnover. Such plasticity may allow for the binding of large substrates whilst permitting the reactive acyl-thioester moieties of CoA charged fatty acids to be correctly positioned within the active site.
Figure 7.
Overlay of HMG-CoA synthase (PDB ID 1TXT) onto OleA. The dimer of OleA is shown in faded cartoon colored as in Figure 3. Three loop elements (L1, L2, and L3) conserved between OleA and HMG-CoA synthase are highlighted, with L2 originating from the neighboring monomer. Residues forming a single active site in each enzyme are drawn in stick. For simplicity, a single active site cysteine (C143) from OleA is drawn. Residues from OleA are colored by monomer and labeled in black font. The E117β (carbon tan) derives from the L2 loop element making hydrogen bonding contacts with the S347 found in the L1 loop. Spatially equivalent residues from HMG-CoA are labeled in yellow font and colored by monomer. The active site of HMG-CoA synthase is formed completely from one monomer (carbon yellow) including the E79 catalytic base that derives from the L3 loop. In HMG-CoA synthase, the position of the E117β aligns with A85β (carbon pale yellow). The overlay was generated in PyMOL using the program SUPER. This figure was produced using PyMOL (http://www.pymol.org/).
Interestingly, most organisms with the oleABCD operon produce a single length olefin product, while in X. campestris multiple length olefins are produced (3). The chain length appears primarily due to the action of OleA, as genetic experiments that switched oleA genes between species took on the olefin profile of the recombinant OleA protein (3). The substrate promiscuity of X. campestris OleA is an unusual trait within the thiolase superfamily. Differences in the alkyl channels of OleA enzymes may explain the diverse product length profiles amongst different OleA proteins.
CONCLUSIONS
Our results support the presence of three major substrate binding channels in OleA that promote the condensation of the two long-chain fatty acyl-CoA substrates. The location of the fatty acid biosynthesis inhibitor cerulenin, in conjunction with a xenon binding site, strongly supports channel A as the additional alkyl binding channel required for OleA catalysis. A glutamic acid residue (Glu117β) from the neighboring monomer likely promotes Claisen condensation by acting as the catalytic base. Future mutagenesis studies of Glu117β and His285 should test their catalytic roles as bases in OleA turnover.
Supplementary Material
Acknowledgments
We thank Jasmine Erickson for assistance with expression and purification of OleA used in these studies. We thank staff at sector 23, GM/CA-CAT, of the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, USA, for assistance. GM/CA-CAT is funded by National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Sciences Grant Y1-GM-1104. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute, and we thank Can Ergenekan for support. We also thank Ed Hoeffner at the Kahlert Structural Biology Laboratory (KSBL) at the University of Minnesota.
This work was supported by National Institutes of Health Grants GM-90260 (C.M.W) and Chemistry Biology Interface Training Grant GM-008700 (J.A.F), the Department of Energy ARPA-E under Award No.DE-AR0000007 (L.P.W), and University of Minnesota Doctoral Dissertation Fellowships (J.A.F) and (B.R.G).
Footnotes
Co-ordinates and structure factors have been deposited in the Protein Data Bank as entries 3ROW (OleA unbound), 3S21 (OleA-cerulenin co-crystal), and 3S23 (OleA-cerulenin co-crystal Xe pressurized).
SUPPORTING INFORMATION AVAILABLE
An α-carbon overlay of all reported OleA structures (Figure S1), and an overlay of OleA-cerulenin with E. coli FabB-cerulenin (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
ABBREVIATIONS: CoA, coenzyme A; Ole, olefin biosynthesis enzyme; Fab, fatty acid biosynthesis enzyme; ACP, acyl carrier protein; HMG-CoA synthase, 3-hydroxy-3-methylglutaryl-CoA synthase; PDB, Protein Data Bank; AU, asymmetric unit; TFZ, translation function Z-score
References
- 1.Li M, Nørgaard H, Warui DM, Booker SJ, Krebs C, Bollinger JM., Jr Conversion of fatty aldehydes to alka(e)nes and formate by a cyanobacterial aldehyde decarbonylase: cryptic redox by an unusual dimetal oxygenase. J Am Chem Soc. 2011;27:6158–6161. doi: 10.1021/ja2013517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008;19:556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Sukovich DJ, Seffernick JL, Richman JE, Gralnick JA, Wackett LP. Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. Appl Environ Microbiol. 2010;76:3850–3862. doi: 10.1128/AEM.00436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wackett LP. Microbial-based motor fuels: science and technology. Microb Biotechnol. 2008;1:211–225. doi: 10.1111/j.1751-7915.2007.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albro PW, Dittmer JC. The biochemistry of long-chain, nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by a cell-free preparation of Sarcina lutea. Biochemistry. 1969;8:3317–3324. doi: 10.1021/bi00836a028. [DOI] [PubMed] [Google Scholar]
- 6.Albro PW, Dittmer JC. The biochemistry of long-chain, nonisoprenoid hydrocarbons. 3. The metabolic relationship of long-chain fatty acids and hydrocarbons and other aspects of hydrocarbon metabolism in Sarcina lutea. Biochemistry. 1969;8:1913–1918. doi: 10.1021/bi00833a022. [DOI] [PubMed] [Google Scholar]
- 7.Albro PW, Dittmer JC. The biochemistry of long-chain, nonisoprenoid hydrocarbonss. II. The incorporation of acetate and the aliphatic chains of isoleucine and valine into fatty acids and hydrocarbon by Sarcina lutea in vivo. Biochemistry. 1969;8:953–959. doi: 10.1021/bi00831a028. [DOI] [PubMed] [Google Scholar]
- 8.Albro PW, Dittmer JC. The biochemistry of long-chain, nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochemistry. 1969;8:394–404. doi: 10.1021/bi00829a055. [DOI] [PubMed] [Google Scholar]
- 9.Tornabene TG, Oro J. 14-C incorporation into the fatty acids and aliphatic hydrocarbons of Sarcina lutea. J Bacteriol. 1967;94:349–358. doi: 10.1128/jb.94.2.349-358.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albro PW, Meehan TD, Dittmer JC. Intermediate steps in the incorporation of fatty acids into long-chain, nonisoprenoid hydrocarbons by lysates of Sarcina lutea. Biochemistry. 1970;9:1893–1898. doi: 10.1021/bi00811a005. [DOI] [PubMed] [Google Scholar]
- 11.Beller HR, Goh EB, Keasling JD. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl Environ Microbiol. 2010;76:1212–1223. doi: 10.1128/AEM.02312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sukovich DJ, Seffernick JL, Richman JE, Hunt KA, Gralnick JA, Wackett LP. Structure, function, and insights into the biosynthesis of a head-to-head hydrocarbon in Shewanella oneidensis strain MR-1. Appl Environ Microbiol. 2010;76:3842–3849. doi: 10.1128/AEM.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frias JA, Richman JE, Erickson JS, Wackett LP. Purification and Characterization of OleA from Xanthomonas campestris and Demonstration of a Non-decarboxylative Claisen Condensation Reaction. J Biol Chem. 2011;286:10930–10938. doi: 10.1074/jbc.M110.216127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath RJ, Rock CO. The Claisen condensation in biology. Nat Prod Rep. 2002;19:581–596. doi: 10.1039/b110221b. [DOI] [PubMed] [Google Scholar]
- 15.Miziorko HM, Lane MD. 3-Hydroxy-3-methylgutaryl-CoA synthase. Participation of acetyl-S-enzyme and enzyme-S-hydroxymethylgutaryl-SCoA intermediates in the reaction. J Biol Chem. 1977;252:1414–1420. [PubMed] [Google Scholar]
- 16.Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J Biol Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- 17.Price AC, Choi KH, Heath RJ, Li Z, White SW, Rock CO. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J Biol Chem. 2001;276:6551–6559. doi: 10.1074/jbc.M007101200. [DOI] [PubMed] [Google Scholar]
- 18.Sachdeva S, Musayev F, Alhamadsheh MM, Neel Scarsdale J, Tonie Wright H, Reynolds KA. Probing reactivity and substrate specificity of both subunits of the dimeric Mycobacterium tuberculosis FabH using alkyl-CoA disulfide inhibitors and acyl-CoA substrates. Bioorg Chem. 2008;36:85–90. doi: 10.1016/j.bioorg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachdeva S, Musayev FN, Alhamadsheh MM, Scarsdale JN, Wright HT, Reynolds KA. Separate entrance and exit portals for ligand traffic in Mycobacterium tuberculosis FabH. Chem Biol. 2008;15:402–412. doi: 10.1016/j.chembiol.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Campobasso N, Patel M, Wilding IE, Kallender H, Rosenberg M, Gwynn MN. Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism. J Biol Chem. 2004;279:44883–44888. doi: 10.1074/jbc.M407882200. [DOI] [PubMed] [Google Scholar]
- 21.Theisen MJ, Misra I, Saadat D, Campobasso N, Miziorko HM, Harrison DH. 3-hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in “real-time”. Proc Natl Acad Sci U S A. 2004;101:16442–16447. doi: 10.1073/pnas.0405809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omura S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev. 1976;40:681–697. doi: 10.1128/br.40.3.681-697.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronnett GV, Kleman AM, Kim EK, Landree LE, Tu Y. Fatty acid metabolism, the central nervous system, and feeding. Obesity (Silver Spring) 2006;14(Suppl 5):201S–207S. doi: 10.1038/oby.2006.309. [DOI] [PubMed] [Google Scholar]
- 24.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haapalainen AM, Merilainen G, Wierenga RK. The thiolase superfamily: condensing enzymes with diverse reaction specificities. Trends Biochem Sci. 2006;31:64–71. doi: 10.1016/j.tibs.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Misra I, Miziorko HM. Evidence for the interaction of avian 3-hydroxy-3-methylglutaryl-CoA synthase histidine 264 with acetoacetyl-CoA. Biochemistry. 1996;35:9610–9616. doi: 10.1021/bi9605797. [DOI] [PubMed] [Google Scholar]
- 31.Chun KY, Vinarov DA, Zajicek J, Miziorko HM. 3-Hydroxy-3-methylglutaryl-CoA synthase. A role for glutamate 95 in general acid/base catalysis of C-C bond formation. J Biol Chem. 2000;275:17946–17953. doi: 10.1074/jbc.M909725199. [DOI] [PubMed] [Google Scholar]
- 32.Kursula P, Ojala J, Lambeir AM, Wierenga RK. The catalytic cycle of biosynthetic thiolase: a conformational journey of an acetyl group through four binding modes and two oxyanion holes. Biochemistry. 2002;41:15543–15556. doi: 10.1021/bi0266232. [DOI] [PubMed] [Google Scholar]
- 33.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.