Abstract
A stepwise addition protocol was developed to display cargo using bacteriophage P22 capsids and the phage decorator (Dec) protein. Three-dimensional image reconstructions of frozen-hydrated samples of P22 particles with nanogold-labeled Dec bound to them revealed the locations of the N- and C- termini of Dec. Each terminus is readily accessible for molecular display through affinity tags such as nickel-nitrilotriacetic acid, providing a total of 240 cargo-binding sites. Dec was shown by circular dichroism to be a β–sheet rich protein, and fluorescence anisotropy binding experiments demonstrated that Dec binds to P22 heads with high (~110 nM) affinity. Dec also binds to P22 nanotubes, which are helically symmetric assemblies that form when the P22 coat protein contains the F170A amino acid substitution. Several classes of tubes with Dec bound to them were visualized by cryo-electron microscopy and their three-dimensional structures were determined by helical reconstruction methods. In all instances, Dec trimers bound to P22 capsids and nanotubes at positions where three neighboring capsomers (oligomers of six coat protein subunits) lie in close proximity to one another. Stable interactions between Dec and P22 allow for the development of robust, nanoscale size, display vehicles.
1. Introduction
“Biomimetics”, or designing materials based on biology has emerged as a means to develop a wide array of substances for use in nanotechnology [1]. In this vein, metals have been used to either bind to and change the surface properties of pre-assembled proteinaceous structures [2], or to direct assembly of protein subunits into complex proteins arrays [3]. Metallization of protein platforms, including viral structures, has been used to produce particles with photonic, optical, and magnetic properties [1, 4]. The highly symmetric capsids of icosahedral viruses make them attractive candidates for molecular display and hence have the potential to aid in the development of new vaccines or nanoscale, cargo delivery vehicles [5–7]. Other methods of molecular display that incorporate a wide range of substrates such as carbon nanotubes, quantum dots, protein cages, and nanoparticles have limitations that diminish their efficacy as nanoplatforms. These include toxicity of the vector to the host, unpredictable loading of cargo from one display vehicle to the next, steric hindrance of the cargo with the display vehicle, and aggregation of chimeric molecules [8, 9]. The size and shape of the nanoparticle can also limit its effectiveness. For example, rod-shaped or helical structures can provide a much larger surface area for modification than an icosahedral structure with a finite number of binding sites [10]. Here we describe the use of a bacteriophage coat (or major capsid) protein that can be modified with cargo when selectively assembled in either of two forms: 1) virus-like particles with icosahedral symmetry and devoid of genome, or 2) rod-shaped “nanotubes” comprised of a helical assembly of the coat protein. This has enabled us to develop a stepwise addition protocol and a highly versatile system with distinct advantages over traditional molecular display methods.
The typical molecular display protocol involves the creation of a protein chimera or fusion, either of which may not fold properly [11]. Hence, a stepwise addition of native proteins that bind with high affinity to partners increases the likelihood that the displayed cargo will retain its physiologically relevant activity. Our protocol exploits the redundancy inherent in the highly symmetric capsids of bacteriophages and further expands this redundancy by adding a trimeric, decoration protein complex (“Dec”) that generates a vehicle with several hundred binding sites for cargo. This approach maximizes template efficiency. Ni-binding chemistry is used to load the cargo, which eliminates the need to create new fusion peptides or chimeras. This in turn provides a generalized template and a robust, high-throughput system.
Viruses that infect bacteria (bacteriophage or phage) are non-pathogenic to humans and can be safely manipulated in a laboratory environment. P22 (family Podoviridae) was chosen for this study because it is a model phage system [12, 13] and its capsid is stable and easy to purify in significant quantity, and because Dec binds selectively to the surface of mature capsids [14, 15]. The P22 virion is a complex, asymmetric structure: it has an icosahedral capsid composed of 415 molecules of the coat protein, gp5 (“gp5” = gene product 5), a 43.5 kbp dsDNA genome, 3–20 molecules each of three “ejection” proteins (gp7, gp16, gp20), a dodecameric “portal” protein (gp1), and a multi-component, tail machine (12, 18, 6, and 3 copies of gp4, gp9, gp10, and gp26, respectively) [12]. To reduce the complexity of the P22 molecular display vehicle, we used in vitro expanded heads (ExH), which are highly stable particles produced by treating P22 precursor capsids (procapsids) with heat and a chemical denaturant. The ExH particle is an empty protein shell comprised solely of coat protein that is arranged as 60 hexameric capsomers (“hexons”), but lacks pentameric capsomers (coat protein “pentons”), as well as the portal protein complex, at the vertices [16, 17]. The burst size of P22 in S. typhimurium (~600–1500 phage per cell [18, 19]) is higher than that for typical phages such as T4 (~150 per E. coli [20]) or φ29 (~570 per B. subtilis [21]). ExH are easily purified in large quantity (~0.5 gm total protein per 18 L of cells [22]). In addition, P22 yields are significantly larger than can be obtained typically with eukaryotic viruses. Previously, icosahedral structures have only shown limited effectiveness as molecular display tools, as sites targeted for modification are often non-discriminatory based on virion symmetry, meaning that all sites in an asymmetric unit are occupied [23]. For example, the phage λ gpD decoration protein, binds the capsid at all three-fold symmetry axes, for a total of 140 binding sites. Alternatively, Dec is known to interact with mature P22 virions in a highly discriminatory fashion, since Dec binds selectively to the capsid at some of the quasi- but none of the strict three-fold sites [15]. This led to a proposal that Dec’s selective binding was likely a consequence of subtle changes in the quaternary structure of the coat protein in assembled virions. This hypothesis is supported by observed differences in binding affinity for the two sites, as reported here. In addition, P22 virus- like particles have been used as nano-containers to encapsidate fluorescent proteins [24] and iron oxide [25]. In this study, we engineered Dec for conjugation with cargo through the use of both polyhistidine tags and reactive sulfhydryl chemistry (by the incorporation of a cysteine) to enable the exterior surface of ExH particles to be labeled and hence improve the versatility of these particles.
2. Materials and Methods
2.1. Purification of P22 ExH and Dec
Procapsid samples comprised of wild-type CP were isolated and purified from P22 infected Salmonella enterica serovar Typhimurium cells as previously described [22]. These were treated with 0.5 M GuHCl to produce shells at a concentration of 1 mg/mL [26], then induced to form expanded, penton-less particles (ExH) by heat treatment at 71°C for 15 minutes [16]. ExH were concentrated to 20 mg/mL using a YM-30 Centricon (MILLIPORE), spun at 12k × g at 4°C and further purified by SEC using Sephacry l S1000 matrix (GE HEALTHCARE) in the presence of 7 M urea. Dialysis until equilibrium was performed against 20 mM sodium phosphate buffer, pH 7.6 at 4 °C and purified ExH sample was concentrated to ~10 mg/mL. Design of Dec expression constructs [14] and purification by affinity-tag chromatography [15] were previously described.
2.2. Dec binding and gold bead conjugation to ExH
The buffer used for the following experiments was at pH 7.8 and included 10 mM Tris-HCl and 125 mM NaCl. ExH (1 mg/mL or 0.06 μM particles) were mixed with Dec at 6 mg/mL (142 μM trimer) for 60 min at room temperature (~20 °C). ExH-Dec complexes were separated from free Dec by SEC using Sephacryl S1000 matrix (GE HEALTHCARE) and concentrated to ~8 mg/mL using a YM100 centricon. Gold conjugation was performed by adding 4 M excess Ni-NTA-nanogold (NANOPROBES) to ExH-Dec complexes and incubating them overnight at 4°C. Unbound gold was separated from gold-labeled ExH by SEC using Sephadex G25 matrix (GE HEALTHCARE). Labeled ExH-Dec-nanogold complexes were concentrated to ~8 mg/mL prior to vitrification.
2.3. Preparation and purification of Dec-bound F170A polyheads
F170A polyheads were co-purified with procapsids as described [27]. Dec at ~1 mg/mL final concentration was added to F170A polyheads (~4 mg/mL final concentration) and incubated at room temperature for 60 minutes.
2.4. Cryo-TEM
Small (3.5 μL) aliquots of ExH complexes (ExH, ExH-DecC-his, ExH-DecC-his-Au or ExH-DecN-his-Au at ~8 mg/mL) or F170A polyheads (~35 mg/mL) were vitrified and examined using standard procedures [28]. Briefly, this involved applying samples to Quantifoil holey grids that had been glow-discharged for ~15 sec in an Emitech K350 evaporation unit. Grids were then blotted with Whatman filter paper for ~5 sec, plunged into liquid ethane, and transferred into a precooled, FEI Polara, multi-specimen holder, which maintained the specimen at liquid nitrogen temperature. Micrographs were recorded on Kodak SO-163 electron-image film at 200 keV in an FEI Polara microscope under minimal-dose conditions (~15 e/Å2) at a nominal magnification of 39,000. The range of objective lens defocus settings used to record each data set is listed in Table 1.
Table 1.
3D reconstruction statistics of icosahedral particles
2.5. 3D image reconstructions of icosahedral particles
Micrographs exhibiting minimal astigmatism and specimen drift were selected for further processing and digitized at 6.35 μm intervals (representing 1.62 Å pixels at the specimen) on a Nikon Supercool 8000 scanner. The program RobEM (http://cryoEM.ucsd.edu/programs.shtm) was used to estimate micrograph defocus and astigmatism, extract individual particle images, and preprocess the images as described. For each specimen, 150 particle images were used as input to the random-model computation procedure to generate an initial 3D density map at ~25 Å resolution [29]. Each map was then used to initiate determination and refinement of particle orientations and origins for the complete set of images using the current version of AUTO3DEM (v4.01.07) [30]. Phases and amplitudes of the particle structure factor data were corrected to compensate for the effects caused by the microscope contrast-transfer function as described [31]. The Fourier Shell Correlation (FSC0.5) criterion was used to estimate the resolutions of all 3D reconstructions (Table 1) [32]. Graphical representations were generated with the RobEM (http://cryoEM.ucsd.edu/programs.shtm) and Chimera [33] visualization software packages.
2.6. Particle boxing, preprocessing, and polyhead reconstruction using IHRSR++
The helixboxer routine in EMAN [34] was used to extract individual polyhead images from digitized film data that displayed minimum astigmatism and specimen drift. Each polyhead image, which was 400 pixels wide and ranged in length from 400 to 8192 pixels, was embedded in a square box of 81922 pixels with the background value of the box set equal to the average pixel density along the original border of the boxed image. These padded images were Fourier transformed using SPIDER [35], and the computed diffraction pattern of each polyhead enabled us to assess polyhead quality and remove particles that were not suitable for the final reconstruction as described previously [36]. Each polyhead image was cut into several overlapping segments and these were further classified on the basis of polyhead diameter as described [36].
Before each polyhead reconstruction was computed, all included image segments were adjusted to correct for the phase reversals caused by the microscope contrast transfer function. Reconstructions were computed using IHRSR++ (version 1.4) [36], which is an enhanced version of IHRSR [37]. For each data set, several different Cn symmetries were applied (Table 1), but in each instance only one yielded a readily interpretable map with recognizable hexons.
2.7. Circular Dichroism
The secondary structure and stability of the N-his and C-his tagged Dec proteins were analyzed by circular dichroism. Protein concentrations were ~0.45 mg/mL after dilution of the concentrated proteins into H2O. Spectra were obtained from 255 to 195 nm with a 1 nm stepsize, bandpass set to 3 nm, and 20 sec averaging per point at 20 °C in a 1 mm path length cuvette. Melting curves were obtained for each protein diluted to 0.25 mg/mL in H2O in a 2 mm path length cuvette. The temperature was ramped at a rate of 0.3 °C/min from 20 to 80 °C and data collected at 218 nm every 0.5 °C with a sampli ng time of 15 sec. The melting temperature is estimated as the midpoint of the melting transition. These data were not fit to any folding model.
2.8. Fluorescence anisotropy
Plasmid-encoded N-his-tagged Dec was first engineered to have an S90C substitution using site-directed mutagenesis with the forward (CTTCTGCATGCGGTTGAGTAGCTAATG) and reverse (GAACCGCATGCAGAAGTGTAGTTAGC) primers to alter the 90th codon from AGC to TGC. The his-tagged S90C variant was purified as described [15] but with a Talon column (ClonTech). The purified S90C protein was labeled with a 20-fold molar excess of Texas Red-maleimide (InVitrogen) after incubation with 2 mM TCEP (Pierce) in 20 mM phosphate buffer, pH 7.6, 100 mM NaCl. After further incubation at room temperature for 2 hours, the non-reacted label was separated from the covalently-linked label by spinning the protein solution sequentially through three 2-mL Zeba Spin desalting columns (Pierce). The extent of labeling was ~ 80% based on the concentration of Dec protein monomers. Thus, statistically, >99% of Dec trimers were labeled with Texas Red. The binding of the Texas Red labeled S90C Dec protein to ExH was confirmed by 1% native agarose gel electrophoresis in Tris acetate EDTA (TAE) buffer (Maniatis). The fluorescence of the Dec protein in the gel was visualized using a BioRad gel documentation system with a UV transilluminator.
The binding affinity of the Texas Red labeled N-his-tagged Dec S90C to ExH was determined by fluorescence anisotropy in an SLM Aminco-Bowman 2 with an automatic polarizer accessory. The excitation was set to 585 nm and the emission to 620 nm. The bandpasses were set to 4 nm. The labeled Dec was diluted into a 1 cm fluorescence cuvette to 0.3 μM in 20 mM phosphate, pH 7.6, and 100 mM NaCl. Aliquots of ExH were added sequentially. The fluorescence anisotropy was read after 15 min incubation with stirring at 20 °C. Each anisotropy measurement was the average of ten separate, eight-sec readings. Each measurement was repeated 5–6 times and averaged. The G-factor was automatically calculated one time per measurement. The dissociation constant was obtained by fitting the data to the standard equation for binding data:
where L is the free ligand expressed in sites ([ExH*60 high affinity sites]), LR is the bound ligand and Rt is the concentration of labeled Dec. The data were fit with the program Kaleidagraph (Synergy Software). The Kd was determined for each assay, which was done in triplicate, and the average Kd and standard deviation determined.
3. Results
3.1. Locations of the N- and C- termini of Dec
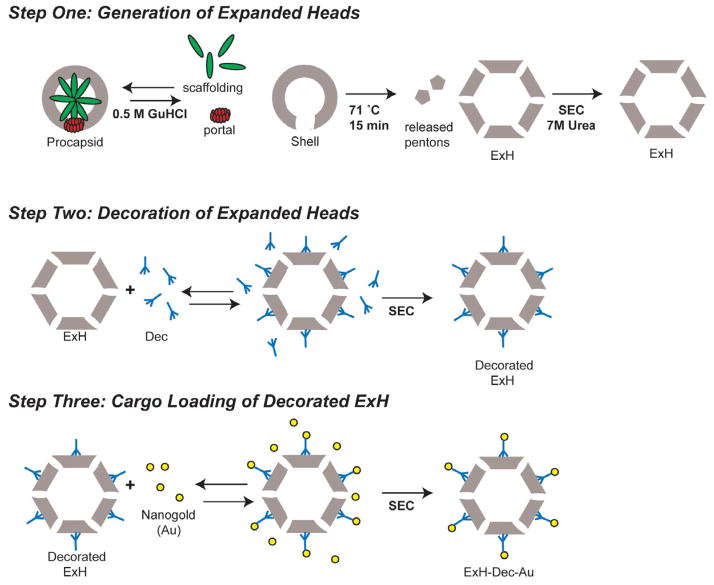
Dec with a hexa-histidine tag at either the N- or C-terminus binds to intact P22 virions [14] and similarly, both bind to ExH. We used size exclusion chromatography to purify ExH-Dec complexes from unbound Dec (Figure 1) and vitrified and imaged these samples using low- dose, cryo-electron microscopy (cryo-EM) methods [28]. Image reconstruction methods were then used to compute three-dimensional density maps of the bound complexes. A cryo- reconstruction of ExH with no Dec bound (EMDB ID 5150) [17] was used to calculate an ExH- Dec minus ExH difference map to identify density features corresponding to Dec (data not shown). Both forms of tagged Dec (hexa-histidine at N- and C-termini) bound to ExH at the same sites, and in the same locations as previously reported for native Dec bound to authentic virions [15]. This thus demonstrates that the terminal affinity tags do not disrupt Dec binding to capsids. The Dec trimers bound to all 80 potential sites, with 60 at high affinity (quasi-three fold between hexons) and the other 20 at lower affinity (strict icosahedral three-fold, see below). Hence, each ExH particle provides 240 binding sites for cargo.
Figure 1. Schematic diagram of the stepwise addition protocol used to label ExH.
For a full description of the purification procedures, see Materials and Methods. Abbreviations: ExH (heat-expanded heads); SEC (size exclusion chromatography).
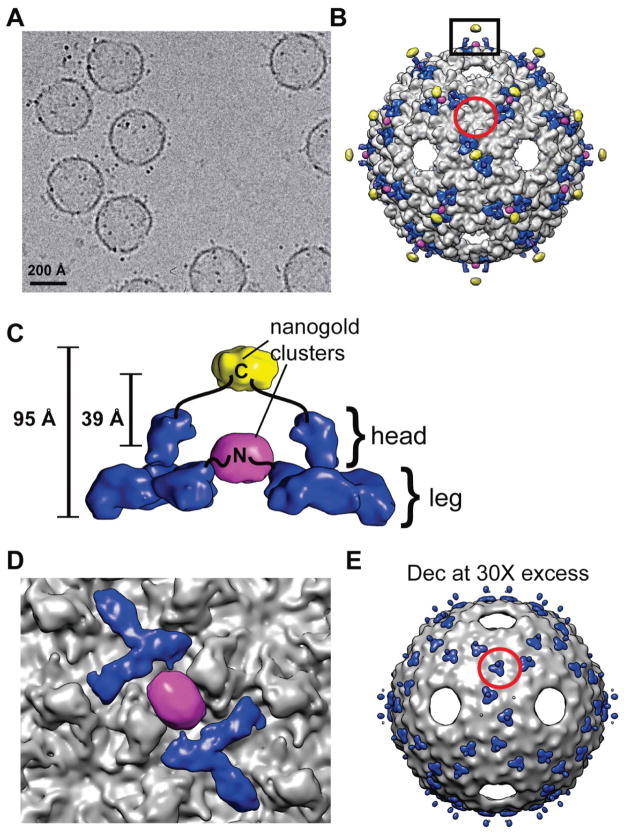
We next determined if the N- and C- terminal hexa-histidine tags could serve as useful sites on Dec to load cargo. Nickel nitriloacetic acid (Ni-NTA) nanogold beads (NANOPROBES) are known to bind efficiently to hexa-histidine tags [38] with little if any non-specific binding [39] and these beads were easily recognized in our cryo-EM images of purified, vitrified specimens of ExH-Dec-nanogold complexes (Figure 2A). Icosahedrally-averaged cryo-reconstructions computed from images of ExH-DecN-his and ExH-DecC-his along with difference map analysis [17, 40], enabled us to locate the bound gold beads, and therefore the N- and C-termini of Dec. This showed that the termini are separated by at least ~39 Å (Figure 2B–D). The C-terminus is located at the tip of the “head” of Dec and the gold beads extend as much as ~95 Å away from the surface of the ExH shell (Figure 2B, C). This exposed position on Dec favors stoichiometric binding of cargo because steric hinderance effects should be significantly reduced. The N-terminus of Dec occurs at the base of the “leg”, and though it lies close to the ExH shell, the nanogold beads bound efficiently, which indicates that other small cargo could bind with minimal steric interference (Figure 2C, D). Gold labeling of each histidine-tagged terminus revealed the location of two distinct binding sites and thus, two spatially distinct locations for the Dec protein termini, and thus potentially two different locations for bound cargo. In contrast, the N- and C- termini of phage λ gpD are in close proximity to one another [41], where a loaded cargo molecule would likely be in a similar location when attached to either termini.
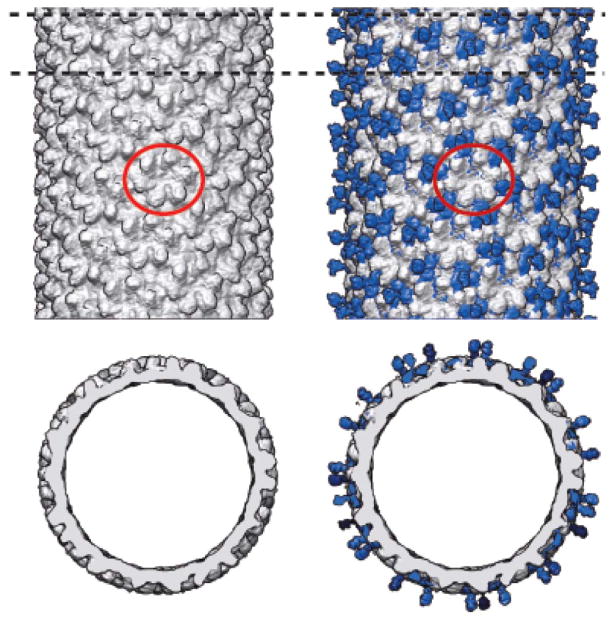
Figure 2. Dec binding to ExH.
A) Micrograph of vitrified sample of ExH-DecC-his to which Ni-NTA nanogold (“Au”) was bound. B) Shaded-surface representation, viewed along a two-fold axis, of a hybrid density map constructed from portions of four separate cryo-reconstructions (ExH, ExH-Dec, ExH-DecN-his-Au, and ExH-DecC-his-Au). ExH appears in greyscale, Dec is rendered blue, and the nanogold clusters are colored yellow (C-his tag) and magenta (N-his tag). The Dec density was obtained from the ExH-Dec minus ExH difference map, and the nanogold densities were obtained from the corresponding ExH-Dec-Au minus ExH-Dec difference maps. The red circle outlines the region near one icosahedral three-fold axis. C) Enlarged view of the region boxed in panel (B), showing the nanogold clusters bound to a pair of neighboring Dec trimers. Black lines schematically represent the linkers that connect the clusters to the N- and C-termini in the leg and head portions of Dec [15], respectively. D) Close-up view of (B), with density beyond a radius of ~320 Å masked out to emphasize and highlight interactions between the Dec leg domain (blue) and the N-terminal, his-tagged nanogold cluster (magenta). E) Similar to (B) for the cryo-reconstruction of the ExH-DecC-his sample incubated with 30-fold excess Dec. This shows Dec trimers bound at strict three-fold sites on the ExH particle (one such trimer is highlighted by the red circle for comparison with panel (B)).
The cryo-reconstruction of the ExH-DecC-his-nanaogold particle showed little or no density connecting the nanogold cluster to Dec and the overall size of the density ascribed to each nanogold cluster was spread over a volume much larger than expected for a bead of 18 Å diameter. These observations indicate that the his tags are flexible and the nanogold beads can adopt a variety of different positions relative to Dec. Ni-NTA nanogold has much more inherent flexibility in its binding compared to mono-maleimido nanogold, which has been used to locate specific cysteine residues [17]. Since each Dec has a hexahistidine tag, it is thereotically possible for up to three Ni-NTA moieties to bind to each Dec monomer. However, given the size of the nanogold bead, steric hindrance likely limits the binding to one nanogold to each hexahistidine tag. Also, nanogold binding was only observed at positions midway between neighboring Dec trimers (Figure 2B–D). This could be explained by either of two possibilities: 1) nanogold binds to all Dec monomers but only in a consistent manner where two tags are in close apposition and constrain the beads to lie within a relatively small volume so the image averaging procedures, which impose icosahedral symmetry, enhance rather than smear out the gold signal, or 2) nanogold only binds when two, closely-juxtaposed tags are present. We believe the first possibility is more likely given that nanogold has been shown to bind to P22 coat proteins at non-neighboring polyhistidine tags [42]. The precise location of bound entities, specifically designed for molecular display, has not always been possible to identify [10]. Therefore, our results with the ExH-Dec complexes are noteable in that the cargo and its positions were clearly observed.
Consistent with previous reports concerning Dec interactions with native P22 virions [15], Dec bound the icosahedral capsid shell of ExH at quasi-three fold axes of symmetry (between hexons), but not at true three-fold symmetry axes (Figure 2B). However, in samples in which an approximately 30 molar excess of Dec was present, a cryo-reconstruction of the ExH-Dec complex showed density corresponding to Dec trimers at both strict and quasi-three-fold symmetry positions (Figure 2E). Alternatively, when free Dec was separated from bound complexes using gel filtration chromatography, density corresponding to Dec was not observed at the strict three-fold symmetry axes (Figure 2B). These data suggest that Dec binds to ExH with two different affinities, which may span several orders of magnitude. Perhaps, this discrimination can be exploited in the future to include multiple types of cargo on the P22 surface through thermodynamic control.
3.2. Dec is a β-sheet rich protein with moderate stability
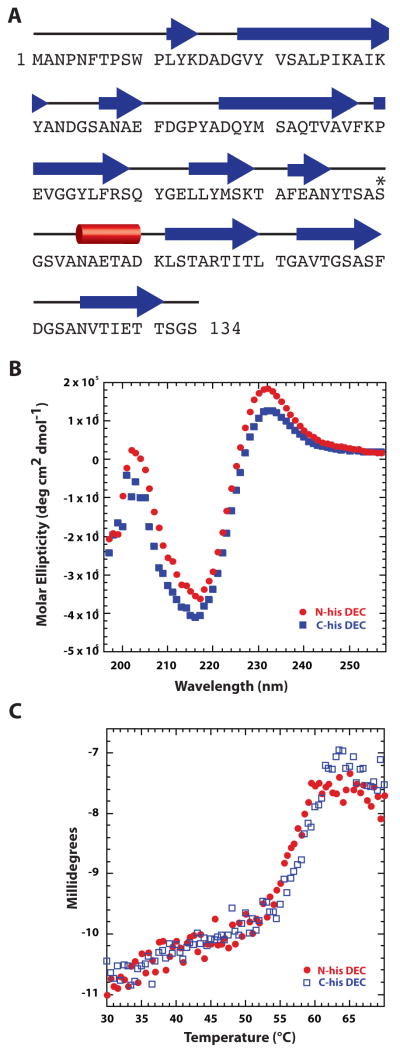
If Dec is to be embraced as a viable cargo carrier, more detailed biophysical characterization of the protein is required. We recorded circular dichroism spectra from N- and C- tagged Dec samples that show Dec is a β-sheet rich protein, which is consistent with its predicted secondary structure based on amino acid sequence (Figure 3A, B). Previous studies have shown that the phage decoration proteins, gpD of λ and hoc of RB49, a T4-like phage, are also β-rich proteins [41, 43]. However, these proteins bear no recognizable sequence similarity to Dec. Since “cement” proteins like Dec can add considerable stability to the phage and virions to which they are bound [14, 44], we hypothesized that Dec itself might be a very stable complex as was demonstrated for the SHP decoration protein of phage 21 [45]. Also, unlike λ gpD, which remains monomeric until it forms trimers upon binding to the λ phage head [46], Dec and SHP are trimers in solution [14, 45]. In contrast to the very stable SHP protein trimers (which require heating for more than 20 min at 95 °C to unfold), Dec trimers have melting temperatures (Tm) of ~57.5 and ~59.0 °C for the N-his and the C-his tagged proteins, respectively, indicative of a protein with only moderate stability. In addition, the tagged Dec trimers were found to be highly soluble, with protein concentrations greater than 60 mg/mL easily attained (data not shown). Although the Tm values for Dec exceed physiologically relevant temperatures (typically ~37 °C for enteric bacteriophage) and therefore Dec most likely remains trimeric and completely and correctly folded in vivo, this is not an exceedingly high Tm for a stabilizing protein [45]. Because Dec adds considerable stability to native phage [14], the contacts that Dec trimers form with the capsid surface must contribute to this increased stability. Interactions with the capsid may also induce changes in Dec conformation and these could contribute as well to overall virion stability. The stability gained by binding to capsids makes Dec a suitable molecule for cargo display.
Figure 3. Dec is a β-rich protein.
A. Secondary structure of Dec predicted by PsiPred [62] shows high β content. Other programs like SABLE [63] gave similar results. Individual β-strands are depicted by blue arrows, and the lone helix is represented by a red cylinder. The asterisk identifes S90, which was engineered to cysteine for anisotropy experiments. B. Circular dichroism spectra of the N- and C-terminal, hexa-histidine-tagged Dec protein. C. Circular dichroism spectra at 218 nm monitored during a thermal melting experiment in which the sample was raised from 30 to 70 °C at a rate of 0.3 °/min.
3.3. Dec binds to capsids with nanomolar affinity
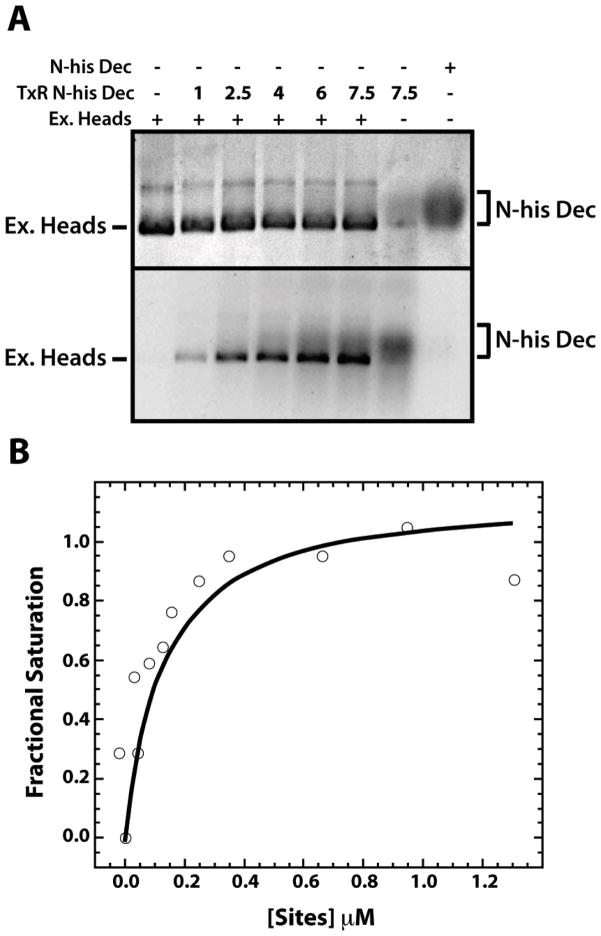
Size exclusion chromatography of bound and free Dec protein followed by SDS-PAGE suggested that the Kd of the ExH-Dec complex was below the sensitivity of such experiments (≤100 nM, data not shown). Therefore, we used fluorescence anisotropy, which is capable of measuring dissociation constants as low as 1 × 10−11 M [47], to determine the affinity of Dec trimers for ExH. To observe an anisotropy signal from Dec, we conjugated it with Texas Red maleimide (Molecular Probes), which fluoresces strongly at 530 nm with a reasonably long lifetime (~4 nsec). Therefore, the change in the rate of rotation of Dec upon binding to ExH was easily detectable. Native Dec has no cysteines to conjugate with this fluorescent probe. Hence, we introduced a cysteine at position 90 (S90C; “DecS90C”) using a QuikChange protocol starting with the N-his-tagged Dec construct [14]. The engineered cysteine at position 90 did not disrupt Dec folding or function, as the purified S90C Dec was soluble, behaved similarly to native Dec, and bound to ExH (Figure 4A). In this native agarose gel, ExH were mixed with increasing concentrations of the Texas Red labeled N-his Dec (TxR N-his Dec) or unlabeled N-his Dec. The fluorescence of the TxR N-his Dec was visualized (Figure 4A lower gel) and then the gel was stained with Coomassie blue (Figure 4A upper gel). Fluorescence produced by the TxR N-his Dec was observed to migrate with the ExH band only when ExH were added to the reaction, but migrated more slowly on the gel in the absence of ExH, consistent with the location of the band corresponding to N-his Dec in the gel stained with Coomassie blue. These data showed that a third site in Dec is readily available for chemical modification (i.e. both the N- and C- termini as well as the cysteine at position 90 can bind cargo). The labeled Dec trimer was titrated with ExH and the binding affinity (of the high-affinity sites, located at the quasi-three fold symmetry axes) was determined to be 110 +/− 70 nM (Figure 4B). Since P22 and phage L coat proteins differ by only four, rather conservative amino acid substitutions [14], the binding affinity of Dec in phage L capsids is likely very similar to that of P22 expanded heads. The affinity of Dec for P22 ExH is similar to that determined for the Hoc protein of phage T4 [48]. We did not determine the affinity of the DecCHis protein for ExH, but we expect similar, if not tighter, binding since the tag lies farther from the ExH surface. In addition to the high affinity that polyhistidine tags have for Ni-NTA (Kd =10−13 M) [49], tagged Dec can bind particles as large as ~50 μm nickel agarose beads [14]. Therefore, Dec bound ExH is a highly efficient template ideally suited for chemical modification.
Figure 4. High affinity binding of Dec trimers to ExH.
A. ExH at 0.027 μM (equivalent to 1.6 μM Dec binding locations, considering only the 60 high affinity sites at quasi-three fold symmetry axes) were mixed with Texas Red labeled, N-his-tagged, S90C Dec (TxR N-his Dec) at increasing μM concentrations, as indicated in the second line of the caption to the gel, at increasing concentrations, as indicated, or with unlabeled, N-his-tagged Dec. Samples were run on a native agarose gel and visualized by Coomassie Blue staining (top) or by fluorescence (excited with a transilluminator at 280 nm) (bottom). B. A representative fluorescence anisotropy assay of the binding of Texas Red labeled, N-his S90C Dec (at a concentration of 0.3 μM), was monitored during a titration with ExH. The abcissa axis gives the μM concentration of ExH multiplied by 60.
3.4. Dec binds to P22 polyheads
Since the nature of the coat protein hexon-hexon interactions clearly affects the affinity of Dec binding to icosahedrally symmetric capsids (i.e. to quasi- versus strict three-fold sites), we questioned whether Dec could bind to other structures that assemble from phage capsid proteins, such as polyheads. These are helical arrays (tubes) of capsid protein oligomers that generally form when an amino acid substitution occurs in the protein or an non-sense mutation knocks out another viral protein [50]. In the case of T4 phage, the native decoration protein, Soc, also binds to T4 polyheads [51]. Cryo-reconstructions of three different P22 polyhead structures, comprised of F170L and F170A variants of the coat protein, were recently reported [36]. Hexons in the F170A polyhead adopt a conformation similar to that seen in mature virions and ExH, but they are arranged in a helical rather than spherical manner and this yields different trimeric associations between neighboring hexons [36]. The F170A polyheads can grow up to 2 μm long [36], which provides a very large surface area for chemical modification. Since most phage decoration proteins, like Dec, only bind the capsid shell between neighboring hexons in the mature (i.e. not procapsid) state [44], we treated purified F170A polyheads with Dec to see if Dec could bind to helical as well as icosahedral assemblies. We examined the structures of the resulting polyhead-Dec particles by cryo-EM and 3D reconstruction (Figures 5 and 6), and the results of this analysis revealed a new mode of interaction between hexons and Dec that can be utilized as an alternative means of molecular display.
Figure 5. Cryo-reconstructions of F170A polyhead in the presence and absence of Dec.
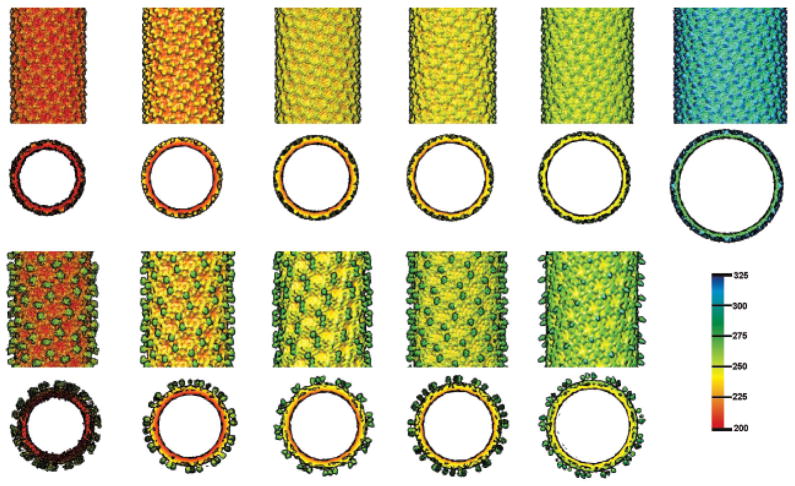
Shaded-surface side (top pair) and axial (bottom pair) views of the cryo-reconstructions of the most abundant form of the F170A polyhead free of Dec (left column), and an overlay (right column) of the F170A polyhead map (grey) and the difference density (blue, polyhead with Dec minus polyhead free of Dec). One hexon is circled in each polyhead to provide a common frame of reference. The bottom row shows a planar slab from each density map and corresponds to the region demarcated by dotted lines in the side views at the top.
Figure 6. Dec binding to different F170A polyheads.
Shaded surface, radially color-cued, representations of the cryo-reconstructionms of six different classes of F170A polyheads, distinguished by their diameter and helical symmetry. The top two rows show side and axial views, respectively, of polyheads without bound Dec and the bottom two rows show corresponding views of five of the six polyheads for which image data of polyhead-Dec complexes were available. Slices parallel to the helix axes show the head domain of Dec extends radially away from the surface of each polyhead. Bar on right shows the radial color scheme used to render all eleven cryo-reconstructions (values given in Å).
An exhaustive search identified six different classes of F170A polyhead, which are distinguished by diameter and helical symmetry (Table 2). Cryo-reconstructions of all these polyheads showed hexons that had essentially the same structure. Reconstructions of F170A polyheads incubated with Dec showed that Dec trimers bind to groups of three neighboring hexons in at least five of the six types of polyhead (Figures 5 and 6). The rarest and largest diameter polyhead (Table 2) was not identified in any of our cryo-EM images of samples with Dec present. In all polyhead-Dec complexes, the density level of the features ascribed to Dec was approximately the same as that for the capsid protein. This suggests that Dec bound to all or nearly all of the available binding sites.
Table 2.
Helical parameters of F170A polyheads solved using an enhanced version of the Iterative Helical Real Space Reconstruction Method (IHRSR++) [36, 37]
| Diameter† (Å) | Cn symmetry | Δϕ (degrees) | ΔZ (Å) | Abundance†† |
|---|---|---|---|---|
| 438 | 2 | 38.6 | 24.7 | 0.071 |
| 463 | 1 | 131.8 | 11.5 | 1.000 |
| 489 | 4 | −57.8 | 42.5 | 0.171 |
| 518* | 3 | 32.3 | 31.5 | 0.326 |
| 530 | 1 | −82.6 | 9.8 | 0.273 |
| 653 | 4 | 24.2 | 31.2 | 0.041 |
Maximum outer diameter of each polyhead
Abundance is calculated by dividing the number of boxed segments corresponding to each helical form by the number of boxed segments from the most abundant form (the box size and shift were fixed for this calculation, although varying box sizes and shifts were used during the reconstruction based on differing ΔZ values).
Previously reported in Parent et al 2010. [36]
Cn, Δφ, and ΔZ are defined in Parent et al 2010. [36]
4. Discussion
The above results and those of many others clearly indicate that decoration proteins bind to capsid surfaces via specific, highly regulated mechanisms. First, these proteins bind to mature particles but not to precursor procapsids [44], likely because the coat protein subunits adopt a different conformation in the fully matured virion [17]. Maturation presumably involves conformational changes in the capsid protein that uncovers or produces the decoration protein binding sites on the capsid surface. Second, as shown here, Dec protein exhibits significant differences in its affinity for sites that have quite similar environments. This suggests that Dec binding to ExH is regulated by the quaternary arrangement of the capsid proteins, which exhibits quite subtle differences at the strict and quasi three-fold sites in P22 ExH [17]. Polyheads however, do not have strict three-fold axes, but are formed instead by hexons arranged as trimers, but these trimeric relationships are not identical to those that occur at the true and quasi three-fold sites in icosahedral structures. Regardless of these differences, interactions among three neighboring, coat protein subunits at the capsomer:capsomer interfaces are clearly necessary for Dec binding. The polyhead data shown here further extend our knowledge of the binding capacity of Dec to include helical arrangements of hexons.
The Dec-phage system represents a useful addition to the existing repertoire of molecular display platforms. Numerous other virion platforms have been reported, such as for bacteriophages M13 [52], Qβ [53], λ [54, 55], T7 [56], and T4 [51], and for the eukaryotic viruses, cowpea mosaic [57] and potato X [7, 10, 11]. However, all of these platforms have made use of pre-assembled structures isolated from infected cells. A major advantage of the P22 system is that the self-assembly of procapsids and polyheads is highly robust and can be controlled in vitro through thermodynamic regulation [27, 58–60] and P22 maturation can be mimicked in vitro through the production of ExH particles[16]. This enables phage-like particles to be assembled in vitro with subsequent, stepwise addition of a display protein onto which a large variety of different cargo can be loaded. In vitro-regulated molecular display also allows non-biological cargo such as metals or polymers to be added, for example, by click chemistry. The interior of P22 ExH has been modified previously [24, 25], and this study, which describes external modification of P22 ExH, therefore increases the versatility of current P22 nanoparticle templates.
5. Conclusions
We have developed a stepwise protocol for adding Dec to P22 heat-expanded heads. The N- and C- termini of Dec were identitifed by means of nano-gold labeling, cryoEM imaging, and three-dimentional reconstruction methods. These decorated, virus-like-particles support the addition of cargo at either terminus of the Dec protein. We characterized Dec via circular dichroism and found that it is predominantly a β-sheet protein of moderate stability. Dec binds to the virus-like particles with nanomolar affinity, and Dec binding to P22 polyheads was characterized by cryoEM and three dimensional reconstruction methods. This work highlights a protocol that removes the need to create fusion proteins for the purpose of nanoscale molecular design, and thus provides a robust platform for cargo binding.
Acknowledgments
We thank R.S. Sinkovits for helpful discussions, M.S. Suhanovsky for help with preliminary experiments, and N.H. Olson for guidance in cryo-TEM. This work was supported in part by NIH grants R37 GM033050 and 1S10 RR-020016 (TSB), GM076661 (CMT), EB001567 (EHE), and AI074825 (SRC), by NIH fellowship F32AI078624 (KNP), and support from UCSD and the Agouron Foundation to establish and maintain cryo-TEM facilities at UCSD (TSB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sarikaya M, Tamerler C, Jen AK, Schulten K, Baneyx F. Molecular biomimetics: nanotechnology through biology. Nat mater. 2003;2:577–85. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 2.Djalali R, Chen YF, Matsui H. Au nanowire fabrication from sequenced histidine-rich peptide. J Am Chem Soc. 2002;124:13660–1. doi: 10.1021/ja028261r. [DOI] [PubMed] [Google Scholar]
- 3.Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS, Tezcan FA. Metal-directed, chemically tunable assembly of one-, two- and three- dimensional crystalline protein arrays. Nat Chem. 2012 doi: 10.1038/nchem.1290. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterji A, Ochoa WF, Ueno T, Lin T, Johnson JE. A virus-based nanoblock with tunable electrostatic properties. Nano Lett. 2005;5:597–602. doi: 10.1021/nl048007s. [DOI] [PubMed] [Google Scholar]
- 5.Douglas T, Young M. Viruses: making friends with old foes. Science. 2006;312:873–5. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 6.Kang S, Uchida M, O’Neil A, Li R, Prevelige PE, Douglas T. Implementation of p22 viral capsids as nanoplatforms. Biomacromolecules. 2010;11:2804–9. doi: 10.1021/bm100877q. [DOI] [PubMed] [Google Scholar]
- 7.Cruz SS, Chapman S, Roberts AG, Roberts IM, Prior DA, Oparka KJ. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci U S A. 1996;93:6286–90. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostarelos K, Bianco A, Prato M. Promises, facts and challenges for carbon nanotubes in imaging and therapeutics. Nat Nanotechnol. 2009;4:627–33. doi: 10.1038/nnano.2009.241. [DOI] [PubMed] [Google Scholar]
- 9.Spenger A, Grabherr R, Tollner L, Katinger H, Ernst W. Altering the surface properties of baculovirus Autographa californica NPV by insertional mutagenesis of the envelope protein gp64. Eur J Biochem. 2002;269:4458–67. doi: 10.1046/j.1432-1033.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- 10.Steinmetz NF, Mertens ME, Taurog RE, Johnson JE, Commandeur U, Fischer R, et al. Potato virus X as a novel platform for potential biomedical applications. Nano Lett. 2010;10:305–12. doi: 10.1021/nl9035753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lico C, Capuano F, Renzone G, Donini M, Marusic C, Scaloni A, et al. Peptide display on Potato virus X: molecular features of the coat protein-fused peptide affecting cell-to-cell and phloem movement of chimeric virus particles. J Gen Virol. 2006;87:3103–12. doi: 10.1099/vir.0.82097-0. [DOI] [PubMed] [Google Scholar]
- 12.Teschke CM, Parent KN. ‘Let the phage do the work’: Using the phage P22 coat protein structure as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–30. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prevelige PE, Jr, King J. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog Med Virol. 1993;40:206–21. [PubMed] [Google Scholar]
- 14.Gilcrease EB, Winn-Stapley DA, Hewitt FC, Joss L, Casjens SR. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J Bacteriol. 2005;187:2050–7. doi: 10.1128/JB.187.6.2050-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang L, Gilcrease EB, Casjens SR, Johnson JE. Highly discriminatory binding of capsid cementing proteins in bacteriophage L. Structure. 2006;14:837–45. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Teschke CM, McGough A, Thuman-Commike PA. Penton release from P22 heat-expanded capsids suggests importance of stabilizing penton-hexon interactions during capsid maturation. Biophys J. 2003;84:2585–92. doi: 10.1016/S0006-3495(03)75063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, et al. P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–410. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aramli LA, Teschke CM. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J Biol Chem. 1999;274:22217–24. doi: 10.1074/jbc.274.32.22217. [DOI] [PubMed] [Google Scholar]
- 19.Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J Mol Biol. 1973;80:669–95. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- 20.Black LW, Brown DT. Head morphologies in bacteriophage T4 head and internal protein mutant infections. J Virol. 1976;17:894–905. doi: 10.1128/jvi.17.3.894-905.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schachtele CF, Oman RW, Anderson DL. Effect of elevated temperature on deoxyribonucleic acid synthesis in bacteriophage phi-29-infected Bacillus amyloliquefaciens. Journal of virology. 1970;6:430–7. doi: 10.1128/jvi.6.4.430-437.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuller MT, King J. Purification of the coat and scaffolding protein from procapsids of bacteriophage P22. Virology. 1981;112:529–47. doi: 10.1016/0042-6822(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 23.Young M, Willits D, Uchida M, Douglas T. Plant viruses as biotemplates for materials and their use in nanotechnology. Annu Rev Phytopathol. 2008;46:361–84. doi: 10.1146/annurev.phyto.032508.131939. [DOI] [PubMed] [Google Scholar]
- 24.O’Neil A, Reichhardt C, Johnson B, Prevelige PE, Douglas T. Genetically programmed in vivo packaging of protein cargo and its controlled release from bacteriophage P22. Angew Chem Int Ed Engl. 2011;50:7425–8. doi: 10.1002/anie.201102036. [DOI] [PubMed] [Google Scholar]
- 25.Reichhardt C, Uchida M, O’Neil A, Li R, Prevelige PE, Douglas T. Templated assembly of organic-inorganic materials using the core shell structure of the P22 bacteriophage. Chem Commun (Camb) 2011;47:6326–8. doi: 10.1039/c1cc11215e. [DOI] [PubMed] [Google Scholar]
- 26.Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–97. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- 27.Suhanovsky MM, Parent KN, Dunn SE, Baker TS, Teschke CM. Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibilty in conformational switching. Mol Microbiol. 2010;77:1568–82. doi: 10.1111/j.1365-2958.2010.07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. [erratum appears in Microbiol Mol Biol Rev 2000 Mar;64(1):237.] Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–25. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan X, Sinkovits RS, Baker TS. AUTO3DEM-an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, et al. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol. 2002;76:12250–8. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–62. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 35.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj Y, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1995;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 36.Parent KN, Sinkovits RS, Suhanovsky MM, Teschke CM, Egelman EH, Baker TS. Cryo-reconstructions of P22 polyheads suggest that phage assembly is nucleated by trimeric interactions among coat proteins. Phys Biol. 2010;7:045004. doi: 10.1088/1478-3975/7/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egelman E. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–34. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 38.Hainfeld JF, Liu W, Halsey CM, Freimuth P, Powell RD. Ni-NTA-gold clusters target His-tagged proteins. J Struct Biol. 1999;127:185–98. doi: 10.1006/jsbi.1999.4149. [DOI] [PubMed] [Google Scholar]
- 39.Buchel C, Morris E, Orlova E, Barber J. Localisation of the PsbH subunit in photosystem II: a new approach using labelling of His-tags with a Ni(2+)-NTA gold cluster and single particle analysis. J Mol Biol. 2001;312:371–9. doi: 10.1006/jmbi.2001.4951. [DOI] [PubMed] [Google Scholar]
- 40.Montesano-Roditis L, Glitz DG, Traut RR, Stewart PL. Cryo-electron microscopic localization of protein L7/L12 within the Escherichia coli 70 S ribosome by difference mapping and Nanogold labeling. J Biol Chem. 2001;276:14117–23. doi: 10.1074/jbc.M008430200. [DOI] [PubMed] [Google Scholar]
- 41.Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, et al. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol. 2000;230:230–7. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 42.Kang S, Lander GC, Johnson JE, Prevelige PE. Development of bacteriophage p22 as a platform for molecular display: genetic and chemical modifications of the procapsid exterior surface. Chembiochem. 2008;9:514–8. doi: 10.1002/cbic.200700555. [DOI] [PubMed] [Google Scholar]
- 43.Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85:8141–8. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prevelige PE., Jr Send for reinforcements! Conserved binding of capsid decoration proteins. Structure. 2008;16:1292–3. doi: 10.1016/j.str.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Forrer P, Chang C, Ott D, Wlodawer A, Pluckthun A. Kinetic stability and crystal structure of the viral capsid protein SHP. J Mol Biol. 2004;344:179–93. doi: 10.1016/j.jmb.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 46.Iwai H, Forrer P, Pluckthun A, Guntert P. NMR solution structure of the monomeric form of the bacteriophage lambda capsid stabilizing protein gpD. J Biomol NMR. 2005;31:351–6. doi: 10.1007/s10858-005-0945-7. [DOI] [PubMed] [Google Scholar]
- 47.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shivachandra SB, Rao M, Janosi L, Sathaliyawala T, Matyas GR, Alving CR, et al. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: a strategy for efficient display of large full-length proteins. Virology. 2006;345:190–8. doi: 10.1016/j.virol.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 49.Casey JL, Keep PA, Chester KA, Robson L, Hawkins RE, Begent RH. Purification of bacterially expressed single chain Fv antibodies for clinical applications using metal chelate chromatography. J Immunol Methods. 1995;179:105–16. doi: 10.1016/0022-1759(94)00278-5. [DOI] [PubMed] [Google Scholar]
- 50.Steven AC, Aebi U, Showe MK. Folding and capsomere morphology of the P23 surface shell of bacteriophage T4 polyheads from mutants in five different head genes. J Mol Biol. 1976;102:373–400. [PubMed] [Google Scholar]
- 51.Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW. Phage display of intact domains at high copy number: a system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 1996;5:1833–43. doi: 10.1002/pro.5560050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnol Adv. 2010;28:849–58. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee D, Liu AP, Voss NR, Schmid SL, Finn MG. Multivalent display and receptor-mediated endocytosis of transferrin on virus-like particles. Chembiochem. 2010;11:1273–9. doi: 10.1002/cbic.201000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sternberg N, Hoess RH. Display of peptides and proteins on the surface of bacteriophage lambda. Proc Natl Acad Sci U S A. 1995;92:1609–13. doi: 10.1073/pnas.92.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunn IS. Assembly of functional bacteriophage lambda virions incorporating C-terminal peptide or protein fusions with the major tail protein. J Mol Biol. 1995;248:497–506. doi: 10.1006/jmbi.1995.0237. [DOI] [PubMed] [Google Scholar]
- 56.Sokoloff AV, Bock I, Zhang G, Sebestyen MG, Wolff JA. The interactions of peptides with the innate immune system studied with use of T7 phage peptide display. Mol Ther. 2000;2:131–9. doi: 10.1006/mthe.2000.0110. [DOI] [PubMed] [Google Scholar]
- 57.Chatterji A, Burns LL, Taylor SS, Lomonossoff GP, Johnson JE, Lin T, et al. Cowpea mosaic virus: from the presentation of antigenic peptides to the display of active biomaterials. Intervirology. 2002;45:362–70. doi: 10.1159/000067929. [DOI] [PubMed] [Google Scholar]
- 58.Parent KN, Doyle SM, Anderson E, Teschke CM. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340:33–45. doi: 10.1016/j.virol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 59.Parent KN, Zlotnick A, Teschke CM. Quantitative analysis of multi-component spherical virus assembly: Scaffolding protein contributes to the global stability of phage P22 procapsids. J Mol Biol. 2006;359:1097–106. doi: 10.1016/j.jmb.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 60.Fuller MT, King J. Regulation of coat protein polymerization by the scaffolding protein of bacteriophage P22. Biophys J. 1980;32:381–401. doi: 10.1016/S0006-3495(80)84963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skerra A, Pluckthun A. Secretion and in vivo folding of the Fab fragment of the antibody McPC603 in Escherichia coli: influence of disulphides and cis-prolines. Protein Eng. 1991;4:971–9. doi: 10.1093/protein/4.8.971. [DOI] [PubMed] [Google Scholar]
- 62.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 63.Adamczak R, Porollo A, Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–75. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]