Abstract
Generation of induced pluripotent stem cells (iPSCs) from somatic cells by defined factors is a mechanism-unknown, yet extremely time-consuming process. Inefficient reprogramming leads to prolonged periods of in vitro iPSC selection, resulting in subtle genetic and epigenetic abnormalities. To facilitate pluripotent reprogramming, we have identified the thyroid hormone triiodothyronine (T3) as an endogenous factor that can enhance reprogramming of human dermal fibroblasts (HDF) and umbilical cord mesenchymal stem cells (UCMSC). This potentiation of iPSC induction is associated with metabolic remodeling activity, including up-regulation of key glycolytic genes, an increase in cell proliferation, and the induction of mesenchymal-epithelial transition (MET). We further identify the activation of the PI3K/AKT signal pathway by T3 as an underlying mechanism for the enhanced conversion to cell pluripotency in this model. These studies demonstrate that T3 enhances metabolic remodeling of donor cells in potentiating cell reprogramming.
Keywords: T3, thyroid hormone, PI3K/AKT signal pathway, Induced pluripotent stem cell, iPSC, reprogramming, metabolic remodeling
1. INTRODUCTION
The transgenic expression of defined factors that are important for maintaining the pluripotent properties of embryonic stem cells can epigenetically reprogram somatic cells to a pluripotent state, resulting in the generation of induced pluripotent stem cells (iPSCs) [1–2]. Human iPSCs display a variety of embryonic stem cell (ESC)-like properties, and are characterized by the capacity for self-renewal and the potential for differentiating into all three germ layers. The features of these stem cells are associated with the specific expression of an array of pluripotency genes (e.g. NANOG, OCT3/4 and SOX2) and a complex transcriptional regulatory circuitry. Most notably, the iPSC technology may surmount technical difficulties in therapeutic cloning to create patient-specific stem cells for regenerative therapy, including Parkinson disease [3], sickle cell anemia [4–5], Huntington disease and other genetic diseases [6]. Thus, iPSCs may provide a versatile and ethical alternative for regenerative medicine and human disease research [7].
However, full reprogramming of human somatic cells is a very inefficient and considerably time-consuming process [8–9], especially when using non-viral methods. During the long period of cell culture and iPSC selection, tremendous cellular stress is created within the defined factor-transfected cells. Prolonged exposure to cellular stress may cause subtle genetic and epigenetic abnormalities in iPSCs, such as de novo generation of copy number variations (CNVs) [10], somatic coding mutations [11], un-erased epigenetic memory [12–14], aberrant DNA methylomes [15], and altered genomic imprinting [16–17]. Obviously, this reprogramming process places a heavy burden on cellular integrity and highlights the importance of discovering new methods to shorten and make more efficient the induction of pluripotency.
Attempts to improve the generation of iPSC cells include the use of DNA methylation inhibitors [18], histone modification inhibitors [19], and inhibition of tumor suppresser genes such as p53 and p16 [20–22]. In addition, a number of small molecules have been identified as being capable of enhancing the reprogramming efficiency [23], including valproic acid (VPA) [19], vitamin C [24], SB431542 and PD0325901 [25]. These small molecules can be characterized as antioxidants, epigenetic modulators and signaling pathway inhibitors. However, the long-term safety of these exogenous chemicals remains unknown.
While the mechanisms underlying the reprogramming process remain mysterious, it has been suggested that the activation of endogenous pluripotency-related genes and certain epigenetic changes are essential for successful iPSC induction [26]. Self-renewal of stem cells requires the down-regulation of senescence-related p53 signaling pathways, the activation of the telomerase system [27], and a very unusual cell cycle characterized by a short G1 phase and a high proportion of cells in S-phase [28–29]. In human epithelial hair follicles, both local and central hormones play key roles in adult stem cell maintenance, proliferation and differentiation [30]. Thus, the microenvironment (or niche), as maintained by endogenous signals (hormones and growth factors), may significantly affect both the behavior of stem cells, as well as the induction of iPSCs. To promote the induction of cell pluripotency, we have attempted to identify endogenous factors that are involved in the maintenance of cell stemness.
2. MATERIALS AND METHODS
2.1. Cell Culture
293T cells were purchased from ATCC and cultured in DMEM (Invitrogen) supplemented with 10% FBS (Hyclone), 0.1 mM non-essential amino acids (Invitrogen), 50 units/ml penicillin, and 50 mg/ml streptomycin (Invitrogen).
Human dermal fibroblasts were cultured from tissue obtained at surgery from a 25 year old healthy male after circumcision in Shenzhen Hospital of Peking University. Human umbilical cord mesenchymal stem cells were cultured from cord tissues collected after caesarean section of a normal pregnancy from Nanshan Hospital of Shenzhen. Primary HDF and UCMSC cells were cultured as previously described [31–32]. Briefly, foreskin and umbilical cord tissues were washed in cold phosphate-buffered saline (PBS) containing penicillin/streptomycin. The tissues were dissected into 0.5–1 mm size pieces and placed in 100 mm dishes containing 3ml FBS. After incubation at 37C, 5% CO2 overnight, 5 ml growth medium were added. Outgrowth of fibroblasts or mesenchymal cells appeared after 5–8 days. HDF or UCMSC cells were cultured in growth medium: DMEM/F12 (Gibco) supplemented with 20% FBS (Hyclone), 0.1 mM beta-mercaptoethanol, L-Glutamine (Invitrogen), 1×10−4 M nonessential amino acids (Invitrogen), 50 units/ml penicillin, and 50 mg/ml streptomycin (Invitrogen). The cells from passages 3 to 5 were used for infection by a retrovirus carrying all of the four defined factors. Human embryonic stem cell line (hESC) H9 and human iPSC were maintained on mitomycin C-inactivated mouse embryonic fibroblasts (MEF) in hES medium: DMEM/F12 (Gibco) supplemented with 20% knockout serum replacement (KSR) (Invitrogen), 10ng/ml b-FGF (PeproTech), 0.1 mM beta-mercaptoethanol, L-Glutamine (Invitrogen), 1×10−4 M nonessential amino acids (Invitrogen), 50 units/ml penicillin, and 50 mg/ml streptomycin (Invitrogen).
2.2. Cell proliferation assays
Cell proliferation was assessed using WST-1 reagent (Roche). OSKM-infected cells were seeded into 96-well plates at 4×103/well and incubated for 24h. The medium was changed to DMEM/F12 supplemented with 20% KSR, and then treated with various concentrations of T3. After 3 days, 100μl WST-1 reagent was added and incubated for 4h. The absorbance at 450nm was read by Multiskan MK3 (Thermo).
2.3. Retroviral Production and iPSC Generation
One day before transfection, 293T cells were plated at 1.2×106 per 60mm dishes (0.1% gelatin coated). Human reprogramming factors pMXs-OCT3/4, pMXs-SOX2, pMXs-KLF4 and pMXs-cMYC (Addgene) were transfected with retrovirus packaging vectors pVPack-VSV-G and pVPack-GP (Stratagene) using Fugene 6 reagent (Roche). For each transfection, the DNA-Fugene 6 mixture contained 400μl Opti-MEM (Invitrogen), 4 μg individual reprogramming factor, 2.5 μg pVPack-GP, 1.5 μg pVPack-VSV-G and 20 μl Fugene 6. After incubation overnight with 293T cells, the media was replaced with 5ml fresh medium. The virus-containing supernatants were collected at 48h and 72h after transfection, and then mixed and filtered through a 0.45-μm cellulose acetate filter (Sartorius). Viral supernatants were concentrated with Amicon Ultra-15 Centrifugal Filter Units (Millipore) at 4C, at 5000rpm for 30 minutes. The concentrated viruses were collected using a side to side washing motion in DMEM, yielding ~ 4ml concentrated retroviruses (48+72h viruses) from four 60mm dishes. We used pMXs-GFP retrovirus as infection control.
The day before infection, HDF or UCMSC cells were seeded into 12-well plates at 3×104 cells per well in serum free medium: DMEM/F12 (Gibco) supplemented with 20% KSR (Invitrogen), 0.1 mM beta-mercaptoethanol, L-Glutamine (Invitrogen), and 1×10−4 M nonessential amino acids (Invitrogen). For viral infection, 0.8ml concentrated retrovirus solution (containing an equal amount of each of the 4 factors), 0.5ml serum free medium and 6 μg/ml polybrene (Sigma) were added to target cells. After incubation for 24h, the cells were washed with PBS and 1 ml fresh growth medium was added. For thyroid hormone treatment, the serum-free medium was supplemented with 5 nM T3. Two days after infection, the media was replaced with growth medium.
Three to five days after infection, morphologically epithelial-like cells appeared. The cells were digested and 3–4 ×104 cells were transferred to 100 mm dishes on mitomycin C-inactivated MEF feeder cells (2×104 /cm2). The media were replaced with hES medium on day 9, and hES-like colonies appeared on day 12 to 14.
2.4. Live cell staining
From day 12 to 16, the hES-like colonies were labeled by live cell staining. The cells were washed with pre-warmed PBS, and then incubated with TRA-1-60 (1:50 dilution, Millipore) and appropriate secondary antibody in phenol red free DMEM (Invitrogen). The cells were washed twice, and the live image was observed in phenol red free media. The TRA-1-60 positive colonies were picked and transferred to 12 well plates with feeder cells.
2.5. Alkaline phosphatase staining and Immunofluorescence
Alkaline phosphatase (AP) staining was performed using the Alkaline Phosphatase Staining kit (Millipore) following the manufacturer’s instruction. The cells were fixed in 4% paraformaldehyde/PBS for 1–2 minutes, rinsed with PBS and then incubated with staining solution in the dark at room temperature. After 15 minutes, colonies of cells expressing AP (red colonies) were recorded using a microscope–mounted camera.
Immunofluorescent staining was used to examine the stem cell markers in iPSC colonies. Briefly, cells were fixed by 4% paraformaldehyde/PBS for 10–15 min and rinsed with PBS, then permeabilized and blocked with 0.1% Triton X-100/PBS containing 3% BSA for 30 min. After incubation with primary antibodies for 1 h at room temperature or overnight at 4C, the samples were washed three times with PBS, and then incubated with secondary antibody for 1 h.
The following antibodies were used: mouse anti-TRA-1-60(1:100 dilution, Millipore), mouse anti-TRA-1-81(1:100 dilution, Millipore), mouse anti-SSEA4 (1:100 dilution, Millipore), mouse anti-Nanog (1:200 dilution, Abcam), Rabbit anti-Nanog (1:100 dilution, Santa Cruz), mouse anti-Oct3/4 (1:100 dilution, Millipore), rabbit anti-Sox2 (1:100 dilution, Millipore), rabbit anti-E-cadherin (1:100 dilution, Cell Signaling), mouse anti-Tuj1 (1:100 dilution, Millipore), goat anti-Sox17 (1:100 dilution, R&D), and mouse anti-Vimentin (1:100 dilution, Santa Cruz). The cell samples were subsequently incubated with Cy3 or Alexa Fluor 488 labeled secondary antibodies for 1 hour. After washing three times with PBS, samples were counterstained with Hoechst 33258 (Invitrogen). Negative controls were stained without the use of primary antibodies. Fluorescence images were acquired with a Zeiss AxioCam Camera.
2.6. Karyotype analysis
For karyotype analysis, iPS cells were maintained on matrigel. The following steps were carried out at Shenzhen Women and Children Hospital, Department of Pathology. Cells were treated with 0.2 μg/mL of colcemid for 3 hours and subsequently harvested by trypsin. After treatment with hypotonic solution and fixation with 3:1 methanol and acetic acid, the samples were analyzed by standard G-banding methodology.
2.7. Real Time-PCR
Total RNA was extracted using TRI Reagent (TaKaRa), and then treated with DNase I (Sigma) to remove genomic DNA contamination [33–34]. M-MLV Reverse Transcriptase (Invitrogen) was used to synthesize complementary DNA from 2 μg total RNA. For non-quantitative RT-PCR analysis, 100–200 ng cDNA were amplified by GoTaq® Green Master Mix (Promega) with DNA Engine Thermal Cycler (BIO-RAD). For quantitative analysis, cDNA samples (no more than 100 ng) were amplified using CFX96TM real-time system (BIO-RAD) by SYBR PrimeScript™ RT-PCR Kit (TaKaRa). PCR primer sequences are shown in Table S1. The mRNA expression level of each gene relative to GAPDH (housekeeping gene) was calculated as previously described [33, 35].
2.8. Bisulfite sequencing analysis
Genomic DNA was extracted using Universal Genomic DNA Extraction Kit Ver.3.0 (TaKaRa). According to the manufacture’s recommendation, 1μg DNA samples were bisulfate-treated with CpGenome Universal DNA Modification Kit (Millipore). The promoter regions of OCT4, NANOG were amplified by PCR; primer sequences are shown in Table S1. PCR products were subcloned into pMD®18-T (TaKaRa), and ten clones of each sample were verified by sequence analysis. The methylation status was analyzed by CpGViewer software.
2.9. In vitro differentiation
For embryoid body (EB) formation, human PSCs were trypsinized by collagenase IV (Invitrogen), and cell clumps were transferred to 60 mm dishes in hES medium without b-FGF. After being maintained in floating culture for 4–6 days, EB were seeded in 0.1% gelatin-coated 6-well plates in DMEM/F12 containing 20% FBS for spontaneous differentiation into mesodermal and endodermal lineages. For neuronal differentiation, EB were cultured in medium containing N2 and B27 supplement (Invitrogen). The differentiated cells were cultured with appropriate antibodies and detected by Immunofluorescence [36].
2.10. Teratoma formation
Human iPS cells were digested with collagenase IV, and resuspended at 5×106 cells/ml in 200 μl matrigel (BD Biosciences) supplemented with 20% FBS. The cell suspension was injected subcutaneously into the dorsal flank of SCID mice. Tumors appeared 6–8 weeks after injection. Teratomas were harvested, fixed in 4% paraformaldehyde, dissected and embedded in paraffin. The sections were stained with hematoxylin and eosin (HE) for histological analysis.
2.11. Activity of the PI3K/AKT pathway
The activation of the phosphatidyl-inositol 3-kinase (PI3K)/AKT pathway was examined by measuring AKT phospho-serine 473, PFKFB2 phospho-serine 466, and AS160 phospho-threonine 642. Western blotting was used to measure phosphoproteins as previously described [33, 37]. Briefly, cells were harvested and proteins were separated by 7.5% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were transferred to polyvinylidene fluoride membranes for immunoblotting with the following antibodies: phospho-AKT Ser473 (#9271, Cell signaling Technology, USA), AKT (#9272, Cell signaling Technology, USA), phospho-AS160 Thr642 (#4288, Cell signaling Technology, USA), p-PFKFB2 Ser466 (SC-32966, Santa Cruz Biotechnologies, USA) and β-actin (ab3280, Abcam, USA).
To study the role of the PI3K/AKT pathway, the OSKM 4F-transduced cells were treated with 10 nM L-triiodothyronine in the presence of 50 μM PI3 kinase-specific inhibitor LY294002 (Cell signaling Technology, USA). Three to five days after treatment, the cells were harvested for western blot analysis, or were transferred to MEF feeder cells (2×104 /cm2) for iPS induction and AP staining of the iPSC colonies.
2.12. Statistical analysis
All data are represented as mean ± SD from 3 independent experiments. Statistical analysis of the data was performed using SPSS13.0, and the significance of differences was examined with Student’s t test. P <0.05 was considered significant.
3. RESULTS
3.1. Identification of T3 as an enhancer of induced cell pluripotency
In order to identify endogenous factors that can enhance the development of cell pluripotency, we tested a variety of chemicals, including hormones, amino acids, vitamins, and growth factors. Using the standard iPSC protocol, we first transfected two human donor cell lines, dermal fibroblasts (HDF) and umbilical cord mesenchymal stem cells (UCMSC), with retroviruses containing four defined transcription factors (Oct4, Sox2, Klf4, and c-Myc). The transfected cells were then incubated with the test substances. Alkaline phosphatase (AP) staining and live cell staining for TRA-1-60 were employed to evaluate the effect of the various substances on cell reprogramming efficiency. On day 14 post-infection, AP and TRA-1-60 positive colonies were selected and the reprogramming efficiency was then assessed in three independent experiments.
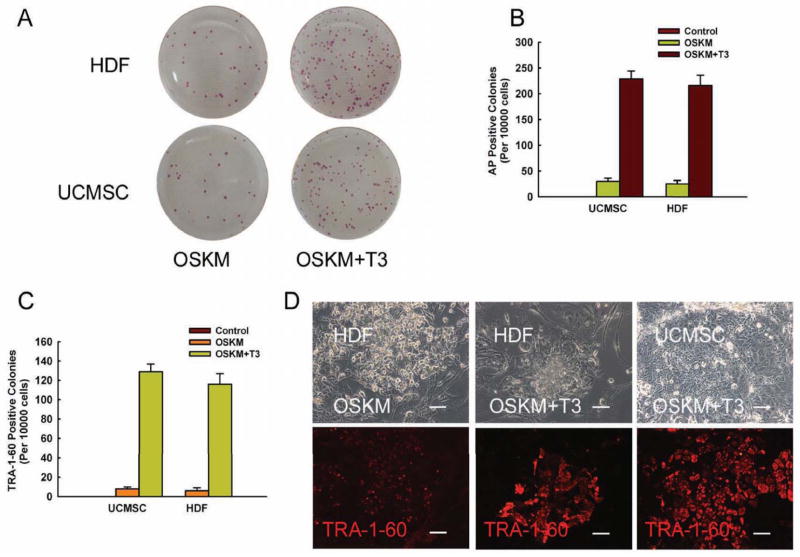
The thyroid hormone T3 was the most potent promoter of iPSC induction. Incubation with T3 at the optimal concentration of 10 nM remarkably increased the number of AP-positive colonies in virally infected HDFs and UCMSCs as compared with controls (Figs.1A–1B). Similar results were also observed in treated cells using TRA-1-60 staining (Fig.1C). Figure 1D shows typical examples of increased TRA-1-60 immunostaining of iPSC colonies in HDF and UCMSC cells. TRA-1-60 positive colonies were placed into 12-well plates for further amplification. For convenience, we use the term “T3-iPSCs” to denote the iPSCs generated from this protocol. A total of more than 30 T3-iPSC lines were obtained.
Figure 1. T3 enhances the formation of alkaline phosphatase and TRA-1-60 positive colonies.
A: Alkaline phosphatase (AP) staining of reprogrammed cells; B: Quantitative analysis of AP-positive colonies; C: Quantitative analysis of TRA-1-60 positive colonies. D: Immunostaining of TRA-1-60 positive colonies; HDF: human dermal fibroblasts; UCMSC: umbilical cord mesenchymal stem cells; OSKM: Oct4-Sox2-Klf4-c-Myc retroviruses.
3.2. Activation of metabolic remodeling by T3 treatment
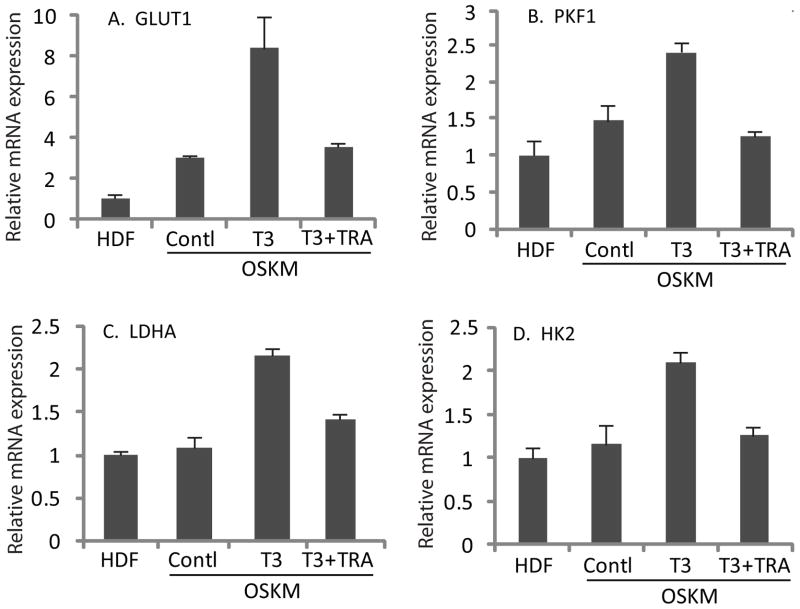
We investigated potential mechanisms underlying the T3-mediated potentiation of iPSC induction. We first examined whether T3 altered metabolic parameters during retrovirus-induced reprogramming. T3 was added to the medium immediately after retrovirus infection for a period of 3 days. On day 4, the treated cells were harvested for quantitative Real-time PCR (Q-PCR). Retroviral transfection of the four iPSC-inducing factors upregulated two genes involved in glucose metabolism, GLUT1 and PFK1. The abundance of mRNA for two key glycolytic enzymes, LDHA and HK2, did not change. However, in the presence of T3, all four glycolytic genes, including GLUT1, PFK1, LDHA and HK2, were significantly upregulated (Fig.2).
Figure 2. Activation of key glycolytic genes by T3-mediated metabolic remodeling.
Following OSKM-retroviral infection, cells were treated with T3 for 3 days and were harvested on day 4 for the expression of GLUT1 (A), PKF1 (B), LDHA (C) and HK2 (D) by Real-time PCR.
To confirm the role of T3 in regulating glucose homeostasis, we incubated a T3-beta nuclear receptor antagonist 1-850 (TRA) [38] together with T3. TRA co-treatment blocked the stimulatory effect of T3 on expression of these genes (Fig.2). Taken together, these data indicated that T3 altered the metabolism of four-factor-transfected cells by increasing their glycolytic status.
3.3. Metabolic remodeling by T3-activated PI3K/AKT signal pathway
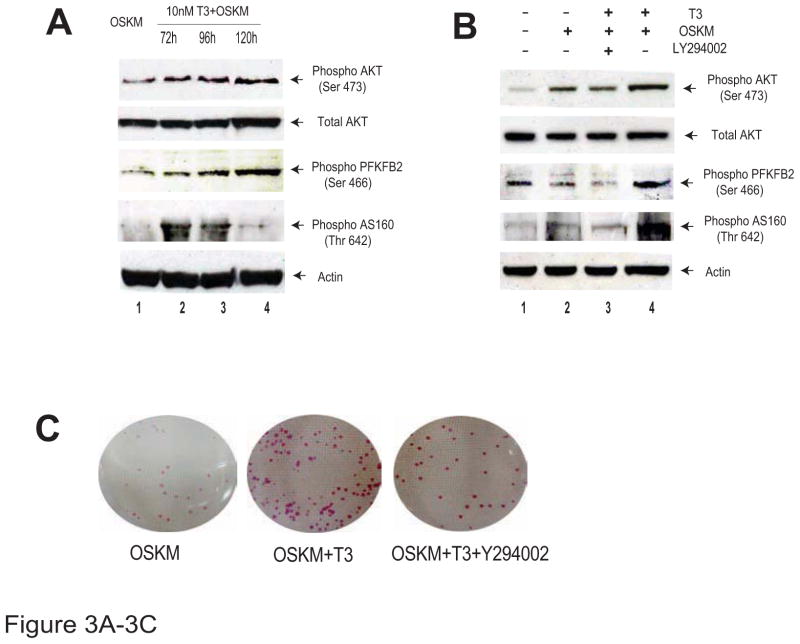
We further examined the molecular mechanisms underlying the activation of glycolysis by thyroid hormone during cell reprogramming. We collected the OSKM/T3-treated cells and examined signal pathways that may participate in glycolytic metabolism. Interestingly, we found that the AKT pathway was significantly activated in a time-dependent manner in parallel with the upregulation of glycolysis (Fig.3A, top panel). The AKT downstream target PFBFB2 was activated by phosphorylation at serine 466 (Fig.3A, panel 3). AS160, another downstream target of the PI3K/AKT pathway, was also activated by phosphorylation at threonine 642 (Fig.3A, panel 4). However, this activation gradually declined with time, suggesting that stimulation of the AS160 pathway was a relatively early event in the activation of glycolysis.
Figure 3. T3 activates metabolic remodeling through the PI3K/AKT signal transduction pathway.
A. Measurement of the PI3K/AKT pathway components by Western blot. Ser 473: phosphorylation at AKT serine 473; Ser 466: phosphorylation at PFKFB2 serine 466; Thr 642: phosphorylation at AS160 threonine 642.
B. Inhibition of T3-mediated PI3K/AKT pathway activation by the PI3K inhibitor LY294002.
C. Inhibition of the PI3k/AKT pathway reduces the formation of the AP-positive iPSC colonies
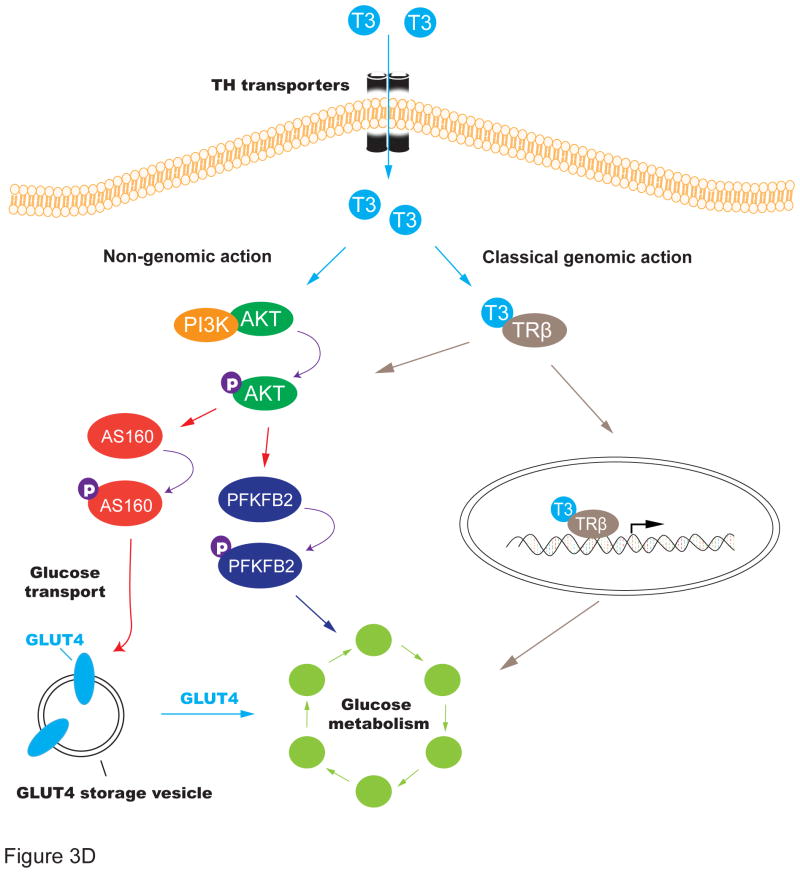
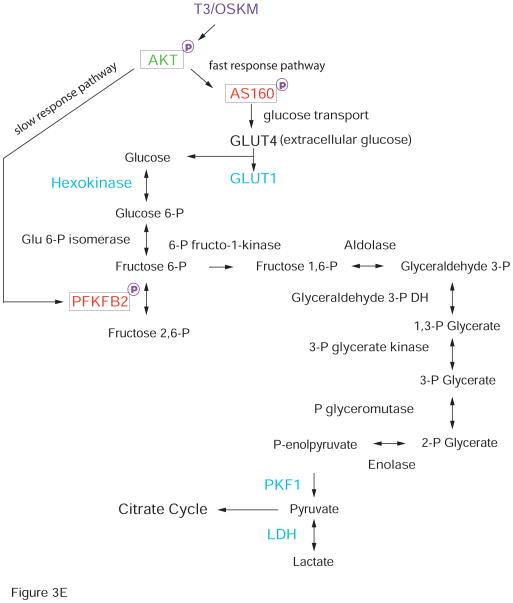
D. Schematic diagram of the T3-mediated activation of the PI3K/AKT pathway
E. Upregulation of the glycolysis pathway by the T3/OSKM treatment.
We further examined the role of the PI3K/AKT pathway by applying LY294002, a specific inhibitor of PI3 kinase (PI3K) [39]. We found that incubation with LY294002 almost completely blocked the T3-mediated activation of AKT and its two downstream targets PFKFB2 and AS160 (Fig.3B). Similarly, treatment of the PI3K inhibitor also abolished the T3-mediated increase in the number of iPSC colonies as measured by AP colony staining (Fig.3C). Together, these data suggest the critical role of the PI3k/AKT pathway in T3-meidated activation of glycolysis during cell reprogramming (Figs. 3D, 3E).
3.4. Promotion of mesenchymal to epithelial transition by metabolic remodeling
Activation of glycolytic metabolism is often associated with plastic transition between epithelium and mesenchyme. It has been suggested that the mesenchymal to epithelial transition (MET) is a critical early event during the reprogramming of mouse somatic cells [40–41]. The upregulation of epithelial related genes E-cadherin and CLDN3 and the downregulation of the mesenchymal gene Snail were considered to be markers of the MET process. We therefore investigated whether T3-mediated metabolic remodeling had any positive effect on MET during cell reprogramming in this OSKM-T3 model.
We used 293T cells as a positive epithelial cell control and uninfected HDFs as a negative control. Following the expression of the four factors, up-regulation of E-cadherin and CLDN3 become evident on post-infection day 6 in comparison to the uninfected control. E-Cadherin and CLDN3 expression was further increased after T3 administration (Fig.4A), and, the expression of Snail1 and Snail2 decreased immediately after reprogramming (Figs. 4B, 4C). Further reduction of Snail1 and Snail2 was observed when T3 treatment was combined with retroviral infection. The regulatory effect of T3 on MET-related genes was significantly inhibited after the addition of a T3 receptor antagonist (Figs. 4A–4C).
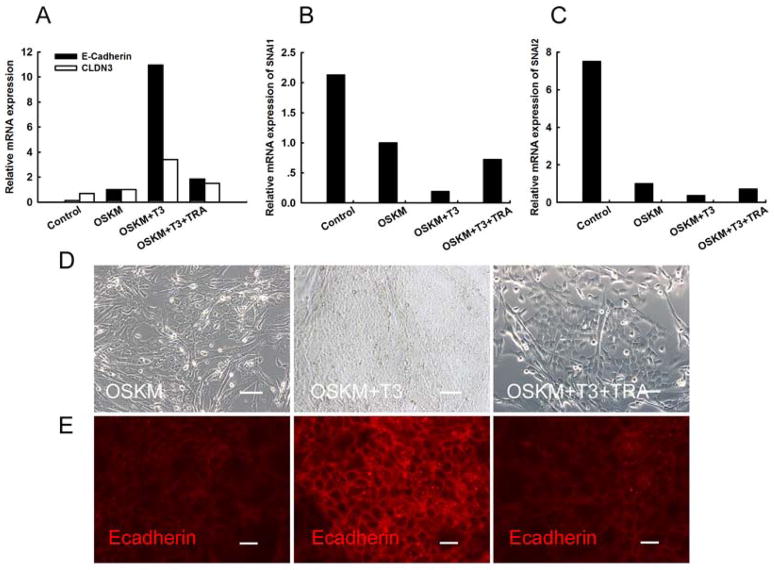
Figure 4. Metabolic remodeling by T3 promotes mesenchymal-to-epithelial transition.
A. Q-PCR quantitation of two epithelial markers, E-cadherin and CLDN3. Addition of T3 receptor antagonist TRA reduces the activation of E-cadherin and CLDN3 by T3 treatment. 293 cells were used as the positive control.
B–C. Down-regulation of Snail1 and Snail2, two mediators of the epithelial-to-mesenchymal transition.
D. Morphological changes of the reprogrammed cells in the presence or absence of T3.
E. Immunostaining of E-cadherin expression in reprogrammed cells in the presence or absence of T3. Note that the T3 receptor antagonist TRA abolishes the T3-mediated activation of E-cadherin.
Infected HDFs also showed morphological changes following the addition of T3 in early cell reprogramming (Fig.4D). Upon treatment with T3, the majority of infected HDFs demonstrated the morphology of typical epithelial cells. The effect of T3 was diminished by co-incubation with the thyroid hormone receptor antagonist.
The reprogrammed cells began to express the epithelial marker E-cadherin, but the expression of E-cadherin was remarkably increased with the addition T3 (Fig.4E). These results suggest that the T3-activated metabolic remodeling is capable of promoting MET through the regulation of some key epithelial and mesenchymal genes.
Active glycolytic flux is correlated with increased proliferation of ESC cells. We thus asked if T3-mediated metabolic remodeling would affect the growth of HDF cells as well. We found that the growth rate of HDF was increased after metabolic remodeling by T3. This effect was maximal at a concentration of 10 nM T3. Accelerated cell growth persisted for only 2 to 3 days following the initial application of T3. After 72 hours, the growth-promoting effect of T3 became less obvious as cells became confluent (Figs.S1A–S1C).
3.5. Expression of pluripotency markers in T3-iPSCs
We then performed a series of assays, including gene expression and DNA methylation analyses, to verify that the T3-iPSC lines obtained were fully reprogrammed and behaved like authentic human iPS cells. The T3-iPSCs generated in this study were morphologically similar to human ES cells and were capable of self-renewal.
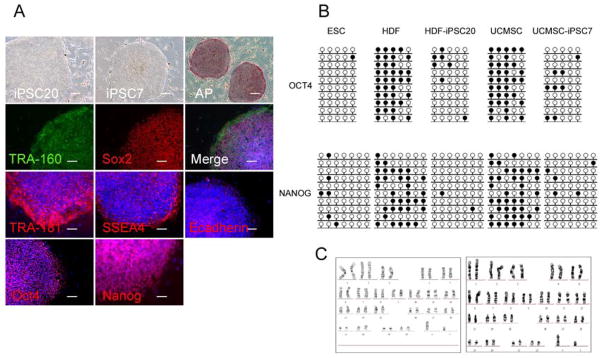
Immunofluorescence analysis confirmed that T3-iPSCs expressed hESC surface markers including TRA-1-60, TRA-1-81, SSEA4, and E-cadherin, as well as nuclear markers such as Oct4, Sox2 and Nanog (Fig.5A). Bisulfite sequencing revealed that the promoter regions of Oct4, Sox2 and Nanog in HDF- and UCMSC-derived T3-iPSC cells became demethylated (Fig.5B), suggesting the reactivation of these pluripotency genes. T3-iPSCs exhibited a normal karyotype after 26 passages (Fig.5C).
Figure 5. Pluripotency of T3-iPS cells.
A: Expression of pluripotent markers. T3-iPS cells derived from HDFs or UCMSCs form compact ES-like colonies and express all major pluripotency-specific markers, including AP, TRA-1-60, Sox2, TRA-1-81, SSEA4, E-Cadeherin, Oct4 and Nanog.
B: DNA demethylation of Oct4 and Nanog promoters in T3-iPS cells as measured by sodium bisulfite sequencing.
C. Karyotype analysis indicates that the iPS cells exhibit no chromosomal abnormities through reprogramming and frequent passaging
3.6. Differentiation potential of T3-iPSCs
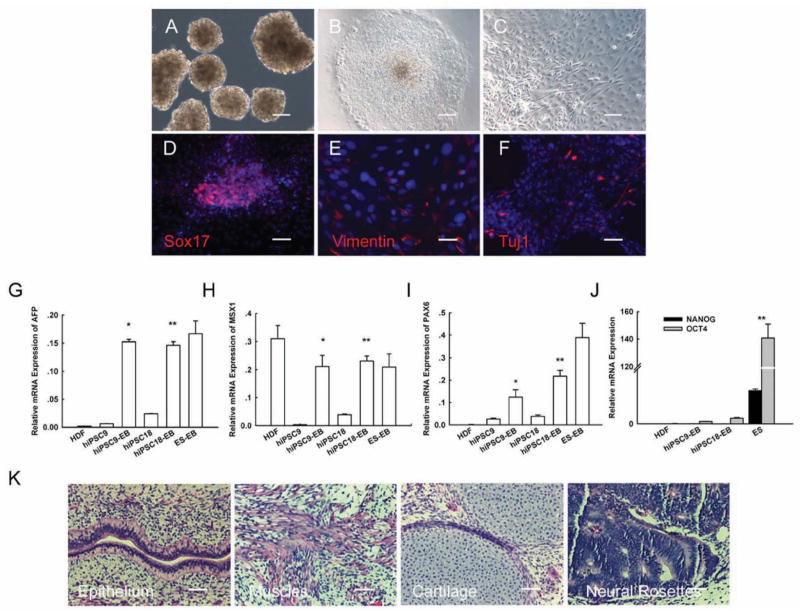
To study the in vitro pluripotency of T3-iPSCs, we generated embryoid bodies (EBs) using the floating cultivation method. T3-iPSCs formed EB-like structures 4–6 days after being cultured in suspension (Fig.6A). EBs were then transferred to FBS-containing medium and were allowed to differentiate spontaneously (Figs. 6B, 3C). Immunofluorescence analyses confirmed that the EBs were able to differentiate into cells of all three germ layers (Figs.6D–6F). Q-RT PCR analysis demonstrated that the expression of markers representing all three germ layers, including AFP (endoderm), MSX1 (mesoderm) and Pax6 (ectoderm), was increased in comparison to the undifferentiated T3-iPSC cells (Figs.6G–6I). At the same time, the expression of pluripotency-related genes was down-regulated (Fig.6J).
Figure 6. Differentiation potential of T3-iPS cells.
A: Formation of embryoid body (EB) from iPS cells at day 8.
B: Adherent culture of EB.
C: Differentiated cells at day 8.
D–F: Immunocytochemical staining of Sox2, Vimentin, and Tuj-1.
G–J: Q-PCR analyses of markers for differentiated three germ layers (AFP, MSX-1, and PAX6), and for pluripotency (Nanog and Oct4).
(K). Three embryonic germ layers of teratoma: epithelium, muscle, cartilage neural rosettes.
The pluripotency of the T3-iPSCs was tested further by in vivo teratoma formation. Cells from seven T3-iPSC lines were injected subcutaneously into immunodeficient (SCID) mice. Tumor formation was observed in all of the injected mice 6–8 weeks post injection. Histological analysis confirmed that T3-iPSCs were able to differentiate into various tissue types of all three germ layers, including epithelium (endoderm), muscle (mesoderm), cartilage (mesoderm) and neural rosettes (ectoderm) (Fig.6K). These results suggest that the T3-iPSCs generated in the present study were pluripotent and had full differentiation potential.
4. DISCUSSION
The low efficiency of the induction of pluripotency in somatic cells, such as fibroblasts, is a major challenge to the translation of laboratory discoveries using iPSCs into patient-tailored regenerative medicine. Moreover, reprogramming is a very stressful process for donor somatic cells. In our early experiments, it was frequently observed that retroviral infection led to signs of senescence, and apoptosis was common among the infected donor cells. Other groups have reported that iPSC induction can be significantly improved by inhibition of apoptosis following the knockdown of tumor suppresser genes (p53, p16) [20–22]. Quite recently, it was found that during the long iPSC induction process, subtle abnormalities occurred at both genetic and epigenetic levels [10–17], thus raising safety concerns over the clinical application of iPSCs.
In this study, we have demonstrated that T3, a natural hormone produced by the thyroid gland, can increase the reprogramming efficiency of the human fibroblast cell line HDF as well as the adult stem cell line UCMSC. Incubating the cells with T3 following the transfection of four iPSC factors significantly boosted reprogramming efficiency (Fig.1). An obvious advantage of this reprogramming regime is that T3 is an endogenous metabolic hormone. Unlike many foreign chemicals or virally delivered factors, T3 treatment should be a very safe approach in generating iPSCs for clinical research.
Most importantly, we found that T3 accelerates iPSC induction through a mechanism fundamentally distinct from those as previously reported for other small molecules used to enhance iPSC production [18–25]. T3 binds to nuclear thyroid hormone receptors, regulating transcription of target genes via the thyroid response elements on the DNA (“genomic action”) [42]. As a catabolic hormone, T3 regulates metabolism by modulating protein synthesis and increasing basal metabolic rate. We have demonstrated that T3 upregulates genes involved in the glycolytic pathway, including GLUT1, PFK1, LDHA, and HK2. Thus, T3 can facilitate the change from mitochondrial oxidative metabolism of somatic cells to anaerobic glycolytic metabolism in support of pluripotent homeostasis. Our results support the concept that metabolic remodeling is a critical early event leading to the T3-accelerated pluripotency. T3 may act as a mediator assisting the essential energetic switch from mitochondrial oxidation to glycolysis, thereby enhancing iPSC induction. In support of the concept of metabolic remodeling, the switch to glycolytic metabolism under mild hypoxic conditions (5%–6% O2) improved the reprogramming efficiency for both mouse and human cells [43]. In addition, highly proliferative cells require the support of elevated glycolytic flux and a high-flux metabolic state from threonine catabolism [44–48].
Increasing evidence indicates that T3 may also mediate some of its biological effects by cell signaling cascades (“non-genomic action”) [49–50]. Phosphatidylinositol-3 kinases (PI3K) constitute a lipid kinase family characterized by their ability to phosphorylate inositol ring 3'-OH groups in inositol phospholipids, initiating the cascade response of the several signaling pathways in response to different stimuli. In this communication, we characterized a T3-dependent signaling cascade leading to upregulation of glycolysis in the OSKM-transduced cells. Following treatment with T3, activated PI3K turns on its downstream target AKT by phosphorylation at serine 473. The active AKT in turns phosphorylates and thus activates two key regulators AS160 and PFKFB2 in glycolysis (Fig.3D). Co-treatment of the PI3K inhibitor LY29402 blocked the activation of the signal pathway (Fig.3B) and the subsequent enhancement of iPSC induction (Fig.3C). Thus, these findings provide new insights into the mechanism underlying T3-potentiated iPSC induction.
Of particular interest is our finding that AS160, a key regulator of glucose transport, responds rapidly to T3. The active form of Thr 642-phophorylated AS160 is significantly upregulated 72 hrs following T3 treatment, but levels soon decline thereafter (Fig.3A). Thus, AS160 may function as an early responding gene of the T3-PI3K-AKT signal pathway, mediating a rapid response to T3 treatment (Fig.3D). These data suggest that in activating the AKT pathway, T3 may accelerate iPSC induction through an alternative non-traditional nongenomic mechanism as previously reported [49–50]. By mobilizing the existing extracellular glucose into the cell, T3 activates the glycolytic metabolism in a rapid manner. Thereafter, the glycolytic activity is then maintained by a second “slow response” pathway through the PFKFB2 enzyme, which converts 6-phospho fructose into 2,6 phospho fructose for glycolysis. This pathway is probably induced through traditional genomic mechanisms of the T3 receptor, although nongenomic actions of T3 cannot be ruled out (Figs. 3D, 3E). Through the tight coordination of these two pathways, T3 activates glycolytic metabolism, providing a novel mechanism for thyroid hormone in enhancing cell pluripotency.
In ESCs, active glycolytic flux is correlated with high levels of proliferation [44]. We also observed that the T3-mediated metabolic remodeling promoted HDF cell growth, providing an advantage for reprogramming of the infected cells. This is particularly true for the stressful process of iPSC selection, during which many virally infected cells undergo apoptosis. In support of this concept, inhibition of apoptosis by blockade of p53 and p16 enhances iPSC induction [20–22].
The mesenchymal-to-epithelial transition (MET) is a critical step in somatic cell reprogramming. Enhanced MET accelerates the reprogramming process. Similarly, the upregulation of E-cadherin, a cell-adhesion-mediating protein, played a key role in promoting MET and improving reprogramming efficiency [40]. T3 has been shown to have a positive regulatory effect on E-cadherin expression in stomach epithelium during metamorphosis [51]. In our experiments, we also noticed that a number of cells assumed a typical epithelial morphology following reprogramming, and this phenomenon became more evident when T3 treatment was combined with the standard retroviral infection (Fig.4D). T3 significantly facilitated MET by upregulating E-cadherin (Figs. 4A, 4E). Our results thus suggest that the MET following T3 metabolic remodeling may also be important in promoting the induction of human iPS cells.
5. CONCLUSION
In summary, the reprogramming process involves complex molecular changes. Numerous studies have been conducted to promote iPSC induction by altering the epigenetic network, using inhibitors of DNA methylation and/or histone acetylation and methylation. In this study, the identification of T3 as a reprogramming enhancer pinpoints for the first time the critical role of metabolic remodeling by endocrine signals in the achievement and maintenance of stemness as well as pluripotency. This finding also sheds light on a new research direction for screening factors and small chemicals to promote iPSC induction, particularly by the activation of the PI3K/AKT signaling pathway.
Supplementary Material
A. Dose-dependent promotion of cell growth by T3. Cell proliferation was measured by WST-1 assay. OSKM-infected cells were treated with various concentrations of T3 for 3 days.
B–C. Time-dependent promotion of cell growth in UCMSC (C) and HDF (D).
Acknowledgments
The authors wish to thank the Guanhui Cui and Jie Qin, the Laboratory of Male Reproductive Medicine and Genetics, Peking University Shenzhen Hospital for tissue specimens. This study was supported by Shenzhen International Collaboration Grant (GJ200807210024A) to X.H.; California Institute of Regenerative Medicine (CIRM) grant (RT2-01942), NIH grant (1R43 CA103553-01), Department of Defense Grant (W81XWH-04-1-0597) to J.F.H., and the Research Service of the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 5.Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci USA. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colman A, Dreesen O. Pluripotent stem cells and disease modeling. Cell Stem Cell. 2009;5:244–247. doi: 10.1016/j.stem.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 11.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 17.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2009;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura T, Mattson MP, Rao MS. Cellular lifespan and senescence signaling in embryonic stem cells. Aging Cell. 2004;3:333–343. doi: 10.1111/j.1474-9728.2004.00134.x. [DOI] [PubMed] [Google Scholar]
- 28.White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 29.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 30.Paus R, Arck P, Tiede S. (Neuro-)endocrinology of epithelial hair follicle stem cells. Mol Cell Endocrinol. 2008;288:38–51. doi: 10.1016/j.mce.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 31.Hu JF, Oruganti H, Vu TH, Hoffman AR. The role of histone acetylation in the allelic expression of the imprinted human insulin-like growth factor II gene. Biochem Biophys Res Commun. 1998;251:403–408. doi: 10.1006/bbrc.1998.9401. [DOI] [PubMed] [Google Scholar]
- 32.Hu JF, Vu TH, Hoffman AR. Genomic deletion of an imprint maintenance element abolishes imprinting of both insulin-like growth factor II and H19. J Biol Chem. 1997;272:20715–20720. doi: 10.1074/jbc.272.33.20715. [DOI] [PubMed] [Google Scholar]
- 33.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex-2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu JF, Vu TH, Hoffman AR. Promoter-specific modulation of insulin-like growth factor II genomic imprinting by inhibitors of DNA methylation. J Biol Chem. 1996;271:18253–18262. doi: 10.1074/jbc.271.30.18253. [DOI] [PubMed] [Google Scholar]
- 35.Chen HL, Li T, Qiu XW, Wu J, Ling JQ, Sun ZH, et al. Correction of aberrant imprinting of IGF2 in human tumors by nuclear transfer-induced epigenetic reprogramming. EMBO J. 2006;25:5329–5338. doi: 10.1038/sj.emboj.7601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu XQ, Pan XH, Wang W, Chen Q, Pang RQ, Cai XM, et al. Transient in vitro epigenetic reprogramming of skin fibroblasts into multipotent cells. Biomaterials. 2009;31:2779–2787. doi: 10.1016/j.biomaterials.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Niu B, Hu JF, Wang H, Ling J, Qian G, et al. Interruption of intrachromosomal looping by CTCF decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J Cell Biol. 2011;193:475–487. doi: 10.1083/jcb.201101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schapira M, Raaka BM, Das S, Fan L, Totrov M, Zhou Z, et al. Discovery of diverse thyroid hormone receptor antagonists by high-throughput docking. Proc Natl Acad Sci U S A. 2003;100:7354–7359. doi: 10.1073/pnas.1131854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 40.Chen T, Yuan D, Wei B, Jiang J, Kang J, Ling K, et al. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Bhargava M, Lei J, Ingbar DH. Nongenomic actions of L-thyroxine and 3,5,3'-triiodo-L-thyronine. Focus on “L-Thyroxine vs. 3,5,3'-triiodo-L-thyronine and cell proliferation: activation of mitogen-activated protein kinase and phosphatidylinositol 3-kinase”. Am J Physiol Cell Physiol. 2009;296:C977–979. doi: 10.1152/ajpcell.00116.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, et al. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 2009;5:140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 47.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis PJ, Leonard JL, Davis FB. Mechanisms of nongenomic actions of thyroid hormone. Front Neuroendocrinol. 2008;29:211–218. doi: 10.1016/j.yfrne.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Cao X, Kambe F, Yamauchi M, Seo H. Thyroid-hormone-dependent activation of the phosphoinositide 3-kinase/Akt cascade requires Src and enhances neuronal survival. Biochem J. 2009;424:201–209. doi: 10.1042/BJ20090643. [DOI] [PubMed] [Google Scholar]
- 51.Izaguirre MF, Casco VH. T3 regulates E-cadherin, and beta- and alpha-catenin expression in the stomach during the metamorphosis of the toad Rhinella arenarum. Biotech Histochem. 2010;85:305–323. doi: 10.3109/10520290903236005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Dose-dependent promotion of cell growth by T3. Cell proliferation was measured by WST-1 assay. OSKM-infected cells were treated with various concentrations of T3 for 3 days.
B–C. Time-dependent promotion of cell growth in UCMSC (C) and HDF (D).