Abstract
The Drosophila Zelda transcription factor plays an important role in regulating transcription at the embryonic maternal-to-zygotic transition. However, expression of zelda continues throughout embryogenesis in cells including the developing CNS and trachea, but little is known about its post-blastoderm functions. In this paper, it is shown that zelda directly controls CNS midline and tracheal expression of the link (CG13333) gene, as well as link blastoderm expression. The link gene contains a 5’ enhancer with multiple Zelda TAGteam binding sites that in vivo mutational studies show are required for link transcription. The link enhancer also has a binding site for the Single-minded: Tango and Trachealess:Tango bHLH-PAS proteins that also influences link midline and tracheal expression. These results provide an example of how a transcription factor (Single-minded or Trachealess) can interact with distinct co-regulatory proteins (Zelda or Sox/POU-homeodomain proteins) to control a similar pattern of expression of different target genes in a mechanistically different manner. While zelda and single-minded midline expression is well-conserved in Drosophila, midline expression of link is not well-conserved. Phylogenetic analysis of link expression suggests that ~60 million years ago, midline expression was nearly or completely absent, and first appeared in the melanogaster group (including D. melanogaster, D. yakuba, and D. erecta) >13 million years ago. The differences in expression are due, in part, to sequence polymorphisms in the link enhancer and likely due to altered binding of multiple transcription factors. Less than 6 million years ago, a second change occurred that resulted in high levels of expression in D. melanogaster. This change may be due to alterations in a putative Zelda binding site. Within the CNS, the zelda gene is alternatively spliced beginning at mid-embryogenesis into transcripts that encode a Zelda isoform missing three zinc fingers from the DNA binding domain. This may result in a protein with altered, possibly non-functional, DNA-binding properties. In summary, Zelda collaborates with bHLH-PAS proteins to directly regulate midline and tracheal expression of an evolutionary dynamic enhancer in the post-blastoderm embryo.
Keywords: CG13333, CNS midline, Drosophila, link, single-minded, zelda
Introduction
The Drosophila zelda (zld or vielfaltig) gene plays an important role in regulating expression of a battery of genes in the blastoderm embryo that control the maternal-to-zygotic transition (Liang et al., 2008). zld encodes a zinc finger transcription factor that can act as transcriptional activator, binding to a set of sequences referred to as TAGteam sites (De Renzis et al., 2007; Liang et al., 2008; ten Bosch et al., 2006). Whole-genome analysis of Zld binding using ChIP-Seq revealed that thousands of these sites are bound by Zld in vivo (Harrison et al., 2011; Nien et al., 2011). It has also been proposed that Zld acts to increase chromatin accessibility for zygotically-expressed transcription factors to bind to its target genes and drive early developmental programs (Harrison et al., 2011). zld is also extensively expressed in the post-blastoderm embryo in the CNS and other cell types (Liang et al., 2008; Staudt et al., 2006). However, its role in controlling post-blastoderm gene expression and development has not been explored. In this paper, we demonstrate that zld activates transcription of CNS midline cell and tracheal expression.
The Drosophila CNS contains a specialized set of neurons and glia that reside at the midline (Wheeler et al., 2006). The single-minded (sim) gene acts as master regulator of CNS midline cell transcription and development (Nambu et al., 1991), and encodes a bHLH-PAS transcription factor that forms a heterodimer with the Tango (Tgo) bHLH-PAS protein (Sonnenfeld et al., 1997). The Sim:Tgo complex activates the transcription of midline-expressed target genes by binding the sequence ACGTG, referred to as a CNS midline element (CME) (Wharton et al., 1994). Midline primordium cells divide and differentiate into midline neurons and two populations of midline glia (MG): anterior midline glia (AMG) and posterior MG (PMG) (Wheeler et al., 2006). Not only does Sim:Tgo control midline primordium formation, but later it interacts with the Ventral veins lacking (Vvl) POU-homeobox protein and Dichaete (D) Sox proteins to control MG transcription (Ma et al., 2000; Sanchez-Soriano and Russell, 1998). Akin to the role of Sim as master regulator of midline transcription, the Trachealess (Trh) bHLH-PAS protein also forms a complex with Tgo and Vvl, binds CMEs on target genes, and acts as a master regulator of tracheal development (Isaac and Andrew, 1996; Sonnenfeld et al., 1997; Wilk et al., 1996; Zelzer and Shilo, 2000). Here, we propose that Sim and Trh collaborate with Zld to control CNS midline and tracheal expression of the link (CG13333) gene.
Increasingly, research on the mechanisms that underpin organismal and evolutionary variation is demonstrating that changes in gene expression commonly play important roles in evolution. Much of this variation is dependent on changes in enhancer sequences, although species differences in regulatory protein function can also be a factor (Gordon and Ruvinsky, 2012). While only beginning to be explored, recent data indicate that expression differences may be common in nervous system-expressed genes (Rebeiz et al., 2011). CNS midline cell gene expression has been particularly well-studied in D. melanogaster (Kearney et al., 2004; Wheeler et al., 2006; Wheeler et al., 2009), and represents a useful system for evolutionary study. In this paper we demonstrate how insights into midline gene regulation and evolution of cis-control regions can be mechanistically achieved.
In the studies described below, we describe a novel role for the Zld transcription factor in regulating post-blastoderm CNS midline cell and tracheal transcription. Zld protein directly activates transcription of the midline and tracheal-expressed link gene, interacting with Sim:Tgo to activate link midline expression and with Trh:Tgo to activate tracheal expression. While zld expression is highly conserved among Drosophila species, link midline expression is present only in species closely-related to D. melanogaster. We propose a two-step model in which binding sites in the link enhancer that promote high expression in MG arise in the lineage leading to D. melanogaster. Finally, we demonstrate that alternatively-spliced forms of zld are generated during embryogenesis, with variants expressed early in development generating a protein with 6 zinc fingers, while a CNS-specific variant encodes proteins lacking the 3 C-terminal zinc fingers, most likely generating a protein with altered or non-functional DNA-binding capabilities.
Materials and methods
Drosophila strains and genetics
The zld mutants, zld681 and vflG0427, were obtained from Christine Rushlow and Gerd Vorbrüggen, respectively (Liang et al., 2008; Staudt et al., 2006). Low levels of zld transcript can be detected by in situ hybridization of zld681 hemizygotes, indicating that this allele may be a strong hypomorph. The Df(1)Exel6253 and Df(1)BSC872 stocks (both deleted for zld) and grainyhead mutant null strain (grhIM) were obtained from the Bloomington Drosophila Stock Center. These mutants were maintained over either P[ftz-lacZ] or P[twi-Gal4] P[UAS-GFP] balancer chromosomes. Homozygous and hemizygous mutant embryos were detected by staining for either: (1) lacZ or GFP expression from balancer chromosomes, (2) zld transcript, or (3) Zld protein. D. simulans, D. sechellia, D. mauritiana, D. erecta, D. yakuba, D. ananassae, D. parabipectinata stocks were obtained from Corbin Jones. The D. pseudoobscura stock was obtained from Karin Pfennig. D. willistoni and D. virilis were obtained from the Drosophila Species Stock Center (La Jolla, CA).
Bioinformatics
Orthologous Drosophila sequences corresponding to the link-5’ fragment were retrieved from the UCSC Genome Browser (genome.ucsc.edu), converted to FastA format using Galaxy (main.g2.bx.psu.edu), aligned using Dialign-TX (Subramanian et al., 2008), and manually adjusted using BioEdit (Hall, 1999). Motif T sites were identified using PhyloGibbs (Siddharthan et al., 2005) and WinDotter (Sonnhammer and Durbin, 1995). Sim and Zld consensus binding sites were annotated using GenePalette (Rebeiz and Posakony, 2004).
Initial predicted gene structures of the zld RA, RB, RC, and RD transcripts were obtained from FlyBase. Transcription start, stop, and splice sites were determined by analysis of ModENCODE RNA-Seq data. Protein domains were predicted using InterProScan. Orthologs of link (CG13333) and zld were identified by reciprocal Protein BLAST searches, and were aligned with Dialign-TX using the STRAP program (Gille and Frommel, 2001).
Analysis of RNA-Seq data to obtain the fraction of zld splice variants was performed on ModENCODE developmental time-course Unique Mapper tracks (Graveley et al., 2011). All reads that overlap ChrX:19672268–19672269 (spanning the 5' splice site) were downloaded in SAM format from the ModENCODE website. Each read was then categorized as "spliced" or "unspliced" based on CIGAR annotation (Li et al., 2009), and the total reads in each category were normalized to the total number of unique reads in the track.
link enhancer cloning and germline transformation
The 285 bp region between Roe1 and link (referred to as link-5’) and the 1197 bp region between link and CG13334 (link-3’) were PCR-amplified from w1118 flies and cloned into the Gateway entry vector pENTR (Invitrogen). Binding site mutants were generated by PCR site-directed mutagenesis and cloned into pCR8 (Invitrogen). Sequences were mutated (underlined residues) as follows: T1 (CAGGTAG > CAAAAAG), T2 (TAGGTGG > TAAAAGG), T3 (CAGGTAG > CAAAAAG), T4 (GAGGTAG > GAAAAAG), and CME (AACGTG > GGATCC). All primer sequences are listed in Table S1. link-5’, link-3’, and link-5’ variants were cloned into pMintgate (Jiang et al., 2010) using Gateway LR Clonase II (Invitrogen). pMintgate constructs were injected into Drosophila embryos that contain the phiC31 destination site attP2 (68A1–B2) (Groth et al., 2004) and possess posteriorly-localized phiC31 integrase.
In situ hybridization and immunostaining
Embryo collection, in situ hybridization, and immunostaining were performed as previously described (Kearney et al., 2004). DGC cDNA clones LD47819 (zld), LD15563 (link), and LP11035 (grh) were used to generate in situ hybridization probes. The D. melanogaster zld α and β probes were amplified from w1118 genomic DNA and cloned into pCR2.1 (Invitrogen). The coding region of EGFP was used to make the GFP probe. The following primary antibodies were used for immunostaining: rabbit anti-GFP (Abcam), mouse anti-Engrailed MAb 4D9 (DSHB) (Patel et al., 1989), guinea pig anti-Sim (Ward et al., 1998), mouse anti-β-galactosidase (Promega), and rat anti-Zld (Chris Rushlow). Alexa Fluor-conjugated secondary antibodies (Invitrogen) were used except for Sim, which was detected using biotinylated goat anti-guinea pig (Vector Laboratories) with streptavidin-HRP (Jackson Laboratories) and tyramide signal amplification (TSA; Perkin Elmer). Fluorescent in situ hybridization was detected using TSA. For in situ hybridization of Drosophila species other than D. melanogaster, orthologous regions were amplified from genomic DNA using degenerate primers and cloned into pCR2.1 or pCR8. Digoxigenin-labeled RNA antisense probes were generated to detect zld and link expression in these species. Confocal image stacks were viewed and processed using ImageJ (Abramoff et al., 2004).
Results
Post-blastoderm expression of zelda in the CNS midline and tracheal cells
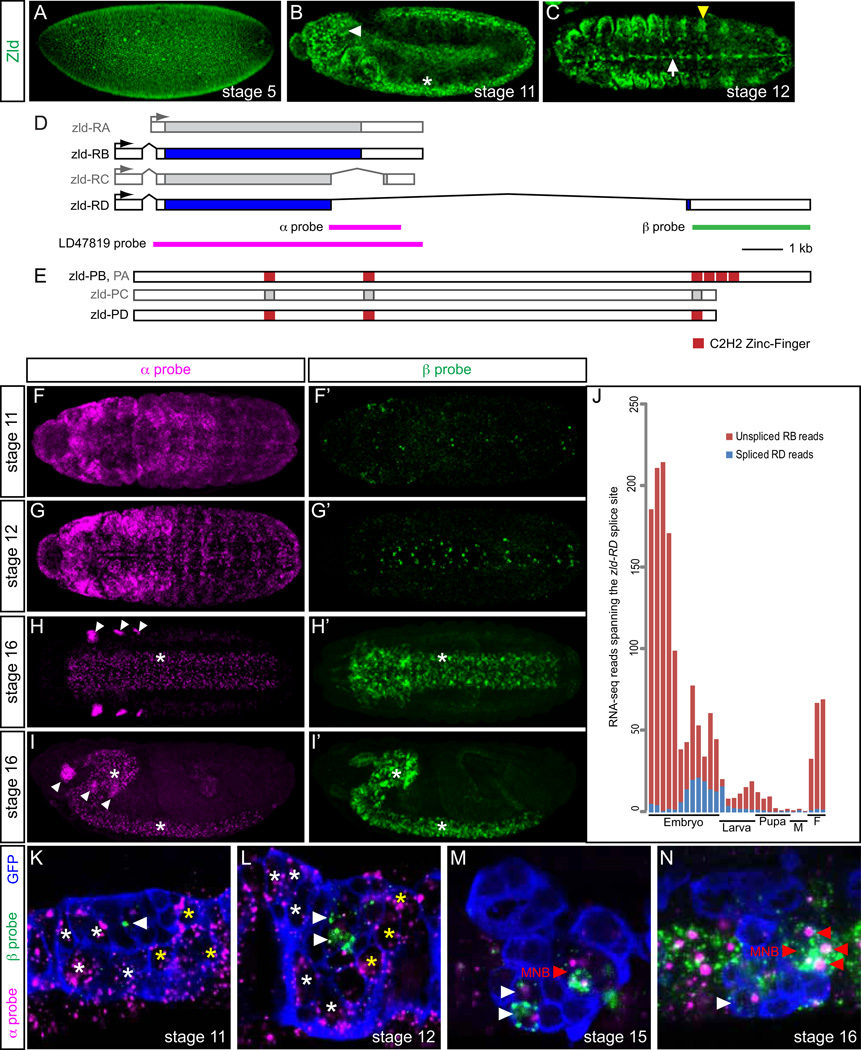
It was previously demonstrated that zld is broadly expressed in the blastoderm (Fig. 1A) and in the developing CNS, including ventral nerve cord (VNC) and brain (Fig. 1B) (Liang et al., 2008; Staudt et al., 2006). To begin investigating potential functions of zld in CNS development, we stained embryos for Zld and noticed strong Zld presence in the CNS midline cells (Fig. 1C). Protein was also detected in ectodermal cells that include the trachea (Fig. 1C). Because of the diversity of midline neuronal and glial cell types, the CNS midline cells are an attractive system to study neural development (Wheeler et al., 2006), so we examined zld RNA expression from stages 11–16 (Fig. S1). At stages 11–13, zld is strongly expressed in AMG, PMG, the median neuroblast (MNB), and iVUM4 with low levels in mVUM4. By stage 14, zld expression is nearly absent in AMG and PMG, but persists in iVUM4 and MNB – this pattern of expression continues at least through stage 17. Midline expression of zld was confirmed by RNA-Seq analysis of purified midline cells, with high levels at stage 11 (122.454 FPKM) and stage 16 (194.927 FPKM) (Joe Fontana and Stephen Crews, pers. comm.). In summary, post-blastoderm expression of zld includes the tracheal primordium, CNS MG, and a subset of midline neurons.
Fig. 1.
zld expression and alternative splicing is dynamic. (A, B) Sagittal views of zld681/FM7c heterozygote embryos stained for Zld protein (green). Anterior left, dorsal up. (A) Zld is broadly present at stage 5. (B) Stage 11 embryo showing Zld in the brain (white arrowhead) and CNS (*). (C) Horizontal view of stage 12 embryo showing Zld in the CNS midline cells (arrow) and trachea (yellow arrowhead). (D) Schematic of zld transcripts. Four alternatively-spliced versions of zld (RA, RB, RC, RD) are shown, as annotated on FlyBase (www.flybase.org). Boxed regions indicate exons, and filled regions are coding sequences. The zld RA and RC transcripts are shaded gray, since they represent rare occurrences or cloning artifacts. The positions of the α, β, and LD47819 cDNA in situ hybridization probes used to detect different zld transcripts are shown. (E) Shown are the 4 predicted Zld proteins (PA, PB, PC, PD). Predicted InterProScan protein domains include 6 zinc finger domains. Three of the four C-terminal zinc-finger domains are absent in PD. The zld-PA form is identical to PB, but is shaded gray since its transcript is rare. The zld-PC protein isoform is shaded gray due to its rarity. (F–I) Maximum projections of zld RNA expression, detected by the zld α probe, showing widespread ectodermal expression at stages 11–12, CNS and brain expression (*) at stage 16, and imaginal disc primordia (arrowheads) at stage 16. The CNS and brain expression resemble nuclear dots, consistent with α probe detection of pre-spliced nuclear RNA corresponding to RD. In contrast, the imaginal disc expression appears to be cytoplasmic, likely corresponding to RB. (F'–I’) Same embryos as (F–I), but detecting zld-RD expression using the zld β probe. Only CNS and brain expression is detected (*), and its non-punctate appearance indicates it is detecting processed mRNA of the RD transcript. (J) RNA-Seq developmental timecourse (ModENCODE) of the abundance of unspliced (RB) and spliced (RD) reads spanning the RD-specific 5' splice site. (K–N) Single midline segments of sim-Gal4 UAS-tau-GFP embryos, detecting both α probe (magenta) and β probes (green). Transcripts in AMG (white *), PMG (yellow *), and midline neurons (arrowheads) are indicated. Midline cells are outlined in blue. (K) zld expression at stage 11 is almost exclusively RB, and is present in AMG and PMG, except for a single midline neuron expressing low levels of RD (arrowhead). (L) During stage 12, both AMG and PMG continue to express RB, while two midline neurons express RD. (M, N) At stages 15 and 16, all zld-expressing cells express RD, including iVUM4, mVUM4 (white arrowheads; only iVUM4 is shown in N), and MNB progeny (red arrowheads; 3 cells are shown in N with a nuclear dot). The large nuclear transcription dots corresponding to the α probe (magenta) co-localize in the same cells with the cytoplasmic β probe (green) staining, consistent with the zld gene producing RD transcripts.
Alternative splicing generates CNS-specific zelda transcripts encoding a protein lacking most of the DNA binding domain
FlyBase (McQuilton et al., 2012) lists 4 different zld gene transcripts (RA, RB, RC, RD) (Fig. 1D) that encode 3 distinct proteins (PA, PB, PC, PD with PA and PB being identical, and PC and PD being nearly identical) (Fig. 1E). Analysis of modENCODE RNA-Seq data (Graveley et al., 2011) provided evidence for only two transcripts, RB and RD. In contrast, there are only single cDNA clones listed in FlyBase corresponding to RA and RC. Consequently, we will refer only to RB and RD and PB and PD, and assume that RA and RC are rare transcripts or cloning artifacts. Most noteworthy is that PD lacks 3 of the 4 Zld C-terminal C2H2 zinc fingers (Fig. 1E) that are sufficient to bind TAGteam sites (Liang et al., 2008). This leaves 3 other C2H2 zinc fingers that are dispersed throughout the protein. Thus, PD may carry-out a biochemical function distinct from PB with respect to target gene transcription. The probe to zld cDNA clone LD47819 used in Fig. S1 detects both the RB and RD transcripts. To investigate which zld transcripts (and proteins) were present in the post-blastoderm embryo and CNS, we generated and analyzed two probes that can recognize zld splice variants (Fig. 1D). The α probe detects both zld mRNAs: as an exonic probe for RB and an intronic probe for RD, reflecting the alternative splicing they undergo. The β probe detects only the RD mRNA transcript.
Detection of zld RNA with the α probe revealed strong expression at stages 11–16 (Fig. 1F–I) that resembled Zld antibody staining (Fig. 1A–C) and hybridization to the long LD47819 cDNA probe (Fig. S1). The α probe detected RNA in the developing epidermis, CNS, brain, and imaginal disc primordia. However, at stages 14–16 the CNS staining appeared punctate (*, Fig. 1H–I), resembling hybridization to unspliced primary RNA (Kosman et al., 2004), a result expected if zld RD is present instead of RB (Fig. 1D). In contrast, the imaginal disc staining resembled spliced mRNA transcripts, similar to the staining in all cell types at earlier stages (arrowheads, Fig. 1H–I). Confirming this interpretation, hybridization to the RD-specific β probe detected low expression in the ventral ectoderm starting at stage 11 (Fig. 1F’), but showed robust CNS expression from stages 12–16 (Fig. 1G’–I’). The RD transcript was not detected in the imaginal disc cells. Thus, zld transcripts at stages 11–12 and in imaginal disc cells at later stages are primarily the RB form, which encodes the PB protein isoform with 6 C2H2 zinc fingers. In contrast, in the CNS, the RB transcripts are reduced, but instead, the RD splice variant is present; these transcripts encode a Zld protein that lacks 3 of the zinc fingers. Analysis of modENCODE developmental timecourse RNA-Seq data (Graveley et al., 2011) are consistent with these observations, in which zld transcripts with the 3’-end unspliced (RB) predominate early, while zld transcripts with 3’ splicing (RD) appear in high numbers in mid-embryogenesis and later (Fig. 1J). The 3’ unspliced transcripts present during late embryogenesis are likely due to imaginal disc expression.
We also analyzed the occurrence of the two zld transcripts at high resolution in CNS midline cells. As described earlier, using the full-length zld probe (LD47819) revealed expression in MG from stages 11–13 and in VUM4 neurons and the MNB from stages 11–16 (Fig. S1). Analysis of zld RNA with the α probe revealed strong expression in MG at stages 11–13 (Fig. 1K, L), but not later (Fig. 1M, N). Expression in VUM4 neurons and the MNB (Wheeler et al., 2006) were present from stages 11–16, although at stages 15–16, the α probe-hybridizing RNA was present as nuclear dots in the midline cells, indicative of RD transcripts (Fig. 1M, N). Consistent with this view, β probe-hybridizing transcripts corresponding to RD were present in VUM4 neurons and MNB progeny at stages 12–16 (Fig. 1M, N). In summary, during stages 11–13, Zld protein with 6 zinc fingers is present broadly in the epidermis and CNS, including MG. After stage 13, the 6 zinc finger Zld isoform is absent or greatly reduced in all cell types, except the imaginal disc primordia. In the CNS, including midline VUM4 and the MNB, Zld protein consists of a 3 zinc finger isoform with potentially altered or non-functional DNA-binding properties.
Drosophila post-blastoderm zelda expression and alternative splicing are conserved
To study the conservation of zld expression in different Drosophila species, we utilized the α probe sequence since it contains stretches of high conservation that are sufficiently long to design primers that can amplify orthologous regions in each species tested. Near identical expression of zld was observed in all 5 species tested (D. melanogaster, D. simulans, D. mauritiana, D. erecta, and D. pseudoobscura) that diverged up to 25 MY ago (Fig. S2A–O). In particular, strong midline expression of zld was observed in all species. It was also apparent that at stage 16, the CNS expression reflected the alternatively-spliced RD transcript since the transcripts were nuclear dots. Similarly, modENCODE RNA-Seq data of Drosophila species, including D. mojavensis and D. virilis, include reads spliced at the RD-specific junction. These data indicate that expression of the RD splice variant is conserved throughout the Drosophila genus.
Midline expression of link is dependent on zelda
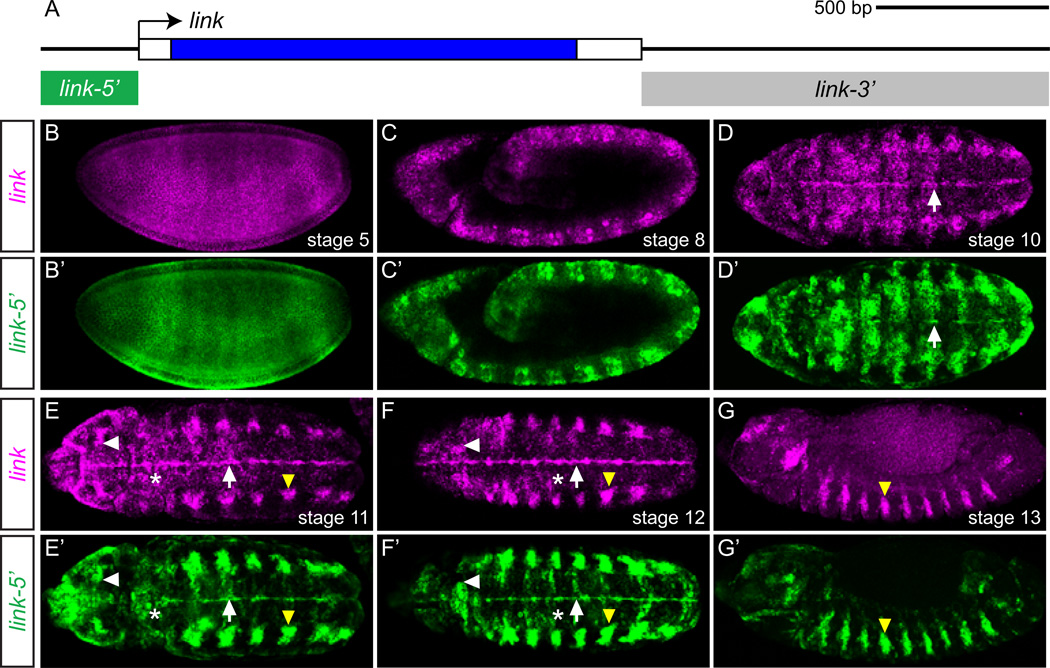
In a separate project to identity and analyze midline enhancers, we studied the expression of the Drosophila CG13333 midline-expressed gene, which we have renamed link (see The Legend of Zelda). The link gene encodes a secreted protein, and is conserved in flies and mosquitoes but is not identifiable in more distant species. The link gene consists of a single exon (Fig. 2A). At stage 5, link is initially expressed ubiquitously (Fig. 2B), but quickly develops anterior-posterior and dorsal-ventral variation, and by stage 8 expression becomes concentrated in the ectoderm in a segmentally-repeated striped pattern (Fig. 2C). At stage 10, expression in the CNS midline cells emerges (Fig. 2D), and by the end of stage 11, expression is apparent in the brain, the tracheal placodes, the lateral CNS, and in the CNS midline primordium cells (Fig. 2E). During stage 12, expression is reduced in the lateral CNS, but continues in the differentiating midline cells (primarily MG; Fig. 5C) and the trachea, as well as in the brain (Fig. 2F). CNS midline and most brain expression ceases by stage 13 (Fig. 2G), while tracheal expression is maintained until stage 15 (not shown).
Fig. 2.
link is expressed in CNS midline cells and a link 5’ 285-bp fragment controls embryonic expression. (A) Schematic of the link locus. Transcribed region is boxed, and coding sequence is filled blue. Arrow indicates 5’-end of the transcription unit. The intergenic regions tested for regulatory function, link-5’ and link-3’, are indicated by boxes below the locus. Scale is indicated above the schematic. (B-G’) Fluorescent in situ hybridization of link-5’ transgenic embryos showing (B–G) endogenous link expression (magenta) compared to (B'–G') GFP expression from the link-5’ transgene in the same embryo. All views are maximum projections, except (B), which shows a single confocal slice. Shown are: (B, B’, C, C', G, G’) sagittal views, and (D, D’, E, E’, F, F') horizontal views. Embryonic stages are indicated in each panel. White arrows indicate midline cells, white arrowheads denote brain cells, yellow arrowheads indicate trachea, and (*) indicates ventral ectoderm.
Fig. 5.
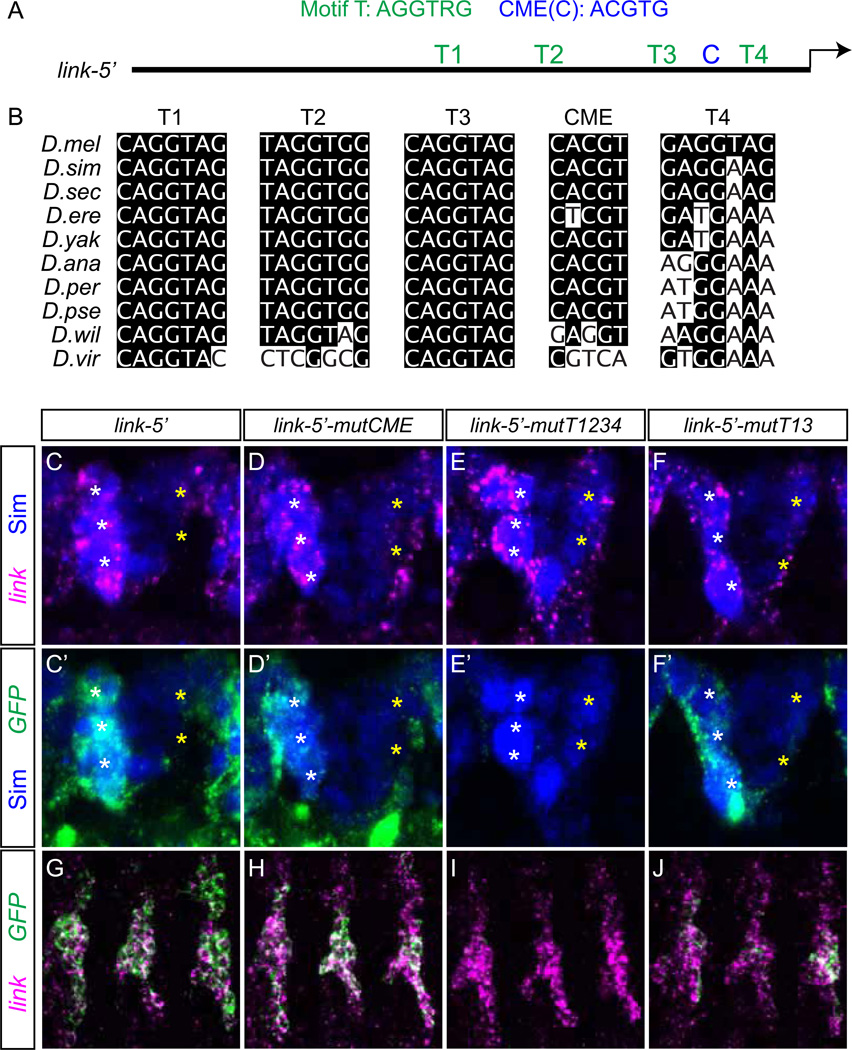
The link Motif-T sites are required for midline and tracheal expression. (A) Schematic of the 285 bp link-5' regulatory region showing the positions of Motif-T (T1–T4) and CME (C) sites. (B) Evolutionary conservation of Motif-T and CME sequences in representative Drosophila species. Nucleotides that match the D. melanogaster (D.mel) sequence are shaded. (C–F) Expression of link is visualized in stage 12 MG, and compared to (C'–F') expression of link-5' transgenes visualized by GFP transcript staining. Sim protein, which is present in MG and a subset of midline neurons, is blue. White * indicate AMG and yellow * indicate PMG. (C, C', D, D’) link-5'-mutCME had a small reduction in MG GFP expression relative to link-5'. (E, E') Mutation of all 4 Motif-T sites (link-5'-mutT1234) resulted in a loss of MG expression. (F, F’) link-5’-mutT13 had a small reduction in MG expression. (G–J) link and GFP expression is visualized in stage 13 tracheal transverse connective branches (3 segments are shown). (G) Co-expression of link and link-5’ in trachea. (H) Mutation of the CME motif caused a small reduction in tracheal expression. (I) Mutation of all Motif-T sites nearly eliminated tracheal GFP expression. (J) Mutation of Motif-T sites 1 and 3 reduced tracheal GFP expression.
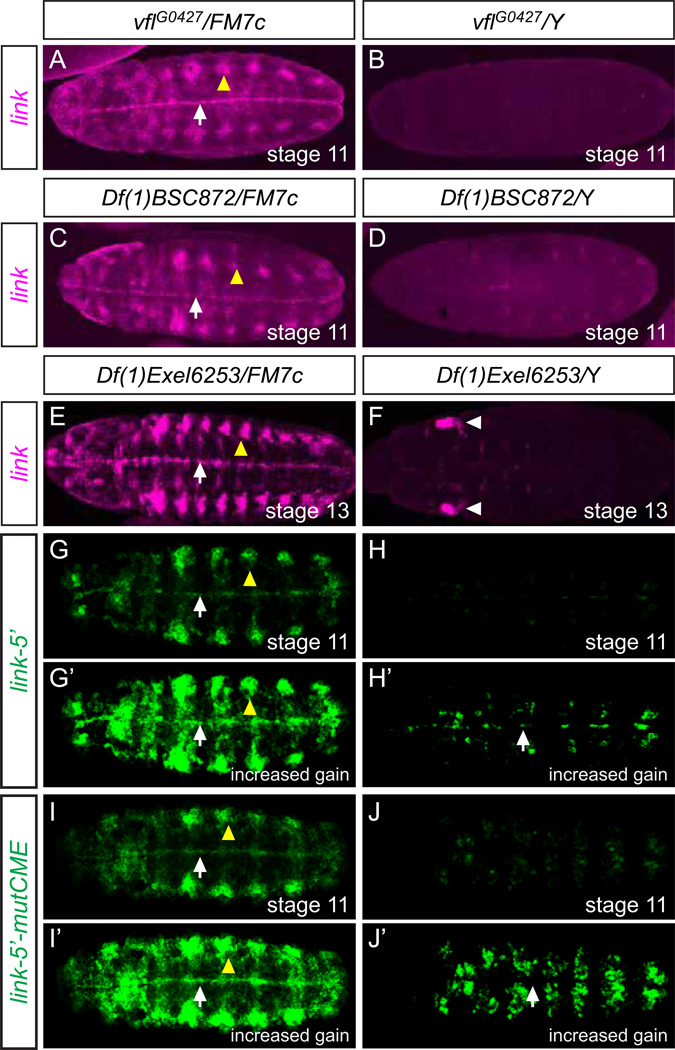
Given the similarity in expression patterns between link and zld, and previous microarray data that early blastoderm (1–2 hr old) link expression is dependent on zld function (Liang et al., 2008), we examined zld mutant embryos for effects on link expression. Our genetic analysis was focused on zld zygotic mutant embryos at embryonic stages 11–13, stages by which maternal zld is likely to be largely depleted. In vflG0427 mutant embryos, link expression was severely reduced (Fig. 3B) compared to wild-type embryos (Fig. 2) and heterozygous (staining control) embryos from the same collection (Fig. 3A). Another hypomorphic zld allele, zld681, also showed a reduction in link expression (data not shown). Consistent with the single-gene mutant results, link midline and tracheal expression was nearly eliminated in two deficiency strains that delete the zld gene, Df(1)BSC872 and Df(1)Exel6253 (Fig. 3C–F). At stage 13, Df(1)Exel6253 (but not vflG0427 or Df(1)BSC872) mutant embryos showed some link expression in the head regions (Fig. 3F) – the reason for this expression is unknown. Only 7 genes, including zld, are deleted in both strains, and zld encodes the only predicted transcription factor. We conclude that zld function is required for embryonic expression of link.
Fig. 3.
zld regulates link embryonic expression. Ventral views, anterior left of maximum projections. Midline cells (white arrows) and tracheal cells (yellow arrowheads) are indicated. (A–F) link expression was assayed in (A) vflG0427/FM7c heterozygous, (B) vflG0427/Y hemizygous, (C) Df(1)BSC872/FM7c heterozygous, (D) Df(1)BSC872/Y hemizygous, (E) Df(1)Exel6253/FM7c heterozygous, and (F) Df(1)Exel6253/Y hemizygous embryos. Heterozygous embryos act as a staining control since they were stained in the same collection as the hemizygous mutant embryos. There is no significant difference in link expression levels between wild-type (Fig. 2) and heterozygous embryos. (B, D, F) With all 3 mutants, midline and tracheal staining was severely reduced. (F) In Df(1)Exel6253 mutant embryos, some head staining (white arrowheads) was detected. (G–H’) link-5’ expression was assayed in Df(1)Exel6253 heterozygous and hemizygous mutant embryos. In G’ and H’, the gain was increased to reveal the weak presence of midline and ventral ectodermal expression and the absence of tracheal expression in H’. (I-J’) link-5’-mutCME was examined in Df(1)Exel6253 heterozygous and hemizygous embryos. Note the complete absence of midline expression, although weak ventral ectoderm expression was present when the gain was increased in J’.
Midline expression of D. melanogaster link is evolutionarily recent
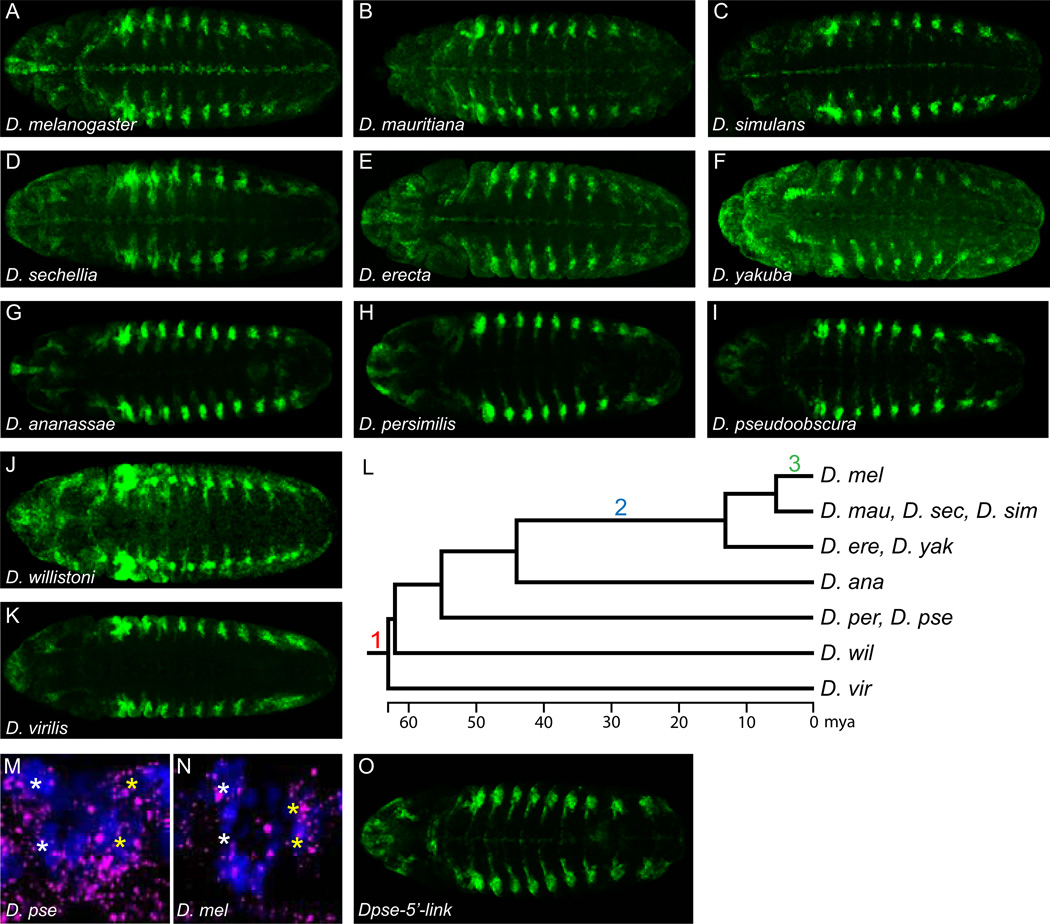
While the embryonic midline expression of zld is well-conserved in our analysis of Drosophila species, the midline expression of link is not well-conserved. Using species-specific link probes, we examined link expression in a number of Drosophila species throughout the Drosophila phylogeny (D. simulans, D. mauritiana, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. persimilis, D. pseudoobscura, D. willistoni, and D. virilis). Broad expression of link in stage 5–10 embryos in these Drosophila species was similar to D. melanogaster (data not shown), but aspects of expression at stage 11 and later differed. Compared to D. melanogaster (Fig. 4A), CNS midline expression was significantly reduced in other members of the melanogaster subgroup consisting of D. mauritiana (Fig. 4B), D. simulans (Fig. 4C) and D. sechellia (Fig. 4D). These three species diverged from D. melanogaster ~5–6 million years ago (mya) (Tamura et al., 2004). Two additional species of the melanogaster subgroup, D. erecta (Fig. 4E) and D. yakuba (Fig. 4F) that diverged from D. melanogaster ~13 mya also showed reduced midline expression. More striking, in 5 more distantly related species (~44–63 MYA divergence) CNS midline expression was nearly or completely absent. These species included: D. ananassae (Fig. 4G), D. persimilis (Fig. 4H), D. pseudoobscura (Fig. 4I), D. willistoni (Fig. 4J), and D. virilis (Fig. 4K). Thus, the high levels of link midline expression observed in D. melanogaster are likely a recently acquired trait that appeared in two steps: appearance of midline expression <44 mya and then upregulation <6 mya exclusively in D. melanogaster (Fig. 4L). In contrast, link tracheal expression was observed in all of the species and has been present for at least ~60 million years (Fig. 4A–K).
Fig. 4.
link midline expression is evolutionarily recent. (A–K) Maximum projections of stage 12 embryos from multiple Drosophila species that were stained for link expression using species-specific probes. (A) D. melanogaster link is prominently expressed in CNS midline cells. (B–F) Compared to D. melanogaster, the closely-related species: (B) D. mauritiana, (C) D. simulans, (D) D. sechellia, (E) D. erecta, and (F) D. yakuba, all have midline expression, but it is considerably reduced relative to tracheal expression. (G–K) In more distantly related species, midline expression is barely detectable over background (D. ananassae, D. persimilis, D. pseudoobscura, D. virilis) or undetectable (D. willistoni). (L) Phylogenetic tree of the Drosophila species tested and a model of link regulatory changes. (1) ~60 mya, link was expressed in trachea but absent from CNS midline cells. (2) In the common ancestor of D. melanogaster and D. erecta/D. yakuba, a regulatory change occurred that promoted midline expression. (3) A second regulatory change occurred after the D. melanogaster lineage split from the D. simulans/D. mauritiana/D. sechellia lineage, resulting in an increase in midline expression in D. melanogaster. (M, N) zld MG (*) expression (magenta) in both (M) D. pseudoobscura and (N) D. melanogaster is robust and comparable in stage 12 segments. Midline cell nuclei are visualized by anti-Sim staining (blue). (O) The Dpse-link-5’ transgene in D. melanogaster drives low levels of midline GFP expression, but robust tracheal expression.
Identification of a link embryonic enhancer
To begin a molecular analysis of link embryonic gene expression, including addressing the questions whether Zld directly regulates link expression and how link midline expression evolved, we sought to identify a link embryonic enhancer. We cloned link flanking sequences into the GFP reporter vector pMintgate (Jiang et al., 2010) and analyzed reporter expression by GFP in situ hybridization and immunodetection. Two fragments were analyzed that encompass the entire intergenic regions: a 285 bp 5’-flanking sequence fragment, link-5’, and an 1197 bp 3’-flanking sequence fragment, link-3’ (Fig. 2A). While GFP expression driven by link-3’ did not reflect any obvious aspect of link endogenous expression (data not shown), GFP expression under the control of link-5’ closely matched endogenous link expression throughout embryogenesis (Fig. 2B'–G'). This indicated that all regulatory sequences required for the embryonic expression of link are contained within link-5'.
Identification of conserved putative transcription binding sites in the link embryonic enhancer
Initially, we took an unbiased approach to identify evolutionarily-conserved over-represented putative transcription factor binding sites in the D. melanogaster link 285 bp link-5’ fragment. Utilizing the PhyloGibbs software program, we identified a conserved sequence motif, AGGTRG (R = A/G), referred to as Motif-T, with four sites in link-5’ (Fig. 5A, B, S3). Two sites were identical to each other, with the sequence CAGGTAG (T1, T3) and were conserved in most sequenced Drosophila species (Fig. 5B). Two additional Motif-T sites in link-5' were related to CAGGTAG with either a single mismatch (GAGGTAG; T4) or two mismatches (TAGGTGG; T2) (Fig. 5A, B). Motif-T sites T1 and T3 match strong sites of the TAGteam heptamers (CAGGTAG, TAGGTAG, CAGGTAA, CAGGCAG) (ten Bosch et al., 2006), which are recognized by the Zld, Grainyhead (Grh), and Bicoid Stability Factor transcription factors (De Renzis et al., 2007; Harrison et al., 2010; Liang et al., 2008). The link T1 and T3 sites were previously recognized as putative Zld binding sites (Liang et al., 2008), and Zelda ChIP-seq detects strong binding to the link 5' region in vivo (Harrison 2011).
The Sim and Trh bHLH-PAS transcription factors are known regulators of midline and tracheal expression, respectively. Both of these proteins form heterodimers with Tgo and bind the CME sequence ACGTG. We identified one CME at the promoter-proximal end of D. melanogaster link-5' (C; Fig. 5A, S3), which is conserved in most Drosophila species from the melanogaster subgroup, but is not present in more distantly related Drosophila species (Fig. 5B). However, a putative compensatory CME is upstream of link in D. willistoni and D. virilis (Fig. S3). Interestingly, D. erecta only has a CME in the 5’-UTR, but not in the 5’ intergenic sequence. In summary, the D. melanogaster link-5’ fragment has two bona fide Zelda TAGteam binding sites (T1, T3) and 1 Sim:Tgo/Trh:Tgo binding site (C), in addition to two binding sites related to Zld TAGteam sites (T2, T4).
The CME binding site is not absolutely required for link midline and tracheal expression
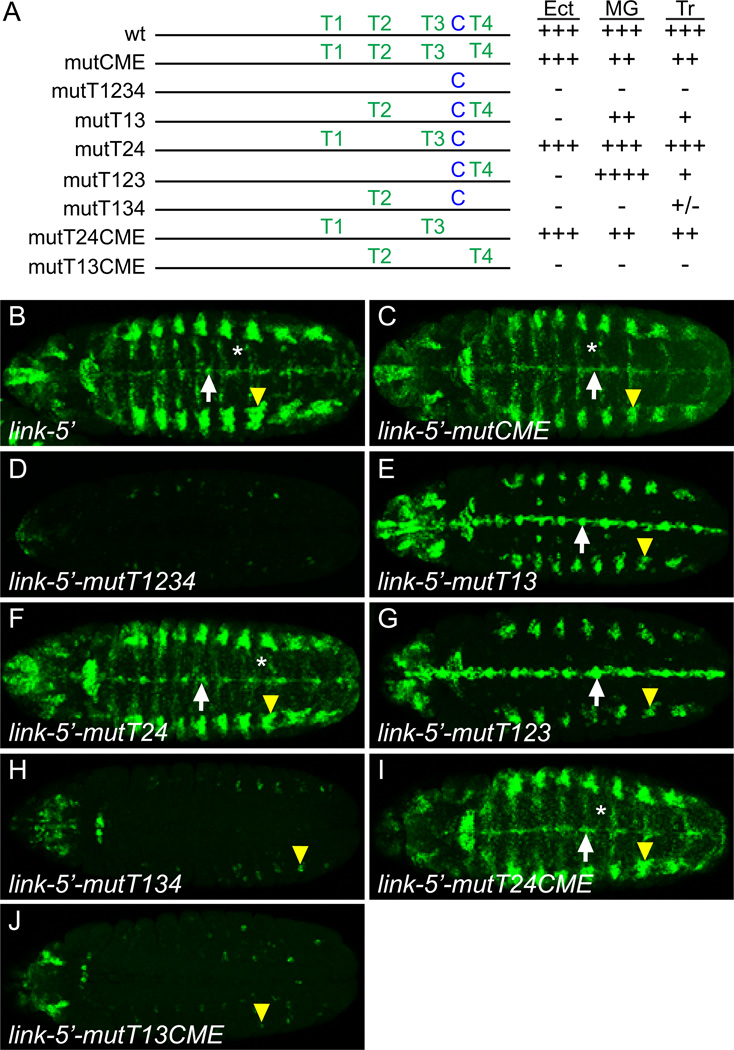
The sim gene plays an important role in regulating CNS midline cell primordium and MG transcription (Nambu et al., 1990), and trh is required for tracheal expression (Isaac and Andrew, 1996; Wilk et al., 1996). When CMEs were mutated in the MG-expressed slit and wrapper genes, all MG expression was abolished (Fulkerson and Estes, 2010; Wharton et al., 1994). Similarly, when CMEs were mutated or deleted in the tracheal-expressed breathless and rhomboid genes, tracheal and midline expression was absent (Ohshiro and Saigo, 1997; Zelzer and Shilo, 2000). To test the requirement of the link CME on midline and tracheal expression, the link-5’ CME was mutated, tested in vivo, and compared to unmutated link-5’ (Fig. 5C, C’, G, 6B). Surprisingly, mutating the CME (link-5’-mutCME) caused only a slight reduction in GFP expression in MG (Fig. 5D, D', 6C) and trachea (Fig. 5H, 6C) (mutational results here and below are summarized in Fig. 6A). This indicated that transcription factors besides Sim:Tgo and Trh:Tgo are necessary for link expression in both MG and trachea.
Fig. 6.
Motif-T and CME sites both contribute to link expression. (A) Schematic of link-5’ transgenes tested for ventral ectodermal (Ect), midline glial (MG), and tracheal (Tr) expression. Observed levels of GFP expression (compared to wild-type levels) are indicated as: wild-type (+++), slightly increased (++++), slightly reduced (++), strongly reduced (+), trace levels (+/−), and absent (−). (B–J) Maximum projection horizontal views of stage 12 embryos showing GFP transcript levels determined by in situ hybridization. Identical settings were used in all cases for microscopy and image processing. The following were used to indicate specific cell types: CNS midline cells (white arrows), trachea (yellow arrowheads), and ventral ectoderm (*).
TAGteam-related sites are required for link expression
To test the role of the Motif-T sites in link expression, all 4 sites were mutated in the link-5’ fragment (link-5’-mutT1234). This resulted in the elimination of nearly all expression in the epidermis, trachea, and MG (Fig. 5E, E’, I, 6D), indicating the importance of Motif-T sites in link embryonic expression. In order to determine which Motif-T sites were contributing to link expression, we mutated either 2 or 3 sites in pairwise combinations. Motif-T sites 1 and 3 are highly conserved, identical to each other, and perfectly match canonical TAGteam sites. When mutated together, link-5’-mutT13 embryonic GFP expression was present, but reduced, in MG and trachea at stage 12 (Fig. 5F, F’, J, 6E) as well as in head structures, but the broad early expression at the maternal-to-zygotic transition (data not shown) and later expression in ventral ectoderm were absent (Fig. 6E). Since sites T1 and T3 together were not absolutely required for midline and tracheal expression, but mutation of sites T1–4 were, we addressed the consequences of mutating sites T2 and T4 (link-5'-mutT24). While this mutant had some slight changes in head and ectodermal expression (Fig. 6F), there were no significant alterations in midline or tracheal expression.
Since the Motif-T double mutants had little effect on midline and tracheal expression, mutations in 3 sites were tested. Mutating sites T1, T2, T3 (link-5’-mutT123) showed strong midline and tracheal GFP expression (Fig. 6G), possibly stronger than the unmutated link-5’, suggesting that the presence of T2 has a slight repressive effect. However, when sites T1, T3, and T4 were mutated (link-5’-mutT134) midline expression was absent and tracheal expression greatly reduced (Fig. 6H). These results indicated that site T4, along with sites T1 and T3, is functional, whereas site T2 does not positively influence link expression. Mutating the CME in addition to sites T2 and T4 (link-5'-mutT24CME) caused only a slight reduction in midline and tracheal expression (Fig. 6I), similar to mutating only the CME. However, when the CME was mutated along with T1 and T3 (link-5’-mutT13CME), midline expression was abolished and tracheal expression was strongly reduced (Fig. 6J). Thus, like site T4, the CME is required for link expression when mutated along with sites T1 and T3. Together, while none of these sites is absolutely required by itself, transcription factors binding Motif-T sites T1, T3, and T4 and the CME together regulate link midline and tracheal expression. These data argue that Zld functions together with Sim and Trh to control link midline and tracheal expression, respectively.
zelda, but not grainyhead, regulates link expression
The mutational analysis of link-5’ implicated TAGteam sites and the CME in regulating link expression. Since both Zld and Grainyhead can bind TAGteam sites (Harrison et al., 2010), we carried-out additional genetic experiments to determine which transcription factor was relevant. Similarly, we sought additional genetic data implicating sim in controlling link expression. When the link-5’ reporter transgene was examined in Df(1)Exel6253 zld hemizygous mutants, all expression was strongly reduced (Fig. 3G, H), although some midline staining was detectable when the gain was increased (Fig. 3G’, H’). However, when link-5’-mutCME was examined in Df(1)Exel6253, the midline staining was completely absent (Fig. 3I–J’). These results support a model in which both zld and sim activate CNS midline expression of link.
The grh gene is expressed during embryogenesis in CNS, epidermal, and tracheal cells (Fig. S4A–C) (Bray et al., 1989; Hemphala et al., 2003). Thus, grh overlaps link expression in these cells. In contrast, whereas link is expressed in MG, grh is only present in the MNB and is not expressed in MG (Fig. S4D–E”). Therefore, grh is unlikely to regulate link expression in MG, but could potentially regulate early embryonic link expression, as well as later epidermal and tracheal expression. However, in embryos homozygous for grhIM, a null allele, link expression resembled wild-type at all developmental stages (data not shown), suggesting that grh does not regulate link expression. Together, the genetic and link-5’ mutational/transgenic studies provide strong evidence that zld, but not grh, directly regulates link expression throughout embryogenesis, via multiple TAGteam-related binding sites, and Sim:Tgo and Trh:Tgo also contribute to link midline and tracheal expression.
While zld regulates CNS midline expression of link, it may not be acting as a global regulator of CNS transcription, since zld mutant embryos did not show a reduction in expression of four additional genes (CG7271, CG8965, escargot, and rhomboid) that are expressed in the CNS, including CNS midline cells (data not shown). Two of these genes (CG7271, escargot) have conserved TAGteam sites and show reduced expression at stage 5 in zld mutants (Liang et al., 2008; Nien et al., 2011).
cis-regulatory alterations in the link enhancer drive evolutionary differences in midline expression
Midline expression of link is a relatively recent occurrence in Drosophila evolution, and the Drosophila species assayed that diverged from D. melanogaster >14 mya expressed little or no link in midline cells. In contrast, midline expression of zld is present in all Drosophila species tested, including D. pseudoobscura (Dpse) (Fig. 4M, N, S2M, N). Similarly, sim is expressed in the midline cells of all arthropods tested, including D. pseudoobscura and D. virilis, as well as mosquito, beetle, and honeybee (Kasai et al., 1998; Zinzen et al., 2006). Since link is not expressed in the midline cells of D. pseudoobscura (Fig. 4I), this suggests that the absence of link midline expression is due to alterations in the link regulatory region and not due to trans-acting differences. To test this, we cloned the upstream region of D. pseudoobscura link (Dpse-link-5') into pMintgate, transformed this construct into D. melanogaster, and assayed expression. When normalized to tracheal expression, Dpse-link-5' midline expression (Fig. 4O) was lower than stage-matched Dmel-link-5' (Fig. 2F’), suggesting that cis-regulatory differences are at least partially responsible for the absence of D. pseudoobscura link midline expression. The Motif-T sites T1, T2, and T3 and the CME are identical between D. melanogaster and D. pseudoobscura, although T4 is not conserved (Fig. 5B). However, mutation of T4 (link-5’-mutT24, link-5’-mutT24CME) did not significantly affect midline expression (Fig. 6F, I), so a combination of T4 and additional diverged sequences may contribute to the alteration in expression.
Discussion
The role of zld in regulating the maternal-to-zygotic transition is extensive, directly activating expression of hundreds of genes. In this paper, we demonstrate that zld has a post-blastoderm role in directly activating expression of link in the CNS midline cells, trachea, and brain. Although zld controls link MG expression, it does not control all MG expression, since CG7271, CG8965, escargot, and rhomboid MG expression was unaffected in zld mutant embryos. Similarly, the well-characterized MG enhancers of the gliolectin, Oatp26f, slit, ventral veins lacking, and wrapper genes do not reveal Zld binding in the embryo, when tested by whole-embryo ChIP (Harrison et al., 2011). Of the 120 genes that were downregulated at least twofold in embryos lacking zld maternal function (Liang et al., 2008), only 4 genes are listed on MidExDB as MG-expressed genes (N=99 genes), indicating no clear enrichment of MG-expressed genes as being zld target genes. Thus, it remains to be seen whether the function of zld in CNS and tracheal development is as widespread and profound as its role in the blastoderm maternal-to-zygotic transition. While zld may not act as a global regulator of CNS transcription, its dynamic expression pattern suggests that it can regulate transcription in a highly temporal and cell-type specific manner in combination with other transcription factors, such as Sim and Trh.
One interesting feature of zld expression is that transcripts are present during embryogenesis as two RNA species that encode two different proteins. In both the blastoderm embryo and during stages 11–12, when zld regulates midline expression, the RB transcript generates a Zld protein containing all 4 C-terminal zinc fingers required for DNA binding (Liang et al., 2008). This is consistent with the role of Zld in activating zen and link transcription by binding TAGteam sites. However, at later embryonic stages zld continues to be expressed in the CNS, but is alternatively spliced into the RD transcript that encodes a protein containing only one of the 4 C-terminal zinc fingers, along with two other N-terminal zinc fingers. Consequently, the PD protein is likely to have different biochemical properties compared to PB, and may be non-functional. In the latter case, termination of zld function in the CNS may be generated by alternative splicing rather than by termination of transcription. Consistent with this view, the zld RD transcript is expressed in midline iVUM4 and MNB progeny neurons and lateral CNS neurons through stage 16, yet link is not expressed in those neuronal cell types. In summary, midline expression of link is due to the midline presence of the Zld PB protein with 4 C-terminal zinc fingers. Even though the Zld PD protein with only 1 C-terminal zinc finger is present in midline and lateral CNS neurons, there is no current evidence that it can activate transcription. The alternative splicing is cell-type specific and not strictly stage-specific, since imaginal disc zld expression in late stage embryos consists of the RB transcript.
In this paper, we describe three aspects of link expression: blastoderm, midline, and trachea expression. Blastoderm expression of link was previously shown to be genetically dependent on zld function (Liang et al., 2008). We demonstrate here that this control is direct, since mutation of the two Motif-T/TAGteam sites T1 and T3 results in an absence of link blastoderm expression. Regulation of link midline and tracheal expression is different: link midline expression is controlled by the combined action of Zld and Sim, and tracheal expression is controlled by Zld and Trh. Sim and Trh are both bHLH-PAS transcription factors that dimerize with Tgo, and bind the same ACGTG (CME) sites (Sonnenfeld et al., 1997; Wharton et al., 1994). While there are subtle differences between link midline and tracheal expression, the basic mechanism of control by Zld/Trh is likely similar to Zld/Sim. Focusing on Sim, it is possible to view link expression as utilizing multiple Zld and Sim:Tgo binding sites in an additive manner with a threshold for expression (Fig. 7A). Mutational studies indicate that the link T1, T3, T4, and CME sites contribute to link midline/tracheal expression. Mutation of T1 and T3 together has little effect on expression, and mutation of the CME (Fig. 7B) or T4 and the CME together has little effect. However, mutation of 3 sites, including T1, T3 and either T4 or CME results in a dramatic loss of link expression.
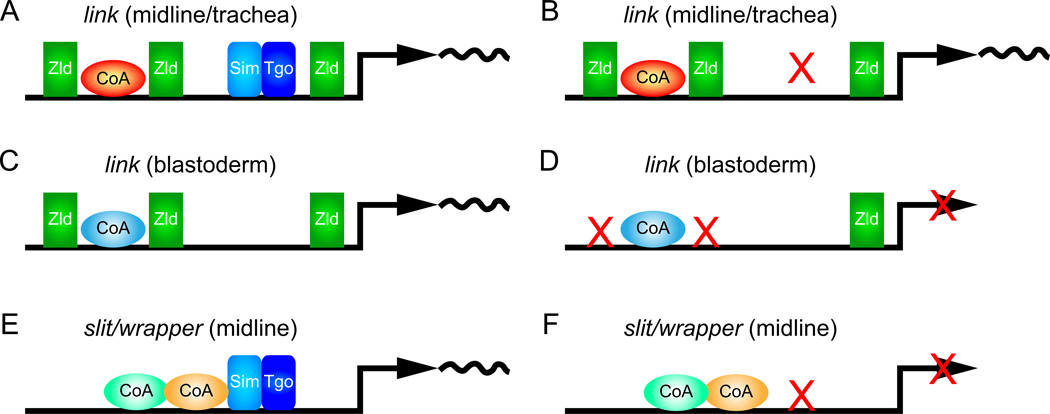
Fig. 7.
Alternate modes of Drosophila MG gene regulation. Schematics are shown representing (A, B) link midline and tracheal expression, (C, D) link blastoderm transcription, and (E, F) slit and wrapper midline expression. (A) The link enhancer has multiple binding sites for Zld (sites T1, T3, and T4 are shown), Sim:Tgo/Trh:Tgo (CME), and predicted additional coactivators (red CoA). Together they result in transcription of link in CNS midline and tracheal cells. (B) When the CME is mutated (X), expression of link is still present. (C) In blastoderm cells, the link enhancer is occupied by Zld and additional coactivators distinct from the link midline/tracheal coactivators (blue CoA). (D) When the TAGteam T1 and T3 sites are mutated, link blastoderm expression is abolished. (E) In the slit and wrapper enhancers, it is proposed that Sim:Tgo forms an activation complex with multiple coactivators. (F) When the Sim:Tgo binding site in the slit and wrapper enhancers is mutated (X), MG expression is abolished.
These results also predict that additional coregulators are required for link expression (Fig. 7A, C). Mutation of T1 and T3 together abolishes link blastoderm expression (Fig. 7D), but not midline/tracheal expression, indicating that the presence of T1 and T3 is not sufficient for transcriptional activation by Zld in all cell types. This suggests that Zld interacts with a blastoderm-specific coregulator to activate link blastoderm expression (Fig. 7C). Similarly, the existence of additional midline/tracheal coregulators is necessary since the presence of 2 TAGteam sites is insufficient for midline/tracheal expression (e.g. zen has 4 TAGteam sites and is not expressed in midline cells, and CG7271 and escargot have multiple TAGteam sites and are regulated in the blastoderm by zld but not in midline cells). Yet, the link-5’ fragment with intact T1 and T3 sites drives strong midline/tracheal expression even when T2, T4, and CME are mutant. This suggests that additional midline/tracheal-expressed coactivators are needed in addition to Zld and Sim/Trh (Fig. 7A). Note that there are a number of well-conserved sequences within the link enhancer in addition to the TAGteam and CME sites (Fig. S3).
Within the midline cells, at stages 11–12, link is prominently expressed in MG. Mechanistically, link MG expression is distinct from other MG-expressed genes, including slit and wrapper. The slit and wrapper MG enhancers have a single CME (similar in number to link) (Fig. 7E), yet mutation of the slit and wrapper CME results in loss of MG expression (Fig. 7F) (Estes et al., 2008; Wharton et al., 1994). This contrasts with link in which mutation of the CME by itself has little effect (Fig. 7B). This result also indicates that the presence of a single CME is insufficient for midline transcriptional activation. Also unlike link, there is no evidence that zld regulates slit and wrapper MG expression, since neither enhancer possess TAGteam sites nor detectably binds Zld in vivo (Harrison et al., 2011), and wrapper expression is not reduced in zld mutant embryos (not shown). However, genetic, biochemical, and mutational studies have provided evidence that Sox proteins (e.g. Dichaete), POU-HD proteins (e.g. Ventral veins lacking), ETS proteins (e.g. Pointed), and poly(T) sequences may act as MG co-activators along with Sim:Tgo (Estes et al., 2008; Ma et al., 2000; Sanchez-Soriano and Russell, 1998). We propose that Sim:Tgo forms a strong association with the slit and wrapper co-activators (Fig. 7E, F), such that when the CME is mutated, the co-activators are either poorly bound or unable to activate transcription on their own. In contrast, Zld and co-activators are able to still activate link MG transcription, even when the CME is mutated. Thus, there are at least two distinct modes of MG enhancers. Each uses Sim:Tgo, but one class employs multiple Zld TAGteam sites to activate link expression along with Sim:Tgo in an additive/threshold manner, whereas the other class (slit and wrapper) is more dependent on an intertwined Sim activation complex. These data further reinforce the view that there exist multiple ways to regulate genes in a similar manner.
The link gene has recently gained midline expression in the melanogaster subgroup, although blastoderm and tracheal expression are stable. Another example of recent evolutionary change in midline expression is the Drosophila α methyl dopa-resistant gene (Wang et al., 1996). Since zld and sim midline expression is well-conserved, the differences are likely due to cis-regulatory changes in the link midline enhancer. This view is supported by the inability of the D. pseudoobscura link regulatory region to drive significant midline expression in D. melanogaster. We propose a two-step model in which ~60 mya, link was weakly or not expressed in midline cells. It acquired midline expression >13 mya, and then <6 mya a second change occurring in the D. melanogaster lineage resulted in increased levels of link midline expression.
The exact alterations that led to the changes in midline expression are unclear. It is unlikely to be due to changes in the T1, T3, and CME sequences since these are identical between D. melanogaster and several species that either lack or have trace levels of midline expression, including D. ananassae, D. persimilis, and D. pseudoobscura. Changes in site T4 are also unlikely to be causative in acquiring midline expression, since it differs significantly in sequence even among species in the melanogaster subgroup that have equivalent link midline expression. Most likely the acquisition of midline expression in the melanogaster subgroup was due to additional uncharacterized sequences in link-5’ sequences. However, the high levels of link expression present in D. melanogaster may be due, in part, to an alteration in T4, since only D. melanogaster T4 contains the TAG nucleotide sequence (Fig. 5B) common among high-affinity TAGteam sites (Liang et al., 2008).
Drosophila Zelda controls link (CG13333) CNS midline and tracheal expression
Zelda collaborates with Single minded and Trachealess
link midline expression recently evolved
Changes in enhancer sequence underlie evolutionary differences in link expression
Alternative splicing generates a Zelda protein with altered DNA binding properties
Supplementary Material
zld is expressed in midline cell neurons and MG. Sagittal views of single confocal segments of wild-type sim-Gal4 UAS-tau-GFP embryos stained with anti-Tau (blue), hybridized in situ to zld (LD47819 cDNA probe; green) and immunostained for En (magenta). (A, A’) Early stage 11 embryo showing the presence of zld in the two recently divided VUM4 neuronal progeny (arrowheads) of the MP4 precursor cell, and strong expression in AMG (white *) and PMG (yellow *). (B, B’) Late stage 11 embryo with strong zld expression in AMG (white *), PMG (yellow *), and MNB (magenta arrowhead). Expression is present in iVUM4 (white arrowhead) and weakly in mVUM4 (yellow arrowhead). (C, C’) Stage 13 embryo showing declining zld expression in AMG and PMG. Expression in MNB is likely present, but the MNB cannot be clearly distinguished from PMG in this image. Expression persists in iVUM4 and mVUM4. (D, D’) At stage 14, expression of zld is absent in AMG and barely detectable in PMG (yellow *). (E, E’) Stage 15 embryo showing zld expression in iVUM4 and MNB with low levels in a single PMG (yellow *). (F, F’) Stage 16 embryos revealed zld expression in iVUM4 and MNB.
Embryonic expression of zld is conserved in multiple Drosophila species. (A–O) Maximum projections of Drosophila embryos hybridized in situ to species-specific zld α probes (magenta). Expression at early and late stage 12, and stage 16 are nearly identical in the 5 species assayed. Note the conserved CNS midline expression in stage 12 embryos as well as the stage 16 CNS expression that are nuclear dots. This indicates the zld splicing patterns described for D. melanogaster are conserved in the other species. (I, L) Enhanced zld ectodermal expression (*) in D. mauritiana and D. erecta compared to other species was observed – this may reflect non-specific staining.
Drosophila species alignment of the link-5’ sequences. The D. melanogaster link-5’ region was aligned to 11 other Drosophila species with Dialign-TX followed by manual adjustment using BioEdit. Sequences present in at least 7 species are boxed. The locations of the conserved Motif-T sites (T1–4) are indicated above the sequence. The conserved CME is also shown above the sequence along with the species-specific CMEs of D. virilis and D. willistoni. Species names are: D. melanogaster (D.mel), D. simulans (D.sim), D. sechellia (D.sec), D. yakuba (D.yak), D. erecta (D.ere), D ananassae (D.ana), D. pseudoobscura (D.pse), D. willistoni (D.wil), D. mojavensis (D.moj), D. grimshawi (D.gri), and D. virilis (D.vir).
grh is expressed in epidermis and trachea, but not MG. Shown are sagittal views of sim-Gal4 UAS-tau-GFP embryos that were hybridized in situ to grh and link probes, and immunostained with anti-tau (blue). (A–C) grh expression is present in ectodermal cells at stage 5, particularly in the dorsal region, and in the epidermis (*) and trachea (yellow arrowheads) at stages 11 and 13. (D, D’) At stage 11, link is expressed in many midline cells, whereas grh is absent in midline cells. (E–E”) At stage 13, link is expressed in AMG (white *) and PMG (yellow *), whereas grh is expressed only in the MNB (white arrowhead).
Acknowledgements
The authors would like to thank Christine Rushlow for kindly providing anti-Zld antibody, zld mutant stocks, and advice, and Gerd Vorbrüggen for vfl stocks. We thank Corbin Jones, Hung-Jui Shih, Sumit Dhole, and Karin Pfennig for Drosophila strains and rearing advice, Nasser Rusan, David Roberts, Mira Pronobis, Derek Applewhite, Kimberly Peters, and Joshua Currie for reagents and advice, and Daniel McKay and Joseph Fontana for help analyzing high-throughput sequencing data. We are grateful to the Bloomington Drosophila Stock Center for providing Drosophila stocks. The project was supported by NRSA postdoctoral awards to JCP (NICHD) and JDW (NINDS), fellowships from the UNC/NIH Developmental Biology Training Program to JCP and JDW (HD046369), and NIH grants R01 NS64264 (NINDS) and R37 RD25251 (NICHD) to STC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Bray SJ, Burke B, Brown NH, Hirsh J. Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev. 1989;3:1130–1145. doi: 10.1101/gad.3.8.1130. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:1036. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes P, Fulkerson E, Zhang Y. Identification of motifs that are conserved in 12 Drosophila species and regulate midline glia vs. neuron expression. Genetics. 2008;178:787–799. doi: 10.1534/genetics.107.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulkerson E, Estes PA. Common motifs shared by conserved enhancers of Drosophila midline glial genes. J. Exp. Zool. B. Mol. Dev. Evol. 2010;316:61–75. doi: 10.1002/jez.b.21382. [DOI] [PubMed] [Google Scholar]
- Gille C, Frommel C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics. 2001;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I. Tempo and mode in evolution of transcriptional regulation. PLoS Genet. 2012;8:e1002432. doi: 10.1371/journal.pgen.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev. Biol. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphala J, Uv A, Cantera R, Bray S, Samakovlis C. Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development. 2003;130:249–258. doi: 10.1242/dev.00218. [DOI] [PubMed] [Google Scholar]
- Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Jiang L, Pearson JC, Crews ST. Diverse modes of Drosophila tracheal fusion cell transcriptional regulation. Mech. Dev. 2010;127:265–280. doi: 10.1016/j.mod.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y, Stahl S, Crews S. Specification of the Drosophila CNS midline cell lineage: direct control of single-minded transcription by dorsal/ventral patterning genes. Gene Expr. 1998;7:171–189. [PMC free article] [PubMed] [Google Scholar]
- Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Certel K, Gao Y, Niemitz E, Mosher J, Mukherjee A, Mutsuddi M, Huseinovic N, Crews ST, Johnson WA, Nambu JR. Functional interactions between Drosophila bHLH/PAS, Sox, POU transcription factors regulate CNS midline expression of the slit gene. J. Neurosci. 2000;20:4596–4605. doi: 10.1523/JNEUROSCI.20-12-04596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J FlyBase Consortium. FlyBase 101--the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu JR, Franks RG, Hu S, Crews ST. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. 1990;63:63–75. doi: 10.1016/0092-8674(90)90288-p. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JL, Wharton KA, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein which acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshiro T, Saigo K. Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development. 1997;124:3975–3986. doi: 10.1242/dev.124.20.3975. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Posakony JW. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev. Biol. 2004;271:431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Natl. Acad. Sci. USA. 2011;108:10036–10043. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Soriano N, Russell S. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- Siddharthan R, Siggia ED, van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput. Biol. 2005;1:e67.0534–e67.0556. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Staudt N, Fellert S, Chung HR, Jackle H, Vorbruggen G. Mutations of the Drosophila zinc finger-encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Mol. Biol. Cell. 2006;17:2356–2365. doi: 10.1091/mbc.E05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian AR, Kaufmann M, Morgenstern B. DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol. Biol. 2008;3:6. doi: 10.1186/1748-7188-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Wang D, Marsh JL, Ayala FJ. Evolutionary changes in the expression pattern of a developmentally essential gene in three Drosophila species. Proc. Natl. Acad. Sci. USA. 1996;93:7103–7107. doi: 10.1073/pnas.93.14.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Mosher JT, Crews ST. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- Wharton JKA, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev. Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Stagg SB, Crews ST. MidExDB: a database of Drosophila CNS midline cell gene expression. BMC Dev. Biol. 2009;9:56. doi: 10.1186/1471-213X-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R, Weizman I, Glazer L, Shilo B. trachealess encodes a bHLH-PAS protein and is a master regulator gene in the Drosophila tracheal system. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Shilo B. Interaction between the bHLH-PAS protein Trachealess and the POU-domain protein Drifter, specifies tracheal cell fates. Mech. Dev. 2000;19:163–173. doi: 10.1016/s0925-4773(99)00295-6. [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Cande J, Ronshaugen M, Papatsenko D, Levine M. Evolution of the ventral midline in insect embryos. Dev. Cell. 2006;11:895–902. doi: 10.1016/j.devcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
zld is expressed in midline cell neurons and MG. Sagittal views of single confocal segments of wild-type sim-Gal4 UAS-tau-GFP embryos stained with anti-Tau (blue), hybridized in situ to zld (LD47819 cDNA probe; green) and immunostained for En (magenta). (A, A’) Early stage 11 embryo showing the presence of zld in the two recently divided VUM4 neuronal progeny (arrowheads) of the MP4 precursor cell, and strong expression in AMG (white *) and PMG (yellow *). (B, B’) Late stage 11 embryo with strong zld expression in AMG (white *), PMG (yellow *), and MNB (magenta arrowhead). Expression is present in iVUM4 (white arrowhead) and weakly in mVUM4 (yellow arrowhead). (C, C’) Stage 13 embryo showing declining zld expression in AMG and PMG. Expression in MNB is likely present, but the MNB cannot be clearly distinguished from PMG in this image. Expression persists in iVUM4 and mVUM4. (D, D’) At stage 14, expression of zld is absent in AMG and barely detectable in PMG (yellow *). (E, E’) Stage 15 embryo showing zld expression in iVUM4 and MNB with low levels in a single PMG (yellow *). (F, F’) Stage 16 embryos revealed zld expression in iVUM4 and MNB.
Embryonic expression of zld is conserved in multiple Drosophila species. (A–O) Maximum projections of Drosophila embryos hybridized in situ to species-specific zld α probes (magenta). Expression at early and late stage 12, and stage 16 are nearly identical in the 5 species assayed. Note the conserved CNS midline expression in stage 12 embryos as well as the stage 16 CNS expression that are nuclear dots. This indicates the zld splicing patterns described for D. melanogaster are conserved in the other species. (I, L) Enhanced zld ectodermal expression (*) in D. mauritiana and D. erecta compared to other species was observed – this may reflect non-specific staining.
Drosophila species alignment of the link-5’ sequences. The D. melanogaster link-5’ region was aligned to 11 other Drosophila species with Dialign-TX followed by manual adjustment using BioEdit. Sequences present in at least 7 species are boxed. The locations of the conserved Motif-T sites (T1–4) are indicated above the sequence. The conserved CME is also shown above the sequence along with the species-specific CMEs of D. virilis and D. willistoni. Species names are: D. melanogaster (D.mel), D. simulans (D.sim), D. sechellia (D.sec), D. yakuba (D.yak), D. erecta (D.ere), D ananassae (D.ana), D. pseudoobscura (D.pse), D. willistoni (D.wil), D. mojavensis (D.moj), D. grimshawi (D.gri), and D. virilis (D.vir).
grh is expressed in epidermis and trachea, but not MG. Shown are sagittal views of sim-Gal4 UAS-tau-GFP embryos that were hybridized in situ to grh and link probes, and immunostained with anti-tau (blue). (A–C) grh expression is present in ectodermal cells at stage 5, particularly in the dorsal region, and in the epidermis (*) and trachea (yellow arrowheads) at stages 11 and 13. (D, D’) At stage 11, link is expressed in many midline cells, whereas grh is absent in midline cells. (E–E”) At stage 13, link is expressed in AMG (white *) and PMG (yellow *), whereas grh is expressed only in the MNB (white arrowhead).